Curated Database and Preliminary AutoML QSAR Model for 5-HT1A Receptor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Database
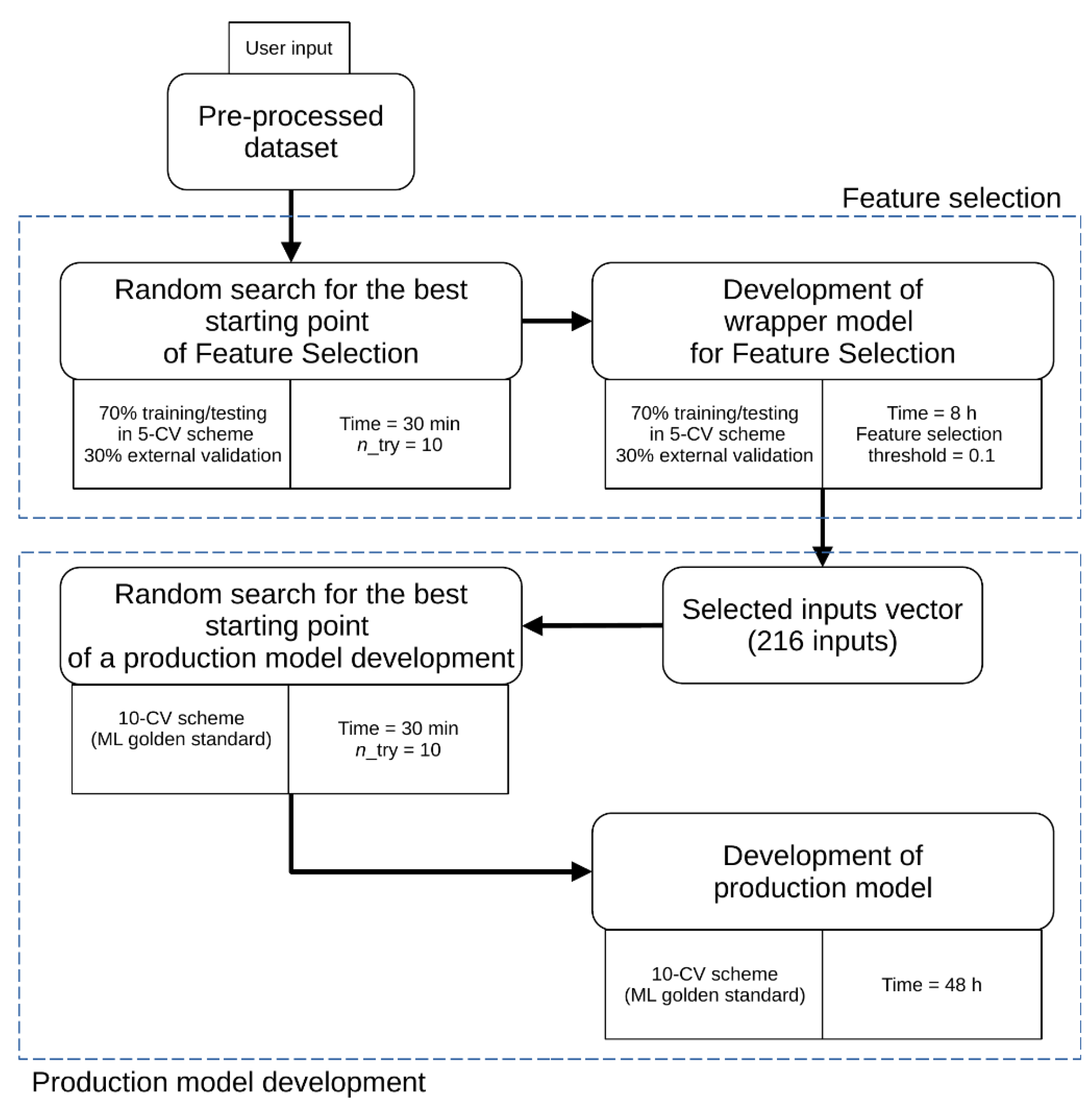
2.2. AutoML Model
3. Results
3.1. Bulding Database
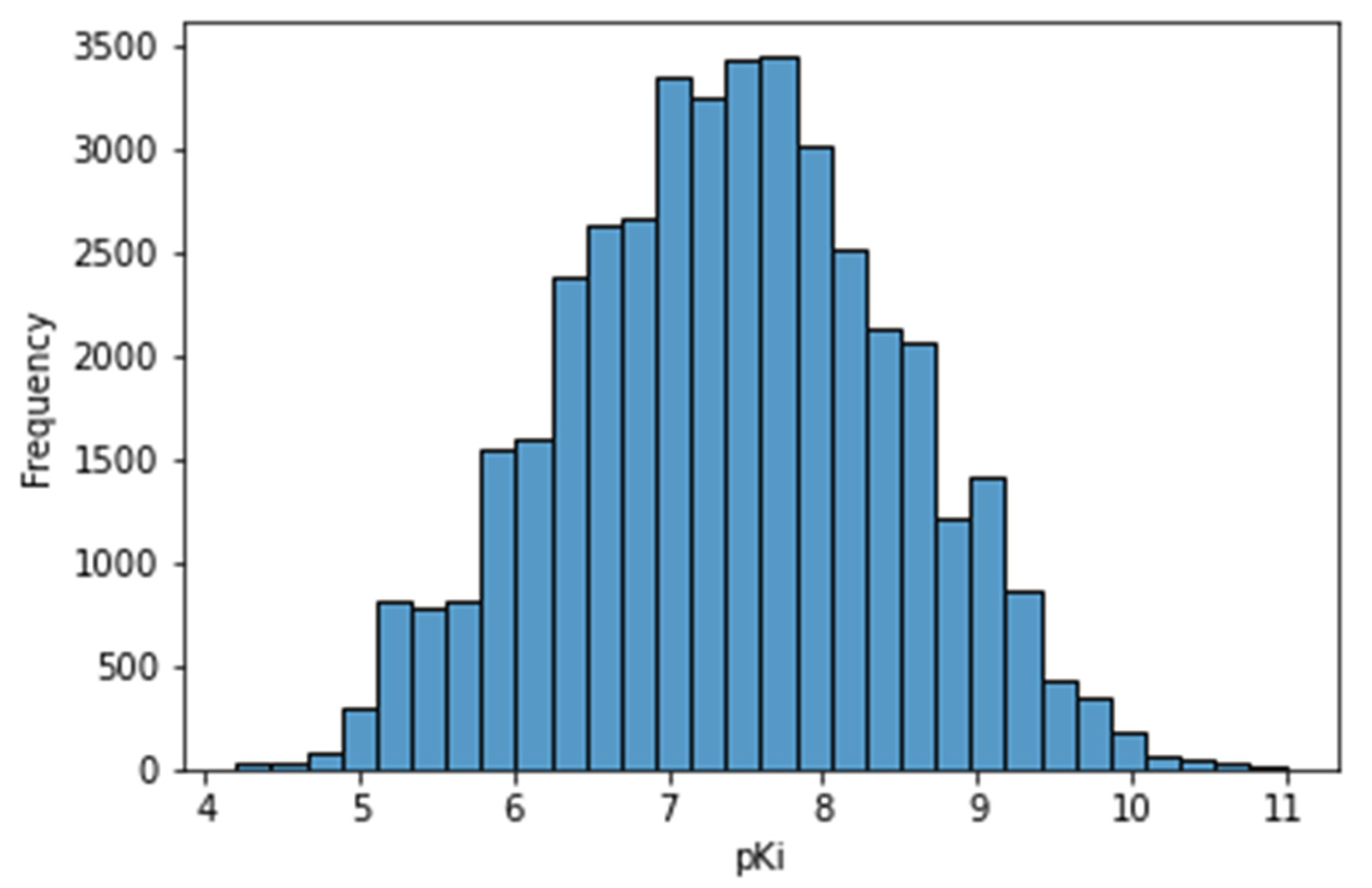
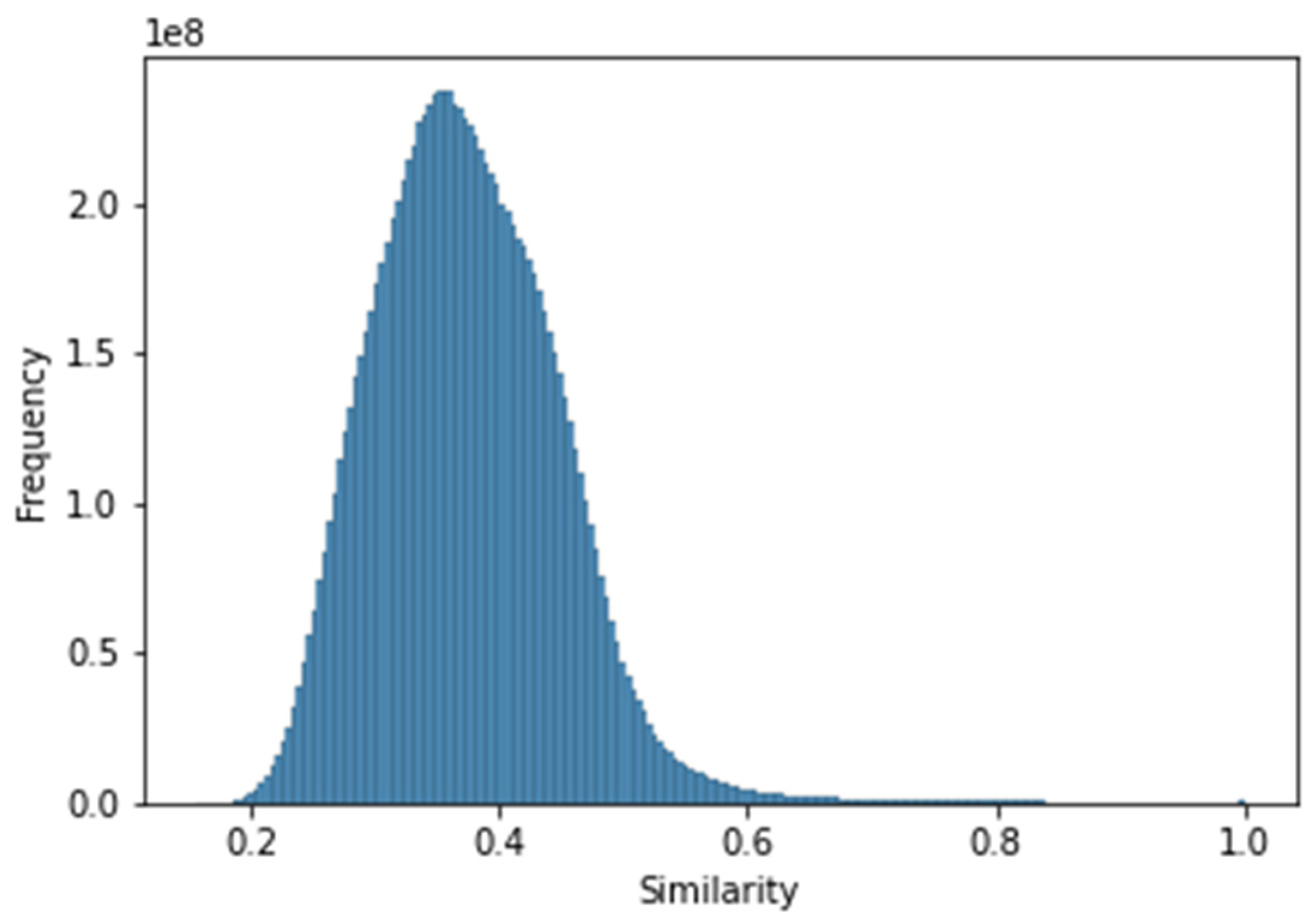
3.2. Exploratory Analysis of the Curated Database
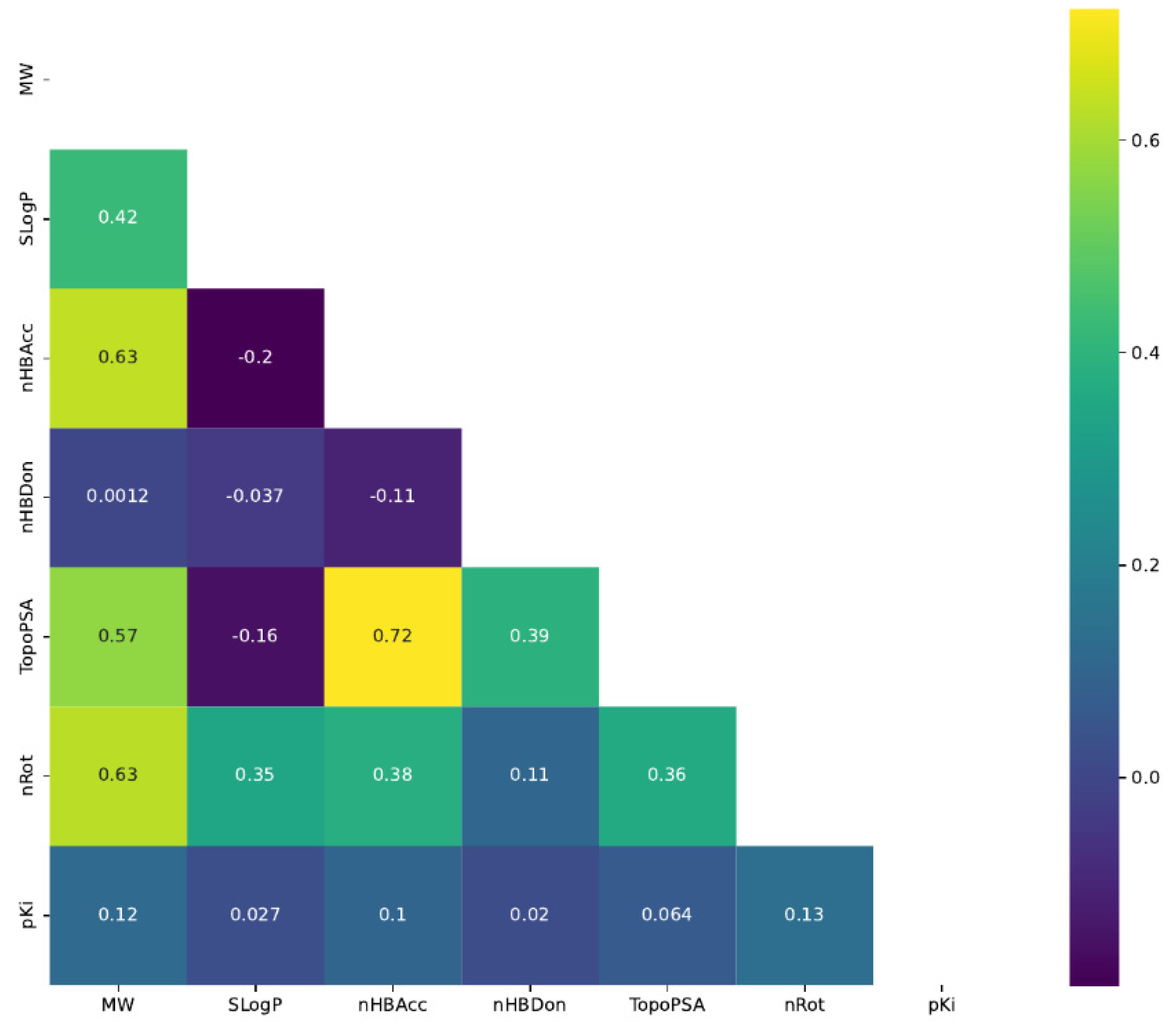
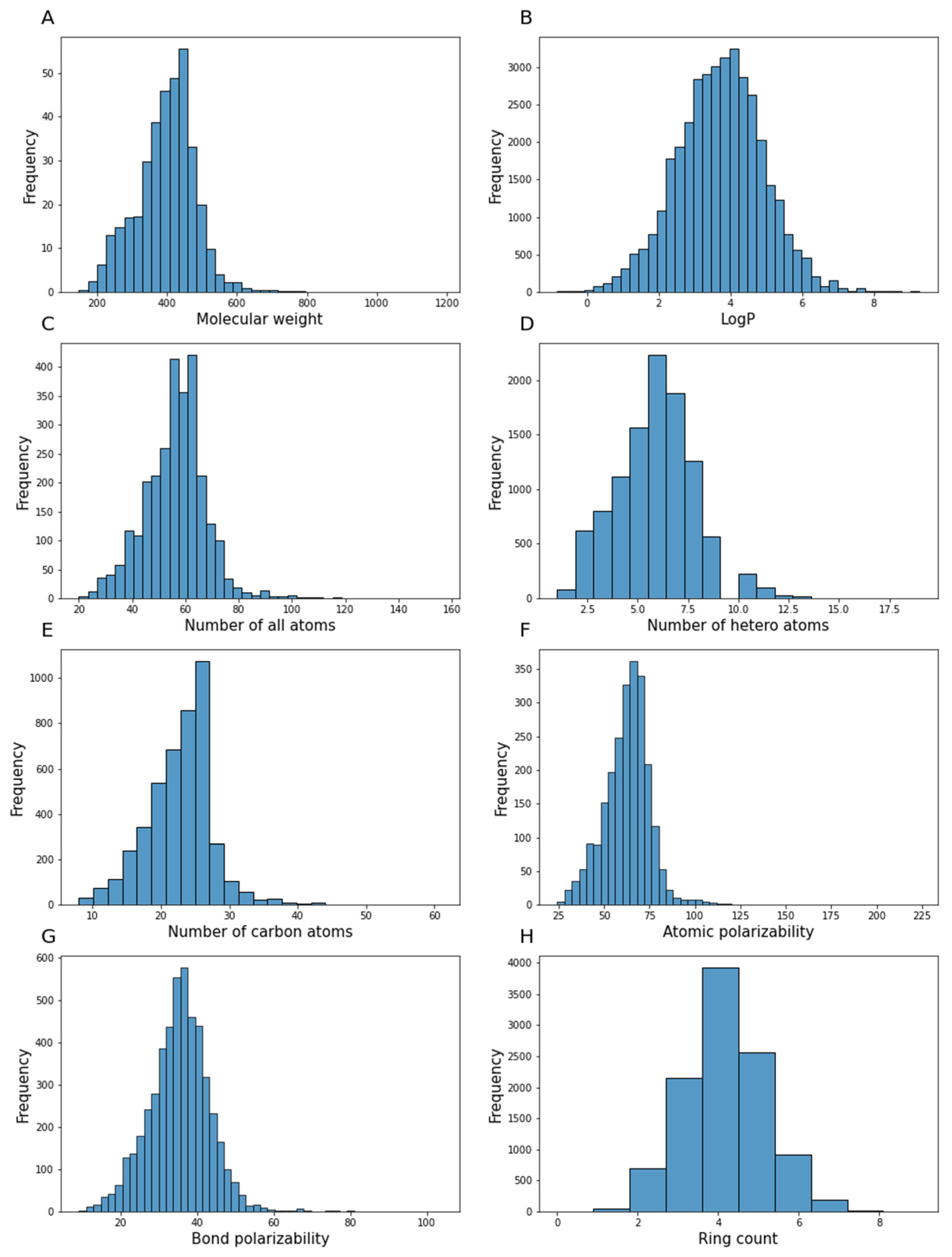
3.2.1. Statistical Exploratory Analysis
3.2.2. Lipinski’s Rule of Five
3.2.3. Drugs Affecting 5-HT1A Receptor
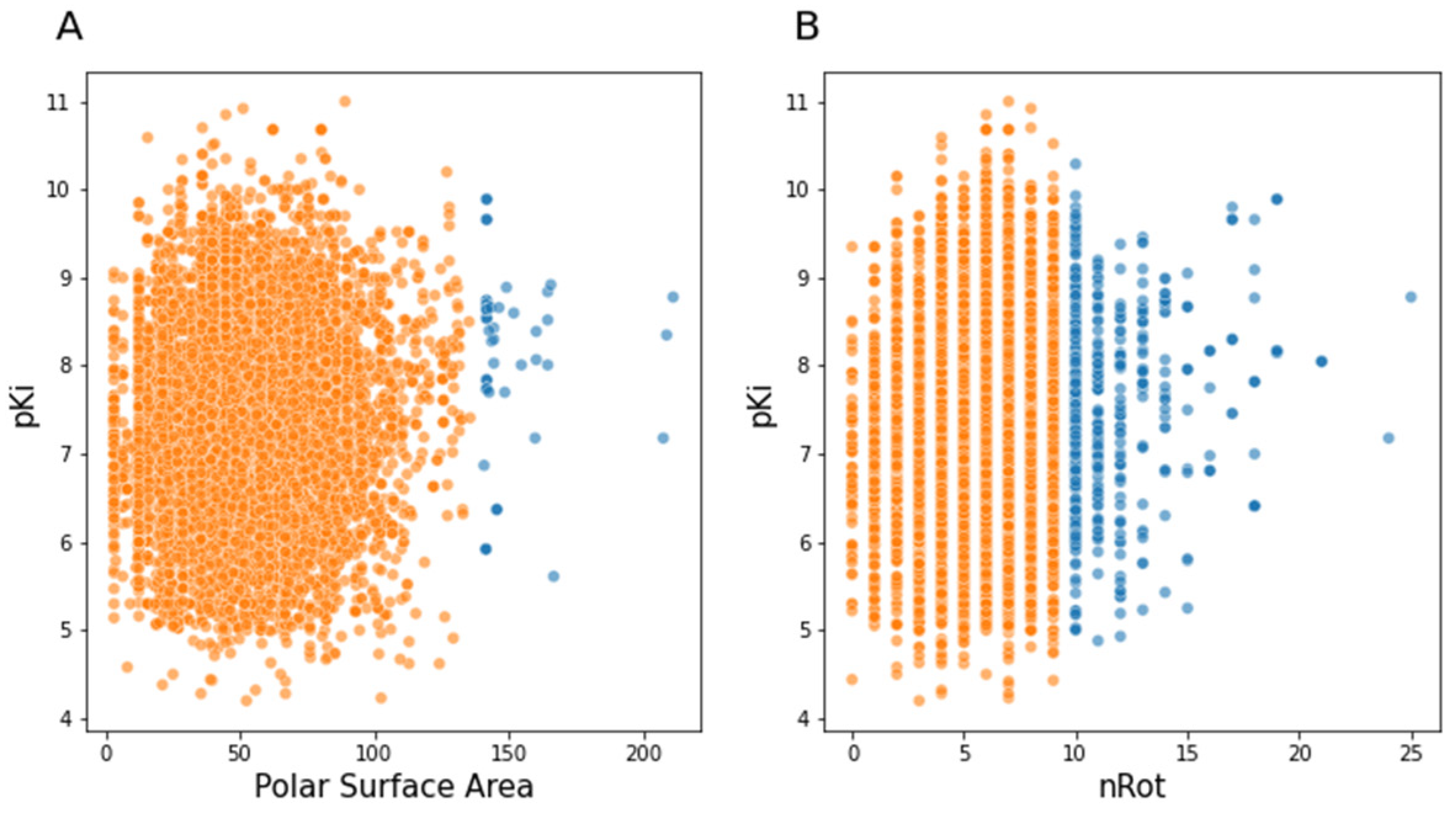
3.2.4. Molecular Descriptors
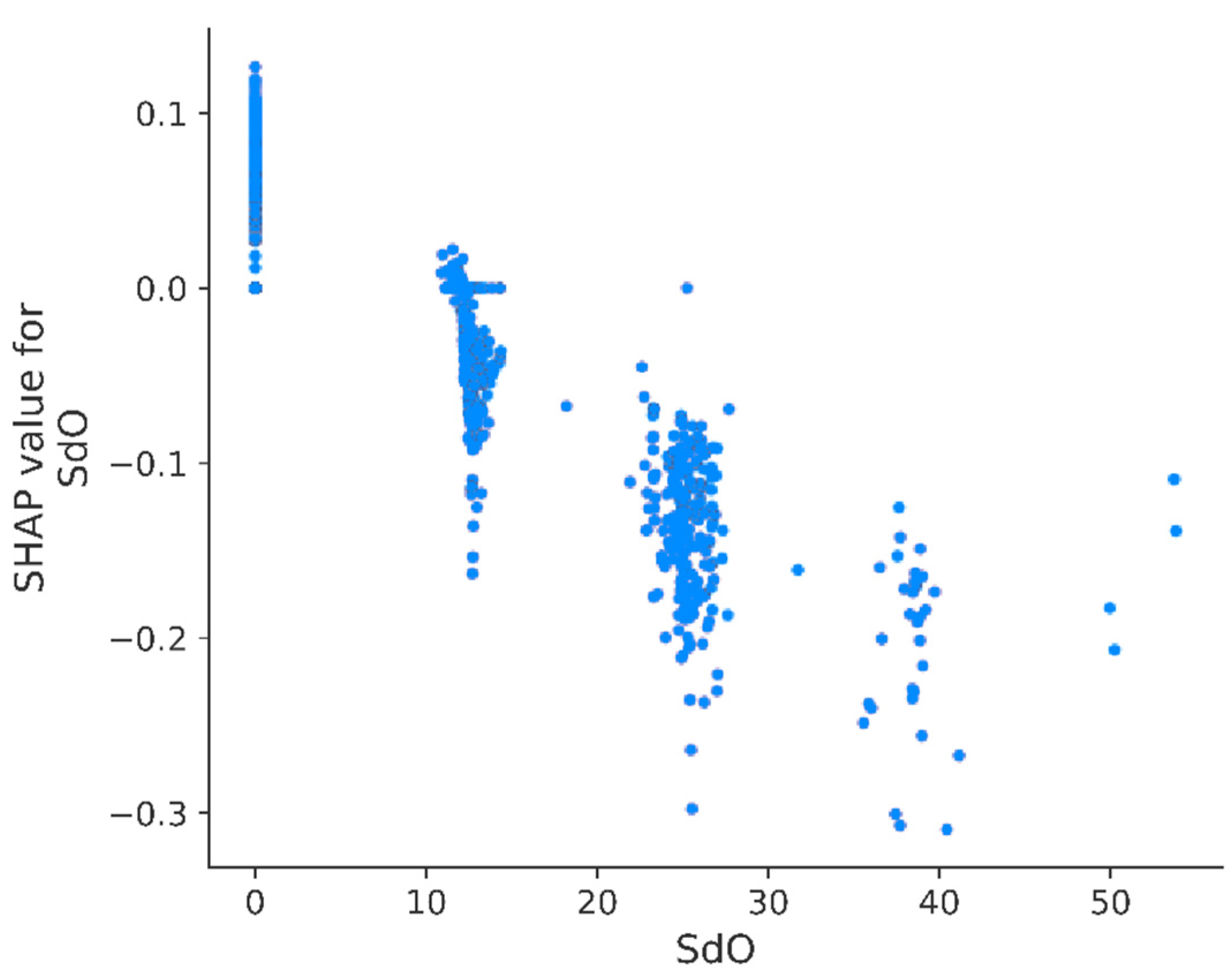
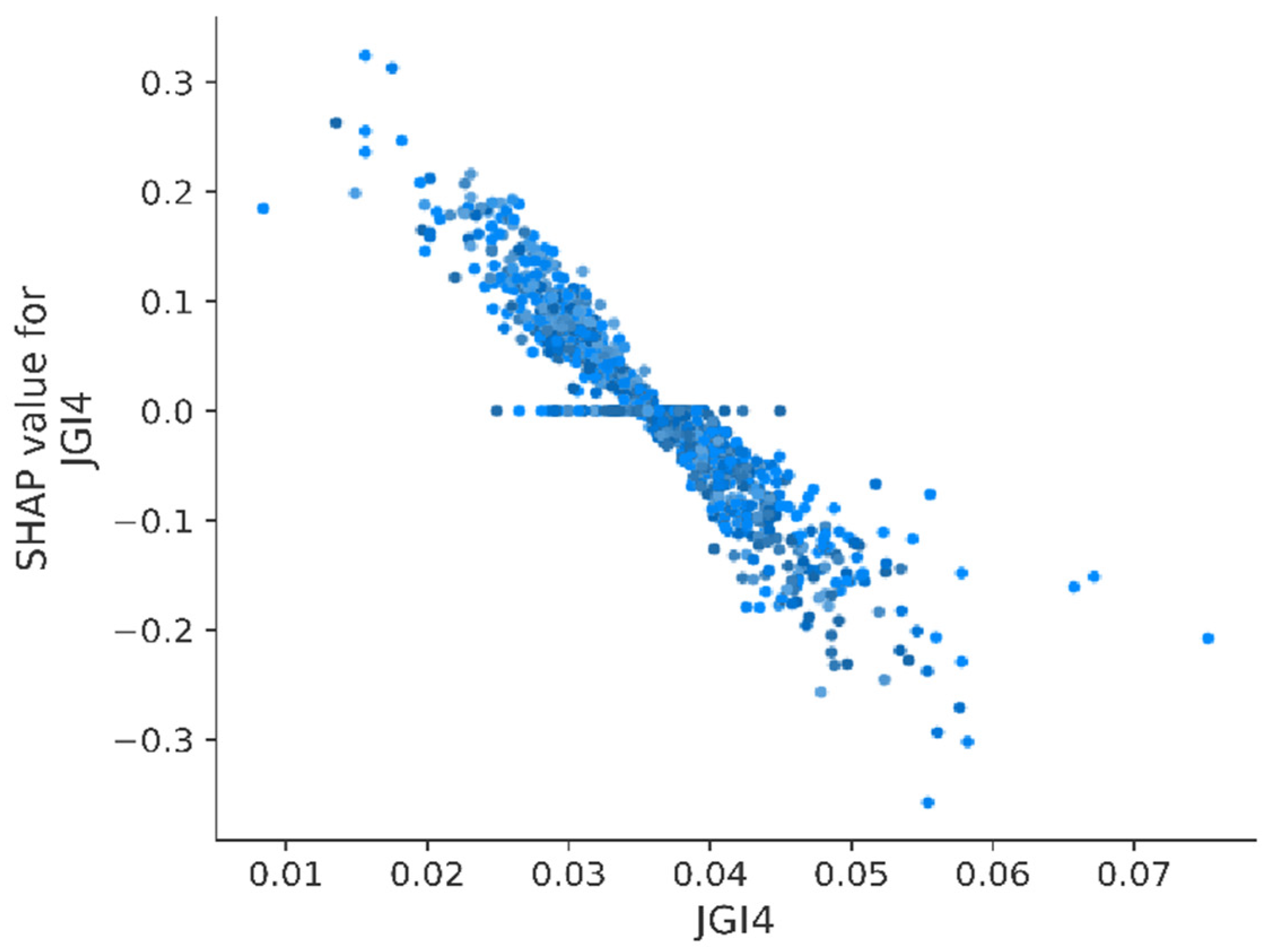
3.3. Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haleem, D.J. Targeting Serotonin1A Receptors for Treating Chronic Pain and Depression. Curr. Neuropharmacol. 2019, 17, 1098–1108. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, Y.; Zhou, C.; Zhang, W.; Gao, S. Classification of 5-HT1A Receptor Ligands on the Basis of Their Binding Affinities by Using PSO-Adaboost-SVM. Int. J. Mol. Sci. 2009, 10, 3316–3337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polter, A.M.; Li, X. 5-HT1A receptor-regulated signal transduction pathways in brain. Cell. Signal. 2010, 22, 1406–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yohn, C.N.; Gergues, M.M.; Samuels, B.A. The role of 5-HT receptors in depression. Mol. Brain 2017, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Carhart-Harris, R.L.; Nutt, D.J. Serotonin and brain function: A tale of two receptors. J. Psychopharmacol. 2017, 31, 1091–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corvino, A.; Fiorino, F.; Severino, B.; Saccone, I.; Frecentese, F.; Perissutti, E.; Di Vaio, P.; Santagada, V.; Caliendo, G.; Magli, E. The Role of 5-HT1A Receptor in Cancer as a New Opportunity in Medicinal Chemistry. Curr. Med. Chem. 2018, 25, 3214–3227. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H. Big Data and Artificial Intelligence Modeling for Drug Discovery. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 573–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2018, 20, 273–286. [Google Scholar] [CrossRef]
- Zhu, X.-L.; Cai, H.-Y.; Xu, Z.; Wang, Y.; Wang, H.-Y.; Zhang, A.; Zhu, W.-L. Classification of 5-HT1A receptor agonists and antagonists using GA-SVM method. Acta Pharmacol. Sin. 2011, 32, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, X.S.; Roth, B.L.; Golbraikh, A.; Tropsha, A. Application of Quantitative Structure–Activity Relationship Models of 5-HT1A Receptor Binding to Virtual Screening Identifies Novel and Potent 5-HT1A Ligands. J. Chem. Inf. Model. 2014, 54, 634–647. [Google Scholar] [CrossRef]
- Sharma, B.K.; Sarbhai, K.; Singh, P. A rationale for the activity profile of arylpiperazinylthioalkyls as 5-HT1A-serotonin and α1-adrenergic receptor ligands. Eur. J. Med. Chem. 2010, 45, 1927–1934. [Google Scholar] [CrossRef]
- Dessalew, N. QSAR study on dual 5-HT1A and 5-HT1B antagonists: An insight into the structural requirement for antidepressant activity. Arch. Pharm. 2008, 341, 314–322. [Google Scholar] [CrossRef]
- Veselinović, A.M.; Milosavljević, J.B.; Toropov, A.A.; Nikolić, G.M. SMILES-based QSAR model for arylpiperazines as high-affinity 5-HT1A receptor ligands using CORAL. Eur. J. Pharm. Sci. 2013, 48, 532–541. [Google Scholar] [CrossRef]
- Weber, K.; Salum, L.B.; Honorio, K.M.; Andricopulo, A.D.; Da Silva, A.B.; Da Silva, A.B.F. Pharmacophore-based 3D QSAR studies on a series of high affinity 5-HT1A receptor ligands. Eur. J. Med. Chem. 2010, 45, 1508–1514. [Google Scholar] [CrossRef]
- Jia, Q.; Cui, X.; Li, L.; Wang, Q.; Liu, Y.; Xia, S.; Ma, P. Quantitative Structure–Activity Relationship for High Affinity 5-HT1A Receptor Ligands Based on Norm Indexes. J. Phys. Chem. B 2015, 119, 15561–15567. [Google Scholar] [CrossRef] [PubMed]
- Govinda, K.; Hassan, M.; Sirimulla, S. KinasepKipred: A Predictive Model for Estimating Ligand-Kinase Inhibitor Constant (pKi). BioRxiv 2019, 798561. [Google Scholar]
- AutoML: Automatic Machine Learning. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-docs/automl.html (accessed on 14 April 2021).
- Elton, D.C.; Boukouvalas, Z.; Fuge, M.; Chung, P.W. Deep learning for molecular design—A review of the state of the art. Mol. Syst. Des. Eng. 2019, 4, 828–849. [Google Scholar] [CrossRef] [Green Version]
- Irwin, J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2004, 45, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; de Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2018, 47, D930–D940. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Pandas. Available online: https://pandas.pydata.org/ (accessed on 30 December 2020).
- Getting Started with the RDKit in Python. Available online: https://www.rdkit.org/docs/GettingStartedInPython.html (accessed on 15 July 2021).
- Moriwaki, H.; Tian, Y.-S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminformatics 2018, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- Szlęk, J. h2o_AutoML_Python, Python Script for AutoML in h2o. 2021. Available online: https://github.com/jszlek/h2o_AutoML_Python (accessed on 10 April 2021).
- LeDell, E.; Poirier, S. H2O AutoML: Scalable automatic machine learning. In Proceedings of the 7th ICML Workshop on Automated Machine Learning, Vienna, Austria, 18 July 2020. [Google Scholar]
- Lundberg, S.M.; Erion, G.G.; Lee, S.I. Consistent Individualized Feature Attribution for Tree Ensembles. arXiv 2018, arXiv:1802.03888. [Google Scholar]
- Lundberg, S.M.; Su-In, L. A Unified Approach to Interpreting Model Predictions. NIPS 2017, arXiv:1705.07874. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Liégeois, J.-F.; Deville, M.; Dilly, S.; Lamy, C.; Mangin, F.; Résimont, M.; Tarazi, F.I. New Pyridobenzoxazepine Derivatives Derived from 5-(4-Methylpiperazin-1-yl)-8-chloro-pyrido[2,3-b][1,5]benzoxazepine (JL13): Chemical Synthesis and Pharmacological Evaluation. J. Med. Chem. 2012, 55, 1572–1582. [Google Scholar] [CrossRef] [PubMed]
- Linnanen, T.; Brisander, M.; Mohell, N.; Johansson, A.M. Serotonergic and dopaminergic activities of rigidified (R)-aporphine derivatives. Bioorganic Med. Chem. Lett. 2001, 11, 367–370. [Google Scholar] [CrossRef]
- Wróbel, M.Z.; Chodkowski, A.; Herold, F.; Marciniak, M.; Dawidowski, M.; Siwek, A.; Starowicz, G.; Stachowicz, K.; Szewczyk, B.; Nowak, G.; et al. Synthesis and biological evaluation of new multi-target 3-(1H-indol-3-yl)pyrrolidine-2,5-dione derivatives with potential antidepressant effect. Eur. J. Med. Chem. 2019, 183, 111736. [Google Scholar] [CrossRef]
- Heier, R.F.; Dolak, L.A.; Duncan, J.N.; Hyslop, D.K.; Lipton, M.F.; Martin, I.J.; Mauragis, M.A.; Piercey, M.F.; Nichols, N.F.; Schreur, P.J.K.D.; et al. Synthesis and Biological Activities of (R)-5,6-Dihydro-N,N-dimethyl-4H-imidazo[4,5,1-ij]quinolin-5-amine and Its Metabolites. J. Med. Chem. 1997, 40, 639–646. [Google Scholar] [CrossRef]
- Staroń, J.; Kurczab, R.; Warszycki, D.; Satała, G.; Krawczyk, M.; Bugno, R.; Lenda, T.; Popik, P.; Hogendorf, A.S.; Higendorf, A.; et al. Virtual screening-driven discovery of dual 5-HT6/5-HT2A receptor ligands with pro-cognitive properties. Eur. J. Med. Chem. 2020, 185, 111857. [Google Scholar] [CrossRef]
- Brindisi, M.; Butini, S.; Franceschini, S.; Brogi, S.; Trotta, F.; Ros, S.; Cagnotto, A.; Salmona, M.; Casagni, A.; Andreassi, M.; et al. Targeting Dopamine D3 and Serotonin 5-HT1A and 5-HT2A Receptors for Developing Effective Antipsychotics: Synthesis, Biological Characterization, and Behavioral Studies. J. Med. Chem. 2014, 57, 9578–9597. [Google Scholar] [CrossRef] [PubMed]
- Zajdel, P.; Kos, T.; Marciniec, K.; Satała, G.; Canale, V.; Kaminski, K.; Holuj, M.; Lenda, T.; Koralewski, R.; Bednarski, M.; et al. Novel multi-target azinesulfonamides of cyclic amine derivatives as potential antipsychotics with pro-social and pro-cognitive effects. Eur. J. Med. Chem. 2018, 145, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Del Bello, F.; Diamanti, E.; Giannella, M.; Mammoli, V.; Marchioro, C.; Mattioli, L.; Titomanlio, F.; Piergentili, A.; Quaglia, W.; Benedetti, G.; et al. Low Doses of Allyphenyline and Cyclomethyline, Effective against Morphine Dependence, Elicit an Antidepressant-like Effect. ACS Med. Chem. Lett. 2012, 3, 535–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, O.M.; Dhanoa, D.S.; Marantz, Y.; Chen, D.; Shacham, S.; Cheruku, S.; Heifetz, A.; Mohanty, P.; Fichman, M.; Sharadendu, A.; et al. An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J. Med. Chem. 2006, 49, 3116–3135. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Robichaud, A.J.; Lee, T.; Tomesch, J.; Yao, W.; Beard, J.D.; Snyder, G.L.; Zhu, H.; Peng, Y.; et al. Discovery of a Tetracyclic Quinoxaline Derivative as a Potent and Orally Active Multifunctional Drug Candidate for the Treatment of Neuropsychiatric and Neurological Disorders. J. Med. Chem. 2014, 57, 2670–2682. [Google Scholar] [CrossRef]
- Krushinski, J.H.; Schaus, J.M.; Thompson, D.C.; Calligaro, D.O.; Nelson, D.L.; Luecke, S.H.; Wainscott, D.B.; Wong, D.T. Indoloxypropanolamine analogues as 5-HT1A receptor antagonists. Bioorganic Med. Chem. Lett. 2007, 17, 5600–5604. [Google Scholar] [CrossRef]
- Żelaszczyk, D.; Jakubczyk, M.; Pytka, K.; Rapacz, A.; Walczak, M.; Janiszewska, P.; Pańczyk, K.; Żmudzki, P.; Słoczyńska, K.; Marona, H.; et al. Design, synthesis and evaluation of activity and pharmacokinetic profile of new derivatives of xanthone and piperazine in the central nervous system. Bioorganic Med. Chem. Lett. 2019, 29, 126679. [Google Scholar] [CrossRef]
- Xu, Y.-C.; Schaus, J.M.; Walker, C.; Krushinski, J.; Adham, N.; Zgombick, J.M.; Liang, S.X.; Kohlman, D.T.; Audia, J.E. N-Methyl-5-tert-butyltryptamine: A Novel, Highly Potent 5-HT1D Receptor Agonist. J. Med. Chem. 1999, 42, 526–531. [Google Scholar] [CrossRef]
- Gu, Z.-S.; Wang, W.-T.; Qian, H.; Zhou, A.-N.; Sun, H.-B.; Zhang, Q.-W.; Li, J.-Q. Synthesis and antidepressant effect of novel aralkyl piperazine and piperidine derivatives targeting SSRI/5-HT1A/5-HT7. Bioorganic Med. Chem. Lett. 2019, 29, 126703. [Google Scholar] [CrossRef]
- Descriptor List. Available online: https://mordred-descriptor.github.io/documentation/master/descriptors.html (accessed on 15 July 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 10 June 2021).
- Lundberg, S. SHAP (SHapley Additive exPlanations). Available online: https://github.com/slundberg/shap (accessed on 10 August 2021).
- Szlęk, J. Model Interpretation. Available online: https://github.com/jszlek/MODEL_INTERPRETATION (accessed on 10 August 2021).
- Bondensgaard, K.; Ankersen, M.; Thøgersen, H.; Hansen, B.S.; Wulff, B.S.; Bywater, R.P. Recognition of Privileged Structures by G-Protein Coupled Receptors. J. Med. Chem. 2004, 47, 888–899. [Google Scholar] [CrossRef]
- Zlatović, M.V.; Sukalovic, V.; Schneider, C.; Roglic, G. Interaction of arylpiperazine ligands with the hydrophobic part of the 5-HT1A receptor binding site. Bioorganic Med. Chem. 2006, 14, 2994–3001. [Google Scholar] [CrossRef]
- Leong, M.K.; Syu, R.-G.; Ding, Y.-L.; Weng, C.-F. Prediction of N-Methyl-D-Aspartate Receptor GluN1-Ligand Binding Affinity by a Novel SVM-Pose/SVM-Score Combinatorial Ensemble Docking Scheme. Sci. Rep. 2017, 7, 40053. [Google Scholar] [CrossRef]
- Nowaczyk, A.; Kulig, K. QSAR studies on a number of pyrrolidin-2-one antiarrhythmic arylpiperazinyls. Med. Chem. Res. 2012, 21, 373–381. [Google Scholar] [CrossRef] [Green Version]
- Warszycki, D.; Mordalski, S.; Kristiansen, K.; Kafel, R.; Sylte, I.; Chilmonczyk, Z.; Bojarski, A.J. A Linear Combination of Pharmacophore Hypotheses as a New Tool in Search of New Active Compounds—An Application for 5-HT1A Receptor Ligands. PLoS ONE 2013, 8, e84510. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Wang, L.; Xie, X.-Q. Ligand Classifier of Adaptively Boosting Ensemble Decision Stumps (LiCABEDS) and Its Application on Modeling Ligand Functionality for 5HT-Subtype GPCR Families. J. Chem. Inf. Model. 2011, 51, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Kurczab, R.; Canale, V.; Zajdel, P.; Bojarski, A.J. An Algorithm to Identify Target-Selective Ligands—A Case Study of 5-HT7/5-HT1A Receptor Selectivity. PLoS ONE 2016, 11, e0156986. [Google Scholar] [CrossRef]
- Kuz’min, V.E.; Polischuk, P.G.; Artemenko, A.G.; Makan, S.Y.; Andronati, S.A. Quantitative structure-affinity relationship of 5-HT1A receptor ligands by the classification tree method. SAR QSAR Environ. Res. 2008, 19, 213–244. [Google Scholar] [CrossRef]





| Feature | Number of Compounds Fulfilling the Rule | Percentage of Curated Database Fulfilling the Rule |
|---|---|---|
| Molecular weight | 8691 | 92.07% |
| LogP | 8190 | 86.76% |
| H-bond acceptors | 9356 | 99.11% |
| H-bond donors | 9403 | 99.61% |
| Complete Lipinski’s rules | 7735 | 81.94% |
| Polar Surface Area | 9382 | 99.39% |
| Rotatable Bonds | 8848 | 93.73% |
| Complete Veber et al.’s rules | 7431 | 78.72% |
| No. | Drug | pKi | The Rule of Five |
|---|---|---|---|
| 1. | Amoxapine | 6.66 | ✓ |
| 2. | Apomorphine | 6.53 | ✓ |
| 3. | Aripiprazole | 8.77 | ✓ |
| 4. | Bromocriptine | 7.62 | MW over limit |
| 5. | Buspirone | 7.46 | ✓ |
| 6. | Cariprazine | 8.59 | ✓ |
| 7. | Clozapine | 6.80 | ✓ |
| 8. | Haloperidol | 5.77 | ✓ |
| 9. | Lisuride | 9.40 | ✓ |
| 10. | Lofexidine | 6.90 | ✓ |
| 11. | Naluzotan | 8.29 | ✓ |
| 12. | Olanzapine | 5.57 | ✓ |
| 13. | Pergolide | 8.40 | ✓ |
| 14. | Pindolol | 7.65 | ✓ |
| 15. | Quetiapine | 6.52 | ✓ |
| 16. | Risperidone | 6.72 | ✓ |
| 17. | Sumatriptan | 6.64 | ✓ |
| 18. | Vilazodone | 9.70 | ✓ |
| 19. | Vortioxetine | 8.02 | ✓ |
| 20. | Ziprasidone | 8.70 | ✓ |
| Chemical Descriptor | Description | Relative Importance | Original Input No |
|---|---|---|---|
| AATSC5d | Averaged and centered Moreau–Broto autocorrelation of lag 5 weighted by sigma electrons | 1.000 | 364 |
| ATSC4d | Centered Moreau–Broto autocorrelation of lag 4 weighted by sigma electrons | 0.930 | 255 |
| SpMAD_Dzi | Spectral mean absolute deviation from Barysz matrix weighted by ionization potential | 0.912 | 761 |
| SlogP_VSA1 | MOE type descriptors using Wildman–Crippen LogP and surface area contribution | 0.614 | 1080 |
| nBondsS | Number of single bonds in non-Kekulized structure | 0.556 | 773 |
| BalabanJ | Balaban’s J index | 0.517 | 665 |
| SsssCH | Sum of sssCH | 0.494 | 921 |
| SssO | Sum of ssO | 0.415 | 940 |
| SssCH2 | Sum of ssCH2 | 0.407 | 917 |
| nBase | Basic group count | 0.384 | 5 |
| Xch-5dv | 5-ordered chi chain weighted by valence electrons | 0.369 | 800 |
| SaaaC | Sum of aaaC | 0.346 | 925 |
| MATS5d | Moran coefficient of lag 5 weighted by sigma electrons | 0.327 | 469 |
| SLogP | Wildman–Crippen LogP | 0.299 | 1229 |
| PEOE_VSA9 | MOE type descriptors using Gasteiger charge and surface area contribution | 0.298 | 1067 |
| AATS6dv | Averaged Moreau–Broto autocorrelation of lag 6 weighted by valence electrons | 0.291 | 140 |
| SdO | Sum of dO | 0.281 | 939 |
| SaasN | Sum of aasN | 0.280 | 936 |
| AATS4i | Averaged Moreau–Broto autocorrelation of lag 4 weighted by ionization potential | 0.278 | 228 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czub, N.; Pacławski, A.; Szlęk, J.; Mendyk, A. Curated Database and Preliminary AutoML QSAR Model for 5-HT1A Receptor. Pharmaceutics 2021, 13, 1711. https://doi.org/10.3390/pharmaceutics13101711
Czub N, Pacławski A, Szlęk J, Mendyk A. Curated Database and Preliminary AutoML QSAR Model for 5-HT1A Receptor. Pharmaceutics. 2021; 13(10):1711. https://doi.org/10.3390/pharmaceutics13101711
Chicago/Turabian StyleCzub, Natalia, Adam Pacławski, Jakub Szlęk, and Aleksander Mendyk. 2021. "Curated Database and Preliminary AutoML QSAR Model for 5-HT1A Receptor" Pharmaceutics 13, no. 10: 1711. https://doi.org/10.3390/pharmaceutics13101711
APA StyleCzub, N., Pacławski, A., Szlęk, J., & Mendyk, A. (2021). Curated Database and Preliminary AutoML QSAR Model for 5-HT1A Receptor. Pharmaceutics, 13(10), 1711. https://doi.org/10.3390/pharmaceutics13101711









