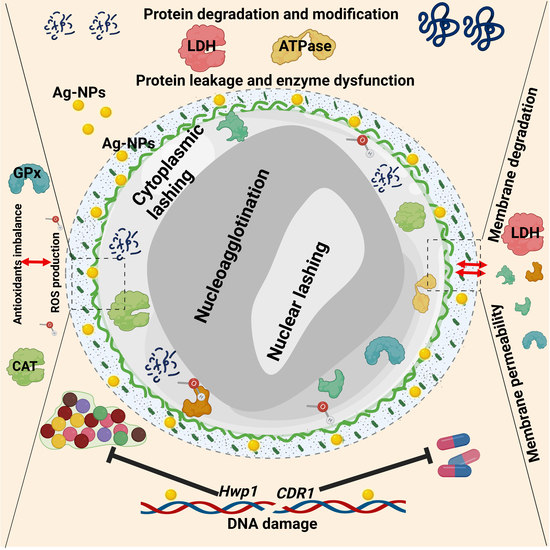
Anticandidal Potential of Two Cyanobacteria-Synthesized Silver Nanoparticles: Effects on Growth, Cell Morphology, and Key Virulence Attributes of Candida albicans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation of NP suspensions
2.2.2. Fungal Cell Culture
2.2.3. Agar Well Diffusion Assay
2.2.4. Measurement of Minimum Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC)
2.2.5. Analysis of LDH Activity
2.2.6. Estimation of ATPase Activity
2.2.7. Estimation of Antioxidant Enzyme Activity
2.2.8. Visualization of C. albicans Cells by TEM
2.2.9. Hwp1 and CDR1 Gene Expression Analysis
2.2.10. Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS–PAGE)
2.3. Statistical Analysis
3. Results
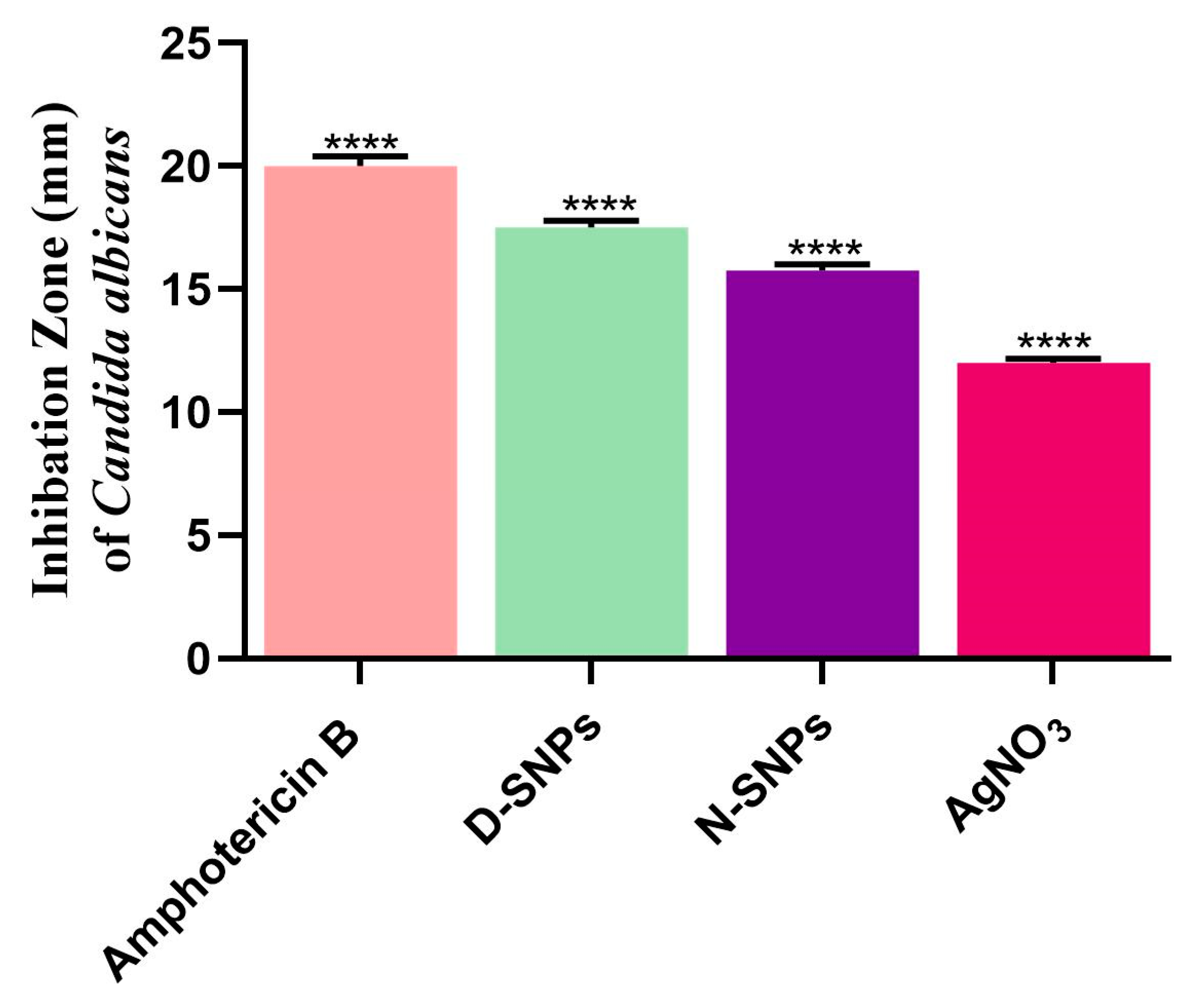
3.1. The Anticandidal Effects of AgNO3 and the Biogenic Silver Nanoparticles
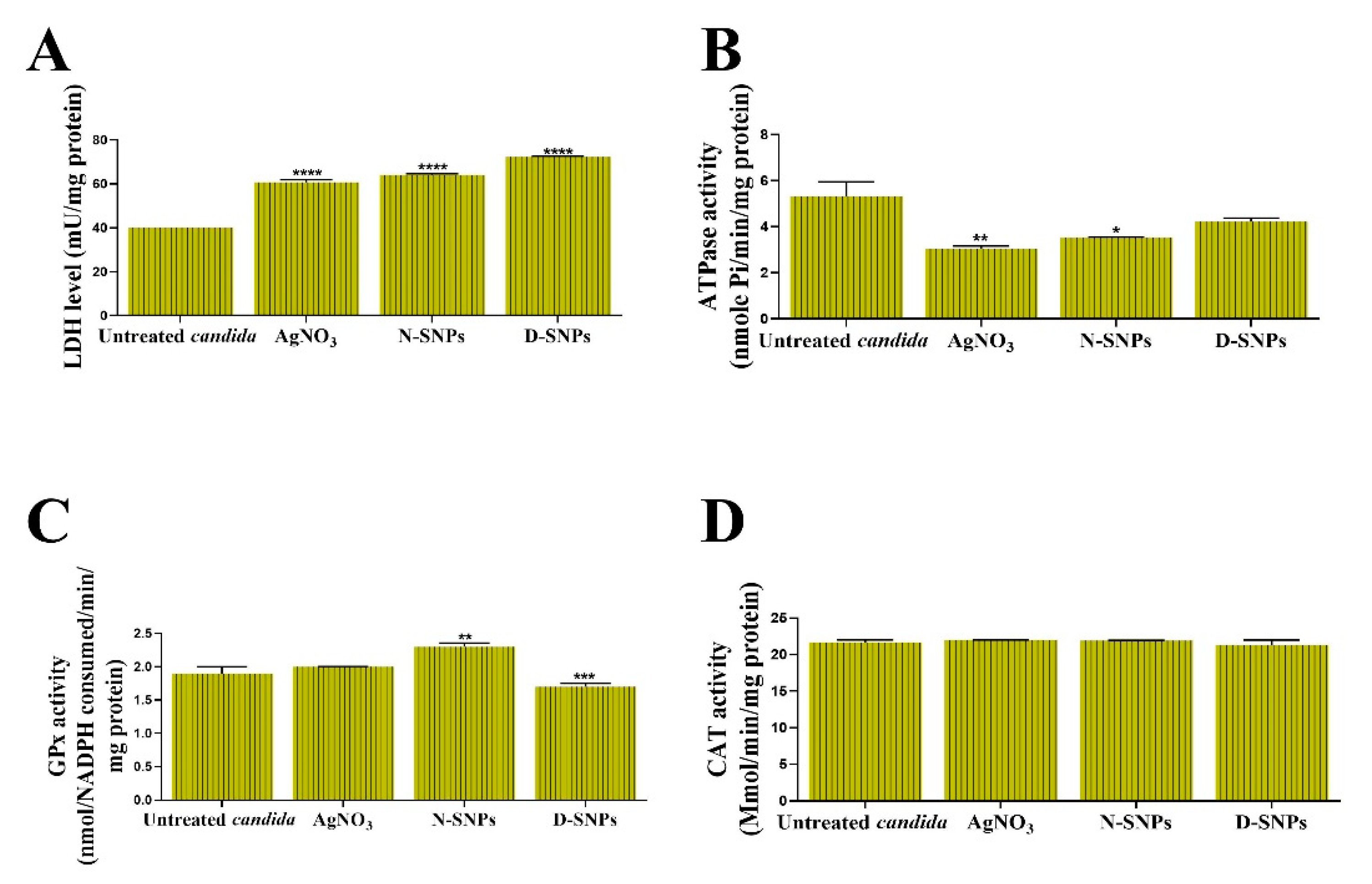
3.2. The Influence of AgNO3 and the Biogenic D-SNPs and N-SNPs on C. albicans Enzyme Activity
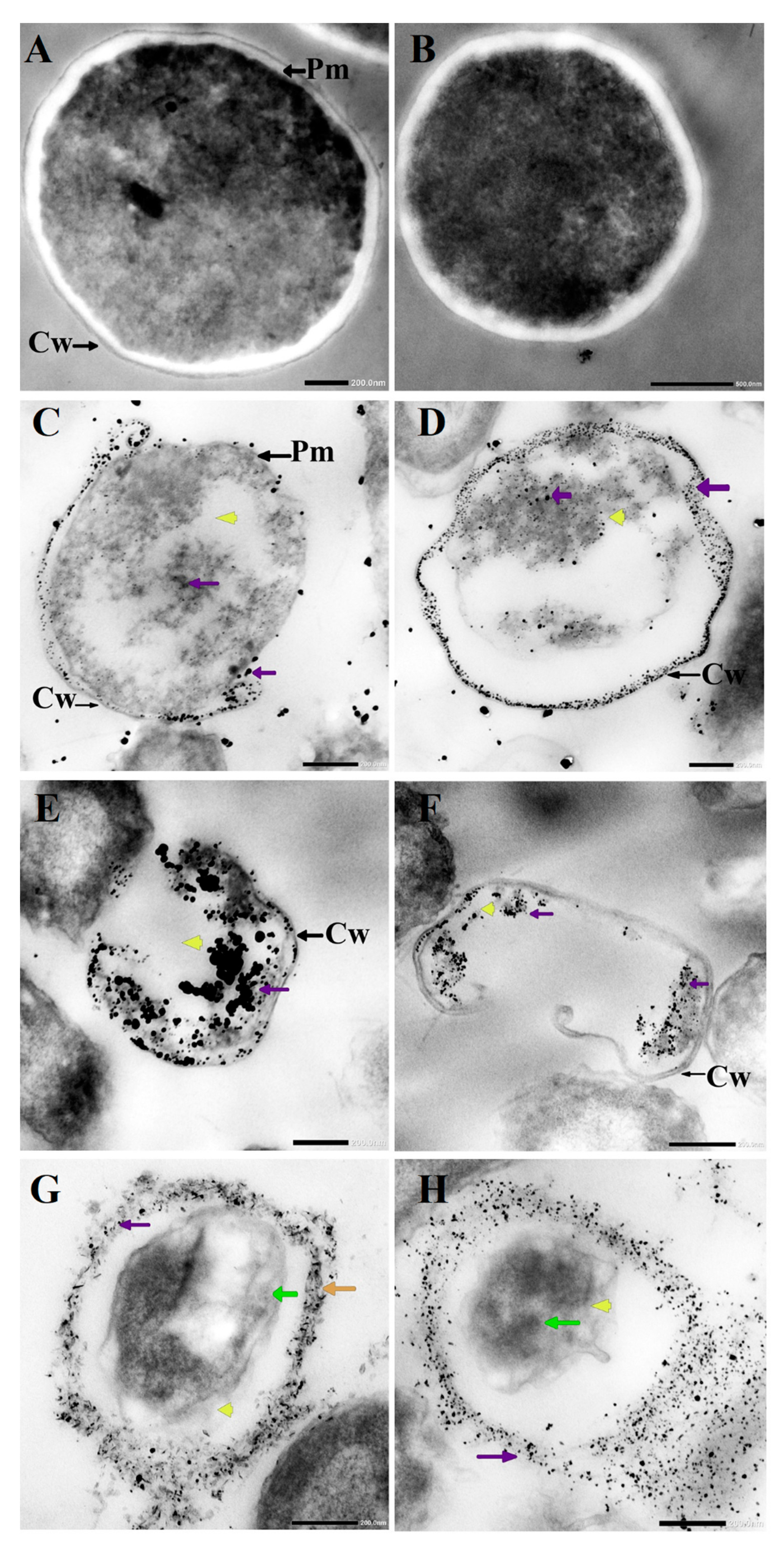
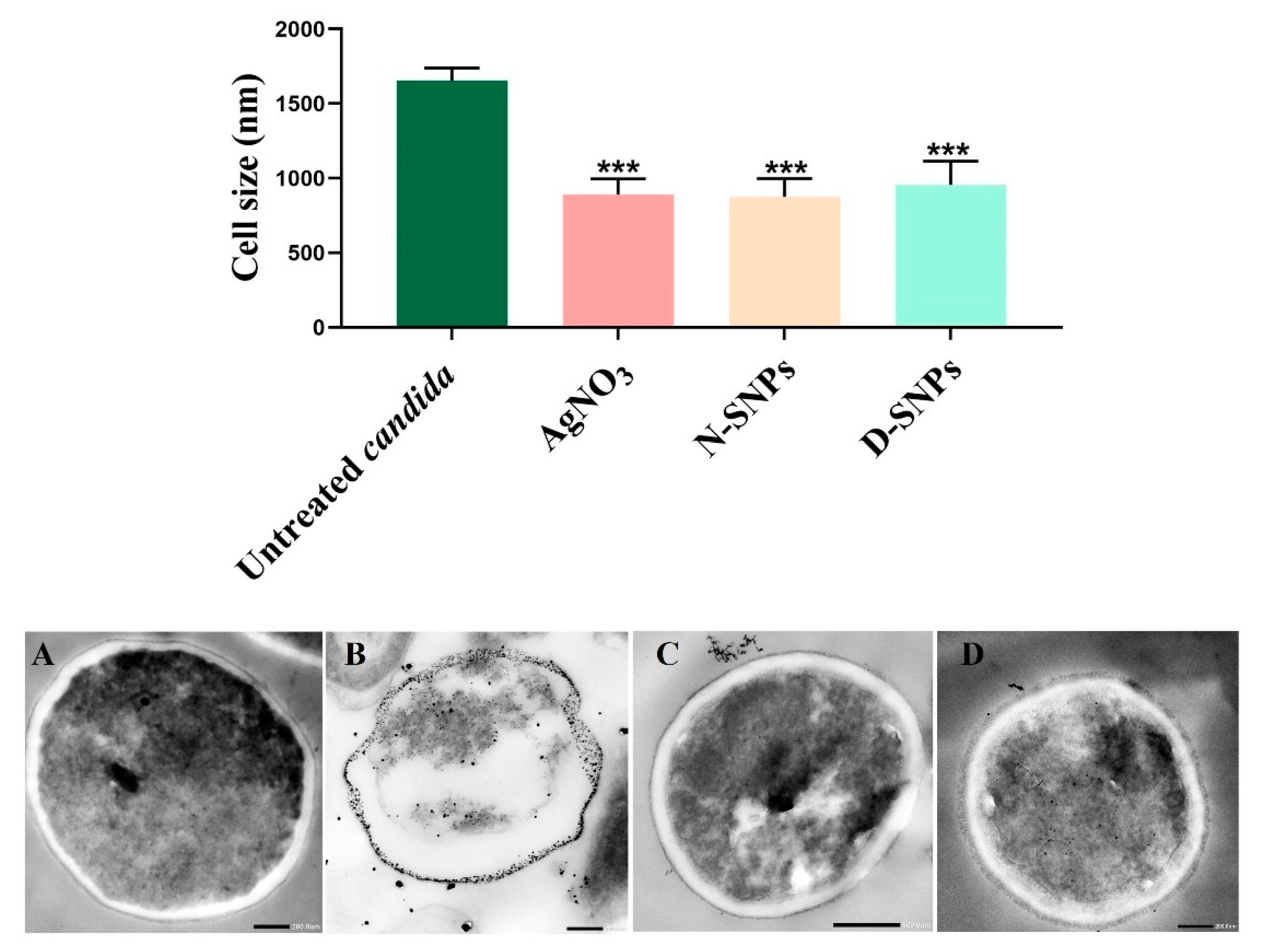
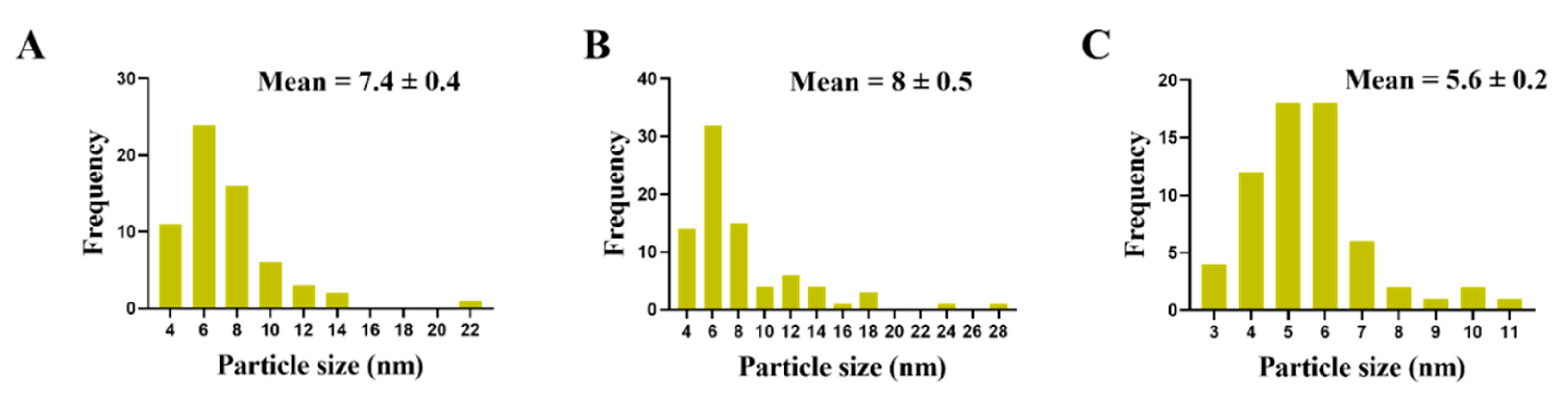
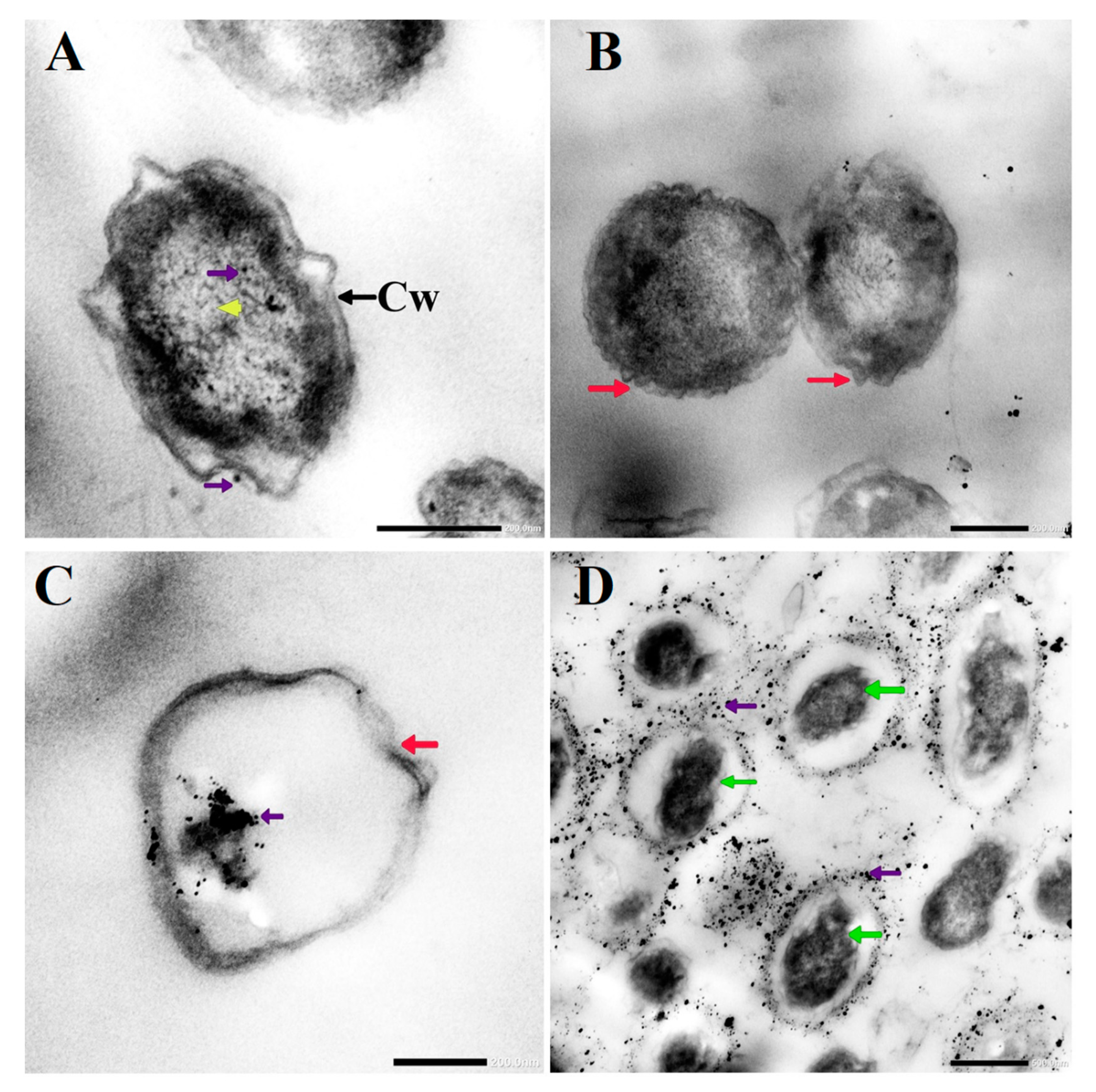
3.3. Morphological Changes in C. albicans Cells Caused by AgNO3, D-SNPs, and N-SNPs
3.4. The qRT-PCR
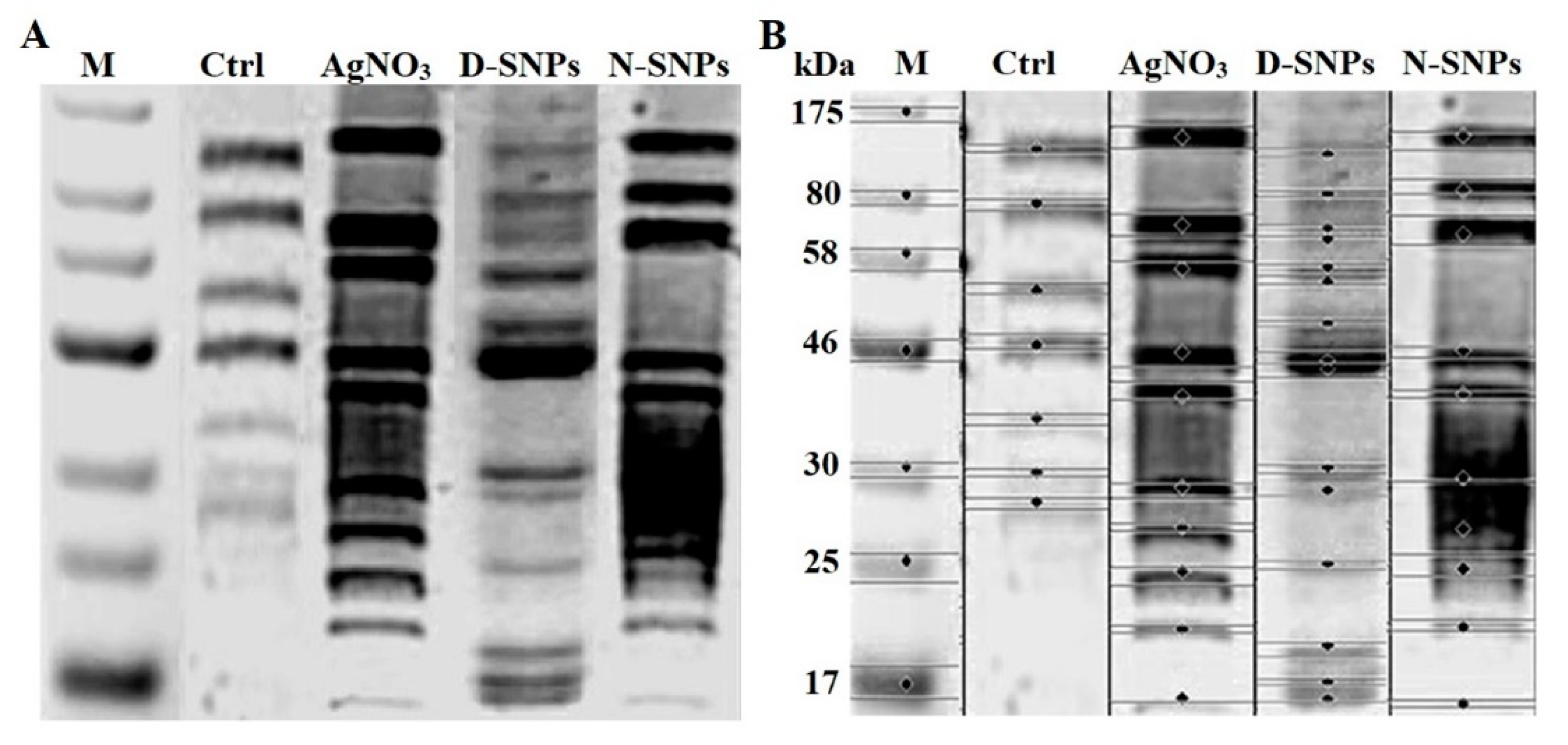
3.5. SDS–PAGE
4. Discussion
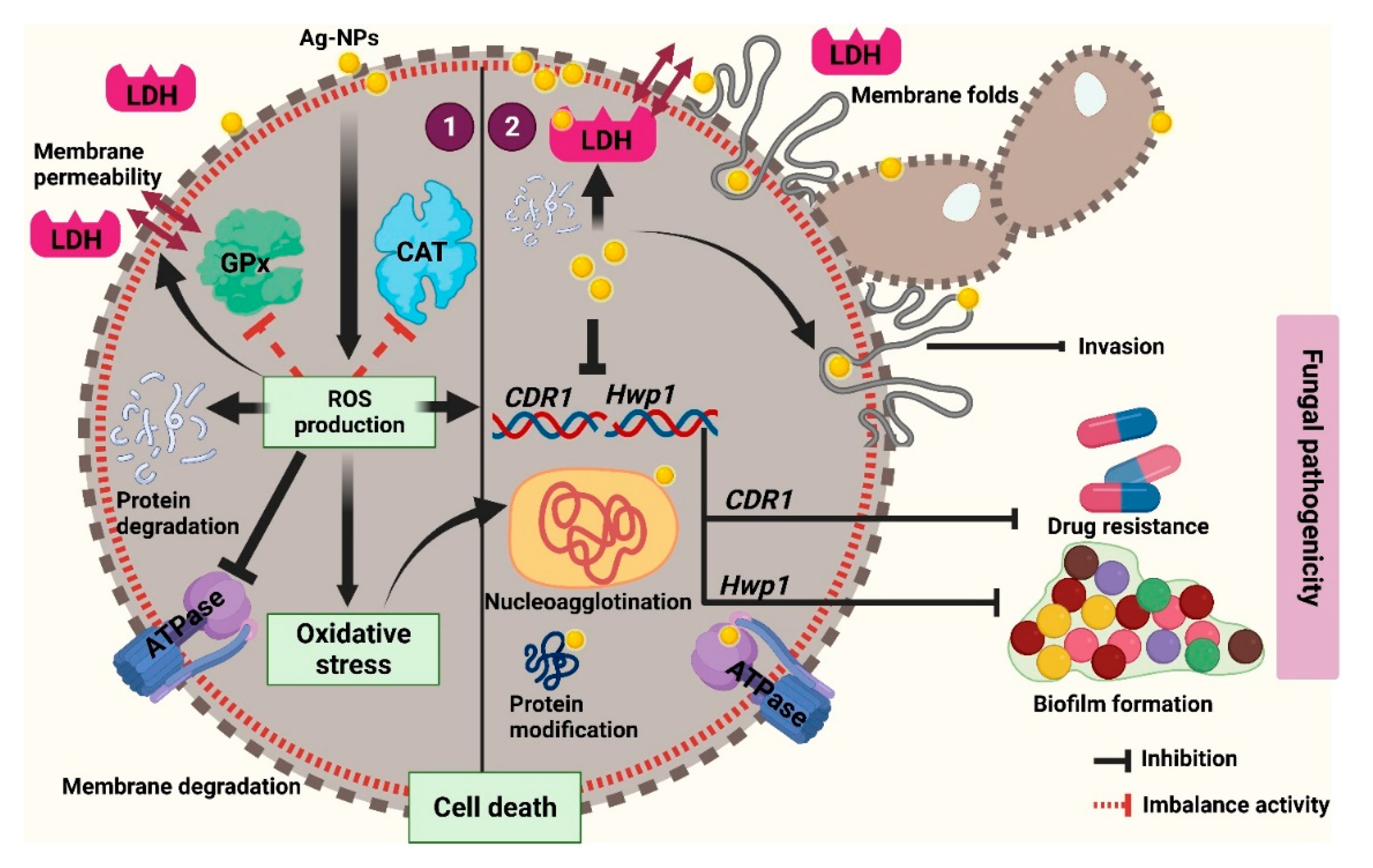
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dadar, M.; Tiwari, R.; Karthik, K.; Chakraborty, S.; Shahali, Y.; Dhama, K. Candida albicans-Biology, molecular characterization, pathogenicity, and advances in diagnosis and control–An update. Microb. Pathog. 2018, 117, 128–138. [Google Scholar] [CrossRef]
- Zhang, X.; Li, T.; Chen, X.; Wang, S.; Liu, Z. Nystatin enhances the immune response against Candida albicans and protects the ultrastructure of the vaginal epithelium in a rat model of vulvovaginal candidiasis. BMC Microbiol. 2018, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.; Sun, H.Y.; Leong, H.N.; Barez, M.Y.C.; Huang, P.Y.; Talwar, D.; Wang, J.H.; Mansor, M.; Wahjuprajitno, B.; Patel, A. Echinocandins in invasive candidiasis. Mycoses 2013, 56, 601–609. [Google Scholar] [CrossRef]
- Goncalves, S.S.; Souza, A.C.R.; Chowdhary, A.; Meis, J.F.; Colombo, A.L. Epidemiology and molecular mechanisms of antifungal resistance in Candida and Aspergillus. Mycoses 2016, 59, 198–219. [Google Scholar] [CrossRef] [PubMed]
- Salci, T.P.; Negri, M.; Abadio, A.K.; Svidzinski, T.I.; Kioshima, É.S. Targeting Candida spp. to develop antifungal agents. Drug Discov. Today 2018, 23, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef]
- Wypij, M.; Jędrzejewski, T.; Trzcińska-Wencel, J.; Ostrowski, M.; Rai, M.; Golińska, P. Green Synthesized Silver Nanoparticles: Antibacterial and Anticancer Activities, Biocompatibility, and Analyses of Surface-Attached Proteins. Front. Microbiol. 2021, 12, 888. [Google Scholar] [CrossRef]
- Hosseini, S.S.; Joshaghani, H.; Shokohi, T.; Ahmadi, A.; Mehrbakhsh, Z. Antifungal activity of ZnO nanoparticles and nystatin and downregulation of SAP1-3 genes expression in fluconazole-resistant Candida albicans isolates from vulvovaginal candidiasis. Infect. Drug Resist. 2020, 13, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dedman, C.J.; Newson, G.C.; Davies, G.-L.; Christie-Oleza, J.A. Mechanisms of silver nanoparticle toxicity on the marine cyanobacterium Prochlorococcus under environmentally-relevant conditions. Sci. Total. Environ. 2020, 747, 141229. [Google Scholar] [CrossRef] [PubMed]
- Lara, H.H.; Romero-Urbina, D.G.; Pierce, C.; Lopez-Ribot, J.L.; Arellano-Jiménez, M.J.; Jose-Yacaman, M. Effect of silver nanoparticles on Candida albicans biofilms: An ultrastructural study. J. Nanobiotechnol. 2015, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Zhang, L. Nanomaterials arising amid antibiotic resistance. Nat. Rev. Microbiol. 2021, 19, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Inam, M.; Foster, J.C.; Gao, J.; Hong, Y.; Du, J.; Dove, A.P.; O’Reilly, R.K. Size and shape affects the antimicrobial activity of quaternized nanoparticles. J. Polym. Sci. Part A Polym. Chem. 2019, 57, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Al-Zaban, M.I. Novel Biogenic Silver Nanoparticle-Induced Reactive Oxygen Species Inhibit the Biofilm Formation and Virulence Activities of Methicillin-Resistant Staphylococcus aureus (MRSA) Strain. Front. Bioeng. Biotechnol. 2020, 8, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Helmlinger, J.; Sengstock, C.; Groß-Heitfeld, C.; Mayer, C.; Schildhauer, T.; Köller, M.; Epple, M. Silver nanoparticles with different size and shape: Equal cytotoxicity, but different antibacterial effects. RSC advances 2016, 6, 18490–18501. [Google Scholar] [CrossRef] [Green Version]
- Dastjerdi, R.; Montazer, M. A review on the application of inorganic nano-structured materials in the modification of textiles: Focus on anti-microbial properties. Colloids Surf. B Biointerfaces 2010, 79, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, V.S.; Mudiam, M.K.R.; Kumar, M.; Dwivedi, S.P.; Singh, S.P.; Prasad, T. Silver nanoparticles induced alterations in multiple cellular targets, which are critical for drug susceptibilities and pathogenicity in fungal pathogen (Candida albicans). Int. J. Nanomed. 2018, 13, 2647. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, L.; Fan, Y.; Feng, Q.; Cui, F.-z. Biocompatibility and toxicity of nanoparticles and nanotubes. J. Nanomater. 2012, 2012, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Muñoz, R.; Meza-Villezcas, A.; Fournier, P.; Soria-Castro, E.; Juarez-Moreno, K.; Gallego-Hernández, A.; Bogdanchikova, N.; Vazquez-Duhalt, R.; Huerta-Saquero, A. Enhancement of antibiotics antimicrobial activity due to the silver nanoparticles impact on the cell membrane. PLoS ONE 2019, 14, e0224904. [Google Scholar] [CrossRef] [Green Version]
- Hamida, R.S.; Ali, M.A.; Redhwan, A.; Bin-Meferij, M.M. Cyanobacteria–A Promising Platform in Green Nanotechnology: A Review on Nanoparticles Fabrication and Their Prospective Applications. Int. J. Nanomed. 2020, 15, 6033–6066. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Abdelmeguid, N.E.; Al-Zaban, M.I.; Baz, L.; Bin-Meferij, M.M. Lichens—A Potential Source for Nanoparticles Fabrication: A Review on Nanoparticles Biosynthesis and Their Prospective Applications. J. Fungi 2021, 7, 291. [Google Scholar] [CrossRef]
- Asmathunisha, N.; Kathiresan, K. A review on biosynthesis of nanoparticles by marine organisms. Colloids Surf. B Biointerfaces 2013, 103, 283–287. [Google Scholar] [CrossRef]
- Zhang, D.; Ma, X.-l.; Gu, Y.; Huang, H.; Zhang, G.-w. Green Synthesis of Metallic Nanoparticles and Their Potential Applications to Treat Cancer. Front. Chem. 2020, 8, 799–817. [Google Scholar] [CrossRef] [PubMed]
- Wahab, M.A.; Li, L.; Li, H.; Abdala, A. Silver Nanoparticle-Based Nanocomposites for Combating Infectious Pathogens: Recent Advances and Future Prospects. Nanomaterials 2021, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Boateng, J.; Catanzano, O. Advanced therapeutic dressings for effective wound healing—A review. J. Pharm. Sci. 2015, 104, 3653–3680. [Google Scholar] [CrossRef] [Green Version]
- Mazurak, V.C.; Burrell, R.E.; Tredget, E.E.; Clandinin, M.T.; Field, C.J. The effect of treating infected skin grafts with Acticoat™ on immune cells. Burns 2007, 33, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Bobo, D.; Robinson, K.J.; Islam, J.; Thurecht, K.J.; Corrie, S.R. Nanoparticle-based medicines: A review of FDA-approved materials and clinical trials to date. Pharm. Res. 2016, 33, 2373–2387. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Al-Zaban, M.I. Lethal Mechanisms of Nostoc-Synthesized Silver Nanoparticles against Different Pathogenic Bacteria. Int. J. Nanomed. 2020, 15, 10499. [Google Scholar] [CrossRef] [PubMed]
- Weir, E.; Lawlor, A.; Whelan, A.; Regan, F. The use of nanoparticles in anti-microbial materials and their characterization. Analyst 2008, 133, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; Ahmad, K.; Baig, M.H.; Lee, E.J.; Choi, I. Mechanistic insights into the antimicrobial actions of metallic nanoparticles and their implications for multidrug resistance. Int. J. Mol. Sci. 2019, 20, 2468. [Google Scholar] [CrossRef] [Green Version]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.s.; Lee, J.; Hwang, J.H.; Kim, K.J.; Lee, D.G. Silver nanoparticles induce apoptotic cell death in Candida albicans through the increase of hydroxyl radicals. FEBS J. 2012, 279, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Bin-Meferij, M.M.; Hamida, R.S. Biofabrication and antitumor activity of silver nanoparticles utilizing novel nostoc sp. Bahar M. Int. J. Nanomed. 2019, 14, 9019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamida, R.S.; Abdelmeguid, N.E.; Ali, M.A.; Bin-Meferij, M.M.; Khalil, M.I. Synthesis of silver nanoparticles using a novel cyanobacteria Desertifilum sp. extract: Their antibacterial and cytotoxicity effects. Int. J. Nanomed. 2020, 15, 49. [Google Scholar] [CrossRef] [Green Version]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Ashrafi, M.; Bayat, M.; Mortazavi, P.; Hashemi, S.J.; Meimandipour, A. Antimicrobial effect of chitosan–silver–copper nanocomposite on Candida albicans. J. Nanostructure Chem. 2020, 10, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: An alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 2017, 18, 569. [Google Scholar] [CrossRef] [Green Version]
- Andrés, M.T.; Fierro, J.F. Antimicrobial mechanism of action of transferrins: Selective inhibition of H+-ATPase. Antimicrob. Agents Chemother. 2010, 54, 4335–4342. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Qasim, M.; Park, C.; Yoo, H.; Choi, D.Y.; Song, H.; Park, C.; Kim, J.-H.; Hong, K. Cytotoxicity and transcriptomic analysis of silver nanoparticles in mouse embryonic fibroblast cells. Int. J. Mol. Sci. 2018, 19, 3618. [Google Scholar] [CrossRef] [Green Version]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Redhwan, A. Cytotoxic effect of green silver nanoparticles against ampicillin-resistant Klebsiella pneumoniae. RSC Adv. 2020, 10, 21136–21146. [Google Scholar] [CrossRef]
- Aslani, P.; Roudbar Mohammadi, S.; Roudbary, M. Novel Formulated Zinc Oxide Nanoparticles Reduce Hwp1 Gene Expression Involved in Biofilm Formation in Candida albicans with Minimum Cytotoxicity Effect on Human Cells. Jundishapur J. Microbiol. 2018, 11, 1–6. [Google Scholar] [CrossRef]
- Parsameher, N.; Rezaei, S.; Khodavasiy, S.; Salari, S.; Hadizade, S.; Kord, M.; Mousavi, S.A.A. Effect of biogenic selenium nanoparticles on ERG11 and CDR1 gene expression in both fluconazole-resistant and-susceptible Candida albicans isolates. Curr. Med. Mycol. 2017, 3, 16. [Google Scholar]
- Mucci, M.J.; Cuestas, M.L.; Landanburu, M.F.; Mujica, M.T. Prevalence of Candida albicans, Candida dubliniensis and Candida africana in pregnant women suffering from vulvovaginal candidiasis in Argentina. Rev. Iberoam. Micol. 2017, 34, 72–76. [Google Scholar] [CrossRef]
- Soliman, H.; Elsayed, A.; Dyaa, A. Antimicrobial activity of silver nanoparticles biosynthesised by Rhodotorula sp. strain ATL72. Egypt. J. Basic Appl. Sci. 2018, 5, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Logeswari, P.; Silambarasan, S.; Abraham, J. Synthesis of silver nanoparticles using plants extract and analysis of their antimicrobial property. J. Saudi Chem. Soc. 2015, 19, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- El-Baz, A.F.; El-Batal, A.I.; Abomosalam, F.M.; Tayel, A.A.; Shetaia, Y.M.; Yang, S.T. Extracellular biosynthesis of anti-Candida silver nanoparticles using Monascus purpureus. J. Basic Microbiol. 2016, 56, 531–540. [Google Scholar] [CrossRef]
- Burd, J.; Usategui-Gomez, M. A colorimetric assay for serum lactate dehydrogenase. Clin. Chim. Acta 1973, 46, 223–227. [Google Scholar] [CrossRef]
- Korshed, P.; Li, L.; Liu, Z.; Wang, T. The molecular mechanisms of the antibacterial effect of picosecond laser generated silver nanoparticles and their toxicity to human cells. PLoS ONE 2016, 11, e0160078. [Google Scholar]
- Singh, P.; Kim, Y.J.; Singh, H.; Wang, C.; Hwang, K.H.; Farh, M.E.-A.; Yang, D.C. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int. J. Nanomed. 2015, 10, 2567. [Google Scholar]
- Bartolommei, G.; Moncelli, M.R.; Tadini-Buoninsegni, F. A method to measure hydrolytic activity of adenosinetriphosphatases (ATPases). PLoS ONE 2013, 8, e58615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano, R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1988, 947, 1–28. [Google Scholar] [CrossRef]
- Monk, B.C.; Niimi, M.; Shepherd, M.G. The Candida albicans plasma membrane and H (+)-ATPase during yeast growth and germ tube formation. J. Bacteriol. 1993, 175, 5566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhao, Y.; Tian, Y.; Zhang, W.; Lü, X.; Jiang, X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 2012, 33, 2327–2333. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fodor, E.; Szabó-Nagy, A.; Erdei, L. The effects of cadmium on the fluidity and H+-ATPase activity of plasma membrane from sunflower and wheat roots. J. Plant Physiol. 1995, 147, 87–92. [Google Scholar] [CrossRef]
- Circu, M.L.; Aw, T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free. Radic. Biol. Med. 2010, 48, 749–762. [Google Scholar] [CrossRef] [Green Version]
- Barros, D.; Pradhan, A.; Mendes, V.M.; Manadas, B.; Santos, P.M.; Pascoal, C.; Cássio, F. Proteomics and antioxidant enzymes reveal different mechanisms of toxicity induced by ionic and nanoparticulate silver in bacteria. Environ. Sci. Nano 2019, 6, 1207–1218. [Google Scholar] [CrossRef]
- Jiang, H.S.; Qiu, X.N.; Li, G.B.; Li, W.; Yin, L.Y. Silver nanoparticles induced accumulation of reactive oxygen species and alteration of antioxidant systems in the aquatic plant Spirodela polyrhiza. Environ. Toxicol. Chem. 2014, 33, 1398–1405. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, H.; Shen, Y.; Zhang, W.; Zhang, L. Antibacterial activity of silver nanoparticles of different particle size against Vibrio Natriegens. PLoS ONE 2019, 14, e0222322. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.; Lee, M.J.; Yun, S.J.; Kim, K.; Choi, I.-H.; Park, S. Silver nanoparticles induce reactive oxygen species-mediated cell cycle delay and synergistic cytotoxicity with 3-bromopyruvate in Candida albicans, but not in Saccharomyces cerevisiae. Int. J. Nanomed. 2019, 14, 4801. [Google Scholar] [CrossRef] [Green Version]
- Choi, O.; Hu, Z. Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ. Sci. Technol. 2008, 42, 4583–4588. [Google Scholar] [CrossRef] [PubMed]
- Salem, O.M.; Hoballah, E.; Ghazi, S.M.; Hanna, S.N. Antimicrobial activity of microalgal extracts with special emphasize on Nostoc sp. Life Sci. J. 2014, 11, 752–758. [Google Scholar]
- Abo-State, M.A.; Shanab, S.M.; Ali, H.E.; Abdullah, M.A. Screening of antimicrobial activity of selected Egyptian cyanobacterial species. J. Ecol. Health Envion. 2015, 3, 7–13. [Google Scholar]
- Panáček, A.; Kolář, M.; Večeřová, R.; Prucek, R.; Soukupova, J.; Kryštof, V.; Hamal, P.; Zbořil, R.; Kvitek, L. Antifungal activity of silver nanoparticles against Candida spp. Biomaterials 2009, 30, 6333–6340. [Google Scholar] [CrossRef] [PubMed]
- Wakshlak, R.B.-K.; Pedahzur, R.; Avnir, D. Antibacterial activity of silver-killed bacteria: The” zombies” effect. Sci. Rep. 2015, 5, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Rahimi, G.; Alizadeh, F.; Khodavandi, A. Mycosynthesis of silver nanoparticles from Candida albicans and its antibacterial activity against Escherichia coli and Staphylococcus aureus. Trop. J. Pharm. Res. 2016, 15, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Chaffin, W.L.; López-Ribot, J.L.; Casanova, M.; Gozalbo, D.; Martínez, J.P. Cell wall and secreted proteins of Candida albicans: Identification, function, and expression. Microbiol. Mol. Biol. Rev. 1998, 62, 130–180. [Google Scholar] [CrossRef] [Green Version]
- Staab, J.F.; Bahn, Y.-S.; Tai, C.-H.; Cook, P.F.; Sundstrom, P. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 2004, 279, 40737–40747. [Google Scholar] [CrossRef] [Green Version]
- Naglik, J.R.; Fostira, F.; Ruprai, J.; Staab, J.F.; Challacombe, S.J.; Sundstrom, P. Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J. Med. Microbiol. 2006, 55, 1323. [Google Scholar] [CrossRef] [PubMed]
- Akins, R.A. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 2005, 43, 285–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rad, K.K.; Falahati, M.; Roudbary, M.; Farahyar, S.; Nami, S. Overexpression of MDR-1 and CDR-2 genes in fluconazole resistance of Candida albicans isolated from patients with vulvovaginal candidiasis. Curr. Med. Mycol. 2016, 2, 24. [Google Scholar]
- Gogoi, S.K.; Gopinath, P.; Paul, A.; Ramesh, A.; Ghosh, S.S.; Chattopadhyay, A. Green fluorescent protein-expressing escherichia c oli as a model system for investigating the antimicrobial activities of silver nanoparticles. Langmuir 2006, 22, 9322–9328. [Google Scholar] [CrossRef] [PubMed]
- Tamiyakul, H.; Roytrakul, S.; Jaresitthikunchai, J.; Phaonakrop, N.; Tanasupawat, S.; Warisnoicharoen, W. Changes in protein patterns of Staphylococcus aureus and Escherichia coli by silver nanoparticles capped with poly (4-styrenesulfonic acid-co-maleic acid) polymer. Asian Biomed. 2019, 13, 39–47. [Google Scholar] [CrossRef] [Green Version]

| Gene | Primers | Reference |
|---|---|---|
| Hwp1 | F: GCT ACC ACT TCA GAA TCATCA TC R: GCA CCT TCA GTC GTA GAG GAC G | [43] |
| CDR1 | F: GGTGCTAATATCCAATGTTGG R: GTAATGGTTCTCTTTCAGCTG | [42] |
| Fungi | Measurements | Treatments | |||
|---|---|---|---|---|---|
| AgNO3 | N-SNPs | D-SNPs | Amphotericin B | ||
| Candida albicans | IZ (mm) | 12 ± 0.17 | 15.8 ± 0.3 | 17.5 ± 0.3 | 20 ± 0.4 |
| MIC (mg/mL) | 1.2 | 1.2 | 1.2 | 0.00625 | |
| MFC (mg/mL) | 1.5 | 1.5 | 1.5 | 0.0125 | |
| MIC/MFC | 0.8 | 0.8 | 0.8 | 0.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamida, R.S.; Ali, M.A.; Goda, D.A.; Redhwan, A. Anticandidal Potential of Two Cyanobacteria-Synthesized Silver Nanoparticles: Effects on Growth, Cell Morphology, and Key Virulence Attributes of Candida albicans. Pharmaceutics 2021, 13, 1688. https://doi.org/10.3390/pharmaceutics13101688
Hamida RS, Ali MA, Goda DA, Redhwan A. Anticandidal Potential of Two Cyanobacteria-Synthesized Silver Nanoparticles: Effects on Growth, Cell Morphology, and Key Virulence Attributes of Candida albicans. Pharmaceutics. 2021; 13(10):1688. https://doi.org/10.3390/pharmaceutics13101688
Chicago/Turabian StyleHamida, Reham Samir, Mohamed Abdelaal Ali, Doaa A. Goda, and Alya Redhwan. 2021. "Anticandidal Potential of Two Cyanobacteria-Synthesized Silver Nanoparticles: Effects on Growth, Cell Morphology, and Key Virulence Attributes of Candida albicans" Pharmaceutics 13, no. 10: 1688. https://doi.org/10.3390/pharmaceutics13101688
APA StyleHamida, R. S., Ali, M. A., Goda, D. A., & Redhwan, A. (2021). Anticandidal Potential of Two Cyanobacteria-Synthesized Silver Nanoparticles: Effects on Growth, Cell Morphology, and Key Virulence Attributes of Candida albicans. Pharmaceutics, 13(10), 1688. https://doi.org/10.3390/pharmaceutics13101688