Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity
Abstract
:1. Introduction
2. Prolamins
3. Celiac Disease (CD)
4. Peptidases that Effectively Hydrolyze Prolamins and Their Immunogenic (Toxic) Peptides
4.1. Hydrolysis of Gluten Proteins and Their Toxic Peptides by Bacterial Peptidases
4.2. Hydrolysis of Gluten Proteins and Their Toxic Peptides by Fungal Peptidases
4.3. Hydrolysis of Gluten Proteins and Their Toxic Peptides by Insect Peptidases
4.4. Hydrolysis of Gluten Proteins and Their Toxic Peptides by Plant Peptidases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farrell, R.J.; Kelly, C.P. Celiac sprue. N. Engl. J. Med. 2002, 346, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Cappello, M.; Morreale, G.C.; Licata, A. Elderly Onset Celiac Disease: A Narrative Review. Clin. Med. Insights Gastroenterol. 2016, 9, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, L.; Molberg, O.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.; Qiao, S.W.; Arentz-Hansen, H.; Molberg, O.; Gray, G.M.; Sollid, L.M.; Khosla, C.J. Identification and analysis of multivalent proteolytically resistant peptides from gluten: Implications for celiac sprue. Proteome Res. 2005, 4, 1732–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopkins, S.; Soon, J.M. Nutritional quality, cost and availability of gluten-free food in England. Br. Food J. 2019, 121, 2867–2882. [Google Scholar] [CrossRef]
- Dunaevsky, Y.E.; Tereshchenkova, V.F.; Oppert, B.; Belozersky, M.A.; Filippova, I.Y.; Elpidina, E.N. Human proline specific peptidases: A comprehensive analysis. Biochim. Biophys. Acta 2020, 1864, 129636. [Google Scholar] [CrossRef]
- Shewry, P.R.; Tatham, A.S. The prolamin storage proteins of cereal seeds: Structure and evolution. Biochem. J. 1990, 267. [Google Scholar] [CrossRef] [Green Version]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef] [Green Version]
- Barak, S.; Mudgil, D.; Khatkar, B.S. Biochemical and functional properties of wheat gliadins: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 357–368. [Google Scholar] [CrossRef]
- Biesiekierski, J.R. What is gluten? J. Gastroenterol. Hepatol. 2017, 32 (Suppl. 1), 78–81. [Google Scholar] [CrossRef] [Green Version]
- Shewry, P.R.; Tatham, A.S.; Forde, J.; Kreis, M.; Miflin, B.J. The classification and nomenclature of wheat gluten proteins: A reassessment. J. Cereal Sci. 1986, 4, 97–106. [Google Scholar] [CrossRef]
- Haush, F.; Shan, L.; Santiago, N.A.; Gary, G.M.; Khosla, C. Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, 996–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuppan, D.; Junker, Y.; Barisani, D. Celiac disease: From pathogenesis to novel therapies. Gastroenterology 2009, 137, 1912–1933. [Google Scholar] [CrossRef]
- Sollid, L.M. Molecular basis of celiac disease. Annu. Rev. Immunol. 2000, 18, 53–81. [Google Scholar] [CrossRef] [Green Version]
- Wieser, H. The precipitating factor in coeliac disease. Baillieres Clin. Gastroenterol. 1995, 9, 191–207. [Google Scholar] [CrossRef]
- Loponen, J. Prolamin Degradation in Sourdoughs. Ph.D. Thesis, University of Helsinki, Helsinki, Finland, 20 December 2006. [Google Scholar]
- Lionetti, E.; Catassi, C. New clues in celiac disease epidemiology, pathogenesis, clinical manifestations, and treatment. Int. Rev. Immunol. 2011, 30, 219–231. [Google Scholar] [CrossRef]
- Lohi, S.; Mustalahti, K.; Kaukinen, K.; Laurila, K.; Collin, P.; Rissane, H.; Lohi, O.; Bravi, E.; Gasparin, M.; Reunanen, A.; et al. Increasing prevalence of coeliac disease over time. Aliment. Pharmacol. Ther. 2007, 26, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Kryszak, D.; Bhatti, B.; Sturgeon, C.; Helzlsouer, K.; Clipp, S.L.; Gelfond, D.; Puppa, E.; Sferruzza, A.; Fasano, A. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann. Med. 2010, 42, 530–538. [Google Scholar] [CrossRef]
- Shumilov, P.V.; Muhina, Y.G.; Netrebenko, O.K.; Ryazanova, O.V.; Schigaleva, N.E.; Kovalenko, A.A.; Levina, E.E.; Ponomareva, A.P. Modern understanding of celiac disease pathogenetic mechanisms: Defining role in course clinical variants. Pediatriya 2016, 95, 110–121. [Google Scholar]
- Grossman, G. Neurological complications of coeliac disease: What is the evidence? Pract. Neurol. 2008, 8, 77–89. [Google Scholar] [CrossRef]
- Ford, R.P. The gluten syndrome: A neurological disease. Med. Hypotheses 2009, 73, 438–440. [Google Scholar] [CrossRef]
- Verdu, E.F.; Armstrong, D.; Murray, J.A. Between celiac disease and irritable bowel syndrome: The “no man’s land” of gluten sensitivity. Am. J. Gastroenterol. 2009, 104, 1587–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Midhagen, C.; Hallert, C. High rate of gastrointestinal symptoms in celiac disease patients living on a gluten-free diet: Controlled study. Am J Gastroenterol. 2003, 98, 2023–2026. [Google Scholar] [CrossRef]
- Ilus, T.; Lähdeaho, M.L.; Salmi, T.; Katri, H.; Jukka, P.; Päivi, S.; Heini, H.; Markku, M.; Pekka, C.; Katri, K. Persistent duodenal intraepithelial lymphocytosis despite a long-term strict gluten-free diet in celiac disease. Am. J. Gastroenterol. 2012, 107, 1563–1569. [Google Scholar] [CrossRef]
- Catassi, C.; Fabiani, E.; Iacono, G.; D’Agate, C.; Francavilla, R.; Biagi, F.; Volta, U.; Accomando, S.; Picarelli, A.; De Vitis, I.; et al. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am. J. Clin. Nutr. 2007, 85, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Hischenhuber, C.; Crevel, R.; Jarry, B.; Maki, M.; Moneret-Vautrin, D.A.; Romano, A.; Troncone, R.; Ward, R. Review article: Safe amounts of gluten for patients with wheat allergy or coeliac disease. Aliment. Pharmacol. Ther. 2006, 23, 559–575. [Google Scholar] [CrossRef]
- Wieser, H.; Koehler, P. Detoxification of gluten by means of enzymatic treatment. J. AOAC Int. 2012, 95, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Goptar, I.A.; Semashko, T.A.; Danilenko, S.A.; Lysogorskaya, E.N.; Oksenoit, E.S.; Zhuzhikov, D.P.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; et al. Cysteine digestive peptidases function as post-glutamine cleaving enzymes in tenebrionid stored-product pests. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 161, 148–154. [Google Scholar] [CrossRef]
- Filippova, I.Y.; Dvoryakova, E.A.; Sokolenko, N.I.; Simonyan, T.R.; Tereshchenkova, V.F.; Zhiganov, N.I.; Dunaevsky, Y.E.; Belozersky, M.A.; Oppert, B.; Elpidina, E.N. New glutamine-containing substrates for the assay of cysteine peptidases from the C1 papain family. Front. Mol. Biosci. 2020, 7, 578758. [Google Scholar] [CrossRef] [PubMed]
- Dvoryakova, E.A.; Vinokurov, K.S.; Tereshchenkova, V.F.; Dunaevsky, Y.E.; Belozersky, M.A.; Oppert, B.; Filippova, I.Y.; Elpidina, E.N. Primary digestive cathepsins L of Tribolium castaneum larvae: Proteomic identification, properties, comparison with human lysosomal cathepsin L. Insect Biochem. Mol. Biol. 2021, submitted. [Google Scholar]
- Wei, G.; Tian, N.; Siezen, R.; Schuppa, D.; Helmerhorst, E.J. Identification of food-grade subtilisins as gluten-degrading enzymes to treat celiac disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G571–G580. [Google Scholar] [CrossRef] [Green Version]
- Bethune, M.T.; Strop, P.; Tang, Y.; Sollid, L.M.; Khosla, C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem. Biol. 2006, 13, 637–647. [Google Scholar] [CrossRef] [Green Version]
- Savvateeva, L.V.; Gorokhovets, N.V.; Makarov, V.A.; Serebryakova, M.V.; Solovyev, A.G.; Morozov, S.Y.; Reddy, V.P.; Zernii, E.Y.; Zamyatnin, A.A., Jr.; Aliev, G. Glutenase and collagenase activities of wheat cysteine protease Triticain-α: Feasibility for enzymatic therapy assays. Int. J. Biochem. Cell. Biol. 2015, 62, 115–124. [Google Scholar] [CrossRef]
- Shan, L.; Marti, T.; Sollid, L.M.; Gray, G.M.; Khosla, C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gass, J.; Ehren, J.; Strohmeier, G.; Isaacs, I.; Khosla, C. Fermentation, purification, formulation, and pharmacological evaluation of a prolyl endopeptidase from Myxococcus xanthus: Implications for celiac sprue therapy. Biotechnol. Bioeng. 2005, 92, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Marti, T.; Molberg, O.; Li, Q.; Gray, G.M.; Khosla, C.; Sollid, L.M. Prolyl endopeptidase-mediated destruction of T cell epitopes in whole gluten: Chemical and immunological characterization. J. Pharmacol. Exp. Ther. 2005, 312, 19–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyle, G.G.; Paaso, B.; Anderson, B.E.; Allen, D.D.; Mart, T.; Li, Q.; Siegel, M.; Khosla, C.; Gray, G.M. Effect of pretreatment of food gluten with prolyl endopeptidase on gluten induced malabsorption in celiac sprue. Clin. Gastroenterol. Hepatol. 2005, 3, 687–694. [Google Scholar] [CrossRef]
- Shan, L.; Mathews, I.; Khosla, C. Structural and mechanistic analysis of two prolylendopeptidases: Role of interdomain dynamics in catalysis and specificity. Proc. Natl. Acad. Sci. USA 2005, 102, 3599–3604. [Google Scholar] [CrossRef] [Green Version]
- Piper, J.L.; Gray, G.M.; Khosla, C. Effect of Prolyl Endopeptidase on Digestive-Resistant Gliadin Peptides in vivo. J. Pharm. Exp. Ther. 2004, 311, 213–219. [Google Scholar] [CrossRef]
- Matysiak-Budnik, T.; Candalh, C.; Cellier, C.; Dugave, C.; Namane, A.; Vidal-Martinez, T.; Cerf-Bensussan, N.; Heyman, M. Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology 2005, 129, 786–796. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Martin, M.C.; Redruello, B.; Del Rio, B.; Ladero, V.; Palanski, B.A.; Khosla, C.; Fernandez, M.; Alvarez, M.A. Generation of food-grade recombinant Lactobacillus casei delivering Myxococcus xanthus prolyl endopeptidase. Appl. Microbiol. Biotechnol. 2014, 98, 6689–6700. [Google Scholar] [CrossRef] [PubMed]
- Shetty, R.; Vestergaard, M.; Jessen, F.; Hagglund, P.; Knorr, V.; Koehler, P.; Prakash, H.S.; Hobley, T.J. Discovery, cloning and characterisation of proline specific prolyl endopeptidase, a gluten degrading thermo-stable enzyme from Sphaerobacter thermophiles. Enzyme Microb. Technol. 2017, 107, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Spicher, G.; Nierle, W. Proteolytic activity of sourdough bacteria. Appl. Microbiol. Biotechnol. 1988, 28, 487–492. [Google Scholar] [CrossRef]
- Gerez, C.L.; Font de Valdez, G.; Rollan, G.C. Functionality of lactic acid bacteria peptidase activities in the hydrolysis of gliadin-like fragments. Lett. Appl. Microbiol. 2008, 47, 427–432. [Google Scholar] [CrossRef]
- Gerez, C.L.; Dallagnol, A.; Rollan, G.; Font de Valdez, G. A combination of two lactic acid bacteria improves the hydrolysis of gliadin during wheat dough fermentation. Food Microbiol. 2012, 32, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Xue, W.; Liu, C.; Tian, Y.; Zhang, K.; Zhu, Z. Screening of Lactic Acid Bacteria and Yeasts from Sourdough as Starter Cultures for Reduced Allergenicity Wheat Products. Foods 2020, 9, 751. [Google Scholar] [CrossRef]
- Katina, K.; Arendt, E.; Liukkonen, K.-H.; Autio, K.; Flander, L.; Poutanen, K. Potential of sourdough for healthier cereal products. Trends Food Sci. Technol. 2005, 16, 104–112. [Google Scholar] [CrossRef]
- Rizzello, C.G.; De Angelis, M.; Di Cagno, R.; Camarca, A.; Silano, M.; Losito, I.; De Vincenzi, M.; De Bari, M.D.; Palmisano, F.; Maurano, F.; et al. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: New perspectives for celiac disease. Appl. Environ. Microbiol. 2007, 73, 4499–4507. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.; Cassone, A.; Rizzello, C.; Gagliardi, F.; Minervini, F.; Calasso, M.; Di Cagno, R.; Francavilla, R.; Gobbetti, M. Mechanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl. Enviroment. Microbiol. 2010, 76, 508–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Sieiro, P.; Redruello, B.; Ladero, V.; Martin, M.C.; Fernandez, M.; Alvarez, M.A. Screening sourdough samples for gliadin-degrading activity revealed Lactobacillus casei strains able to individually metabolize the coeliac-disease-related 33-mer peptide. Can. J. Microbiol. 2016, 62, 422–430. [Google Scholar] [CrossRef]
- Stefanska, I.; Piasecka-Jozwiak, K.; Kotyrba, D.; Kolenda, M.; Stecka, K.M. Selection of lactic acid bacteria strains for the hydrolysis of allergenic proteins of wheat flour. J. Sci. Food Agric. 2016, 96, 3897–3905. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, N.E.C.; Esteves, F.G.; dos Santos-Pinto, J.R.A.; De Paula, C.P.; Da Cunha, A.F.; Malavazi, I.; Palma, M.S.; Rodrigues-Filho, E. Digestion of intact gluten proteins by Bifidobacterium species: Reduction of cytotoxicity and proinflammatory responses. J. Agric. Food Chem. 2020, 68, 4485–4492. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, A.; Cerrone, R.; Capobianco, D.; Filardo, S.; Mancini, P.; Fanelli, S.; Mastromarino, P.; Mosca, L. A probiotic preparation hydrolyzes gliadin and protects intestinal cells from the toxicity of pro-inflammatory peptides. Nutrients 2020, 12, 495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzozowski, B.; Stasiewicz, K.; Ostolski, M.; Adamczak, M. Reducing Immunoreactivity of Gliadins and Coeliac-Toxic Peptides Using Peptidases from L. acidophilus 5e2 and A. niger. Catalysts 2020, 10, 923. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Sun, X.; Salih, E.; Oppenheim, F.G. Identification of Lys-Pro-Gln as a novel cleavage site specificity of saliva-associated proteases. J. Biol. Chem. 2008, 283, 19957–19966. [Google Scholar] [CrossRef] [Green Version]
- Messana, I.; Cabras, T.; Pisano, E.; Sanna, M.T.; Olianas, A.; Manconi, B.; Pellegrini, M.; Paludetti, G.; Scarano, E.; Fiorita, A.; et al. Trafficking and postsecretory events responsible for the formation of secreted human salivary peptides: A proteomics approach. Mol. Cell. Proteomics. 2008, 7, 911–926. [Google Scholar] [CrossRef] [Green Version]
- Helmerhorst, E.J.; Zamakhchari, M.; Schuppan, D.; Oppenheim, F.G. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PLoS ONE 2010, 5, e13264. [Google Scholar] [CrossRef] [Green Version]
- Zamakhchari, M.; Wei, G.; Dewhirst, F.; Lee, J.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS ONE 2011, 6, e24455. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Feo, M.; Wei, G.; Blumenkranz, G.; Dewhirst, F.E.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin. Microbiol. Infect. 2013, 19, E386–E394. [Google Scholar] [CrossRef] [Green Version]
- Darwish, G.; Helmerhorst, E.J.; Schuppan, D.; Oppenheim, F.G.; Wei, G. Pharmaceutically modified subtilisins withstand acidic conditions and effectively degrade gluten in vivo. Sci. Rep. 2019, 9, 7505. [Google Scholar] [CrossRef] [Green Version]
- Tian, N.; Faller, L.; Leffler, D.A.; Kelly, C.P.; Hansen, J.; Bosch, J.A.; Wei, G.; Paster, B.J.; Schuppan, D.; Helmerhorst, E.J. Salivary gluten degradation and oral microbial profiles in healthy individuals and celiac disease patients. Appl. Environ. Microbiol. 2017, 83, e03330–e03316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caminero, A.; Galipeau, H.J.; McCarville, J.L.; Johnston, C.W.; Bernier, S.P.; Russell, A.K.; Jury, J.; Herran, A.R.; Casqueiro, J.; Tye-Din, J.A.; et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 2016, 151, 670–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caminero, A.; McCarville, J.L.; Galipeau, H.J.; Deraison, C.; Bernier, S.P.; Constante, M.; Rolland, C.; Meisel, M.; Murray, J.A.; Yu, X.B.; et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease activated receptor-2. Nat. Commun. 2019, 10, 1198. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.R.; Stanley, E.J.; Wolf, S.; Toland, A.; Wu, S.J.; Hadidi, D.; Mills, J.H.; Baker, D.; Pultz, I.S.; Siegel, J.B. Computational design of an α-gliadin peptidase. J. Am. Chem. Soc. 2012, 134, 20513–20520. [Google Scholar] [CrossRef]
- Wolf, C.; Siegel, J.B.; Tinberg, C.; Camarca, A.; Gianfrani, C.; Paski, S.; Guan, R.; Montelione, G.; Baker, D.; Pultz, I.S. Engineering of Kuma030: A gliadin peptidase that rapidly degrades immunogenic gliadin peptides in gastric conditions. J. Am. Chem. Soc. 2015, 137, 13106–13113. [Google Scholar] [CrossRef] [Green Version]
- Gass, J.; Bethune, M.T.; Siegel, M.; Spencer, A.; Khosla, C. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology 2007, 133, 472–480. [Google Scholar] [CrossRef] [Green Version]
- Lahdeaho, M.L.; Kaukinen, K.; Laurila, K.; Vuotikka, P.; Koivurova, O.P.; Karja-Lahdensuu, T.; Marcantonio, A.; Adelman, D.C.; Mäki, M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.A.; Kelly, C.P.; Green, P.H.R.; Marcantonio, A.; Wu, T.-T.; Mäki, M.; Adelman, D.C. No difference between latiglutenase and placebo in reducing villous atrophy or improving symptoms in patients with symptomatic celiac disease. Gastroenterology 2017, 152, 787–798.e2. [Google Scholar] [CrossRef] [Green Version]
- Syage, J.A.; Murray, J.A.; Green, P.H.R.; Khosla, C. Latiglutenase improves symptoms in seropositive celiac disease patients while on a gluten-free diet. Dig. Dis. Sci. 2017, 62, 2428–2432. [Google Scholar] [CrossRef]
- Syage, J.A.; Green, P.H.R.; Khosla, C.; Adelman, D.C.; Sealey-Voyksner, J.A.; Murray, J.A. Latiglutenase treatment for celiac disease: Symptom and quality of life improvement for seropositive patients on a gluten-free diet. GastroHep. 2019, 1, 293–301. [Google Scholar] [CrossRef]
- ImmunogenX. Available online: https://immunogenx.com (accessed on 20 February 2021).
- Celiac Disease Foundation. Available online: https://celiac.org (accessed on 20 February 2021).
- Stepniak, D.; Spaenij-Dekking, L.; Mitea, C.; Moester, M.; de Ru, A.; Baak-Pablo, R.; van Veelen, P.; Edens, L.; Koning, F. Highly efficient gluten degradation with a newly identified prolyl endoprotease: Implications for celiac disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G621–G629. [Google Scholar] [CrossRef] [PubMed]
- Mitea, C.; Havenaar, R.; Drijfhout, J.W.; Edens, L.; Dekking, L.; Koning, F. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: Implications for coeliac disease. Gut 2008, 57, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Salden, B.N.; Monserrat, V.; Troost, F.J.; Bruins, M.J.; Edens, L.; Bartholome, R.; Haenen, G.R.; Winkens, B.; Koning, F.; Masclee, A.A. Randomised clinical study: Aspergillus niger-derived enzyme digests gluten in the stomach of healthy volunteers. Aliment. Pharmacol. Ther. 2015, 42, 273–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konig, J.; Holster, S.; Bruins, M.J.; Brummer, R.J. Randomized clinical trial: Effective gluten degradation by Aspergillus niger-derived enzyme in a complex meal setting. Sci. Rep. 2017, 7, 13100. [Google Scholar] [CrossRef] [PubMed]
- Montserrat, V.; Bruins, M.J.; Edens, L.; Koning, F. Influence of dietary components on Aspergillus niger prolyl endoprotease mediated gluten degradation. Food Chem. 2015, 174, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Tack, G.J.; Van De Water, J.M.; Bruins, M.J.; Edens, L.; Kooy-Winkelaar, E.M.C.; van Bergen, J.; Koning, F.; Bonnet, P.; von Blomberg, B.M.E.; Schreurs, M.W.J.; et al. Consumption of gluten with gluten-degrading enzyme by celiac patients: A pilot-study. World J. Gastroenterol. 2013, 19, 5837–5847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, K.D.; Rashtak, S.; Lahr, B.D.; Melton, L.J.; Krause, P.K.; Maggi, K.; Talley, N.J.; Murray, J.A. Screening for celiac disease in a North American population: Sequential serology and gastrointestinal symptoms. Am. J. Gastroenterol. 2011, 106, 1333–1339. [Google Scholar] [CrossRef] [Green Version]
- DSM. Available online: https://www.dsm.com (accessed on 20 February 2021).
- Krishnareddy, S.; Stier, K.; Recanati, M.; Lebwohl, B.; Green, P.H. Commercially available glutenases: A potential hazard in coeliac disease. Ther. Adv. Gastroenterol. 2017, 10, 473–481. [Google Scholar] [CrossRef]
- Schulz, K.; Giesler, L.; Linke, D.; Berger, R.G. A prolyl endopeptidase from Flammulina velutipes for the possible degradation of celiac disease provoking toxic peptides in cereal proteins. Process Biochem. 2018, 73, 47–55. [Google Scholar] [CrossRef]
- Eugster, P.J.; Salamin, K.; Grouzmann, E.; Monod, M. Production and characterization of two major Aspergillus oryzae secreted prolyl endopeptidases able to efficiently digest proline-rich peptides of gliadin. Microbiology 2015, 161, 2277–2288. [Google Scholar] [CrossRef]
- Blinkovsky, A.; Byun, T.; Brown, K.; Golightly, E.; Klotz, A. A non-specific aminopeptidase from Aspergillus. Biochim. Biophys. Acta 2000, 1480, 171–181. [Google Scholar] [CrossRef]
- Byun, T.; Kofod, L.; Blinkovsky, A. Synergistic action of an X-prolyl dipeptidyl aminopeptidase and a non-specific aminopeptidase in protein hydrolysis. J. Agric. Food Chem. 2001, 49, 2061–2063. [Google Scholar] [CrossRef]
- Ehren, J.; Moron, B.; Martin, E.; Bethune, M.T.; Gray, G.M.; Khosla, C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS ONE 2009, 4, e6313. [Google Scholar] [CrossRef] [Green Version]
- Vora, H.; McIntire, J.; Kumar, P.; Deshpande, M.; Khosla, C. A scalable manufacturing process for pro-EP-B2, a cysteine protease from barley indicated for celiac sprue. Biotechnol. Bioeng. 2007, 98, 177–185. [Google Scholar] [CrossRef]
- Goptar, I.A.; Filippova, I.Y.; Lysogorskaya, E.N.; Oksenoit, E.S.; Vinokurov, K.S.; Zhuzhikov, D.P.; Bulushova, N.V.; Zalunin, I.A.; Dunaevsky, Y.E.; Belozersky, M.A.; et al. Localization of post-proline cleaving peptidases in Tenebrio molitor larval midgut. Biochimie 2008, 90, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Goptar, A.; Koulemzina, I.A.; Filippova, I.Y.; Lysogorskaya, E.N.; Oksenoit, E.S.; Zhuzhikov, D.P.; Dunaevsky, Y.E.; Belozersky, M.A.; Elpidina, E.N. Properties of Post-Proline Cleaving Enzymes from Tenebrio molitor. Russ. J. Bioorg. Chem. 2008, 34, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Goptar, I.A.; Shagin, D.A.; Shagina, I.A.; Mudrik, E.S.; Smirnova, Y.A.; Zhuzhikov, D.P.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; et al. A digestive prolyl carboxypeptidase in Tenebrio molitor larvae. Insect Biochem. Mol. Biol. 2013, 43, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Tereshchenkova, V.F.; Goptar, I.A.; Kulemzina, I.A.; Zhuzhikov, D.P.; Serebryakova, M.V.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; Elpidina, E.N. Dipeptidyl peptidase 4—An important digestive peptidase in Tenebrio molitor larvae. Insect Biochem. Mol. Biol. 2016, 76, 38–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tereshchenkova, V.F.; Goptar, I.A.; Zhuzhikov, D.P.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; Elpidina, E.N. Prolidase is a critical enzyme for complete gliadin digestion in Tenebrio molitor larvae. Arch. Insect Biochem. Physiol. 2017, 95, e21395. [Google Scholar] [CrossRef] [PubMed]
- Tereshchenkova, V.F.; Klyachko, Е.V.; Benevolensky, S.V.; Belozersky, М.А.; Dunaevsky, Y.Е.; Filippova, I.Y.; Elpidina, Е.N. Preparation and Purification of Recombinant Dipeptidyl Peptidase 4 Tenebrio molitor. Appl. Biochem. Microbiol. 2019, 55, 231–236. [Google Scholar] [CrossRef]
- Elpidina, E.N.; Goptar, I.A. Digestive peptidases in Tenebrio molitor and possibility of use to treat celiac disease. Entomol. Res. 2007, 37, 139–147. [Google Scholar] [CrossRef]
- Mika, N.; Gorshkov, V.; Spengler, B.; Zorn, H.; Rühl, M. Characterization of novel insect associated peptidases for hydrolysis of food proteins. Eur. Food Res. Technol. 2015, 240, 431–439. [Google Scholar] [CrossRef]
- Gass, J.; Vora, H.; Bethune, M.T.; Gray, G.M.; Khosla, C. Effect of barley endoprotease EP-B2 on gluten digestion in the intact rat. J. Pharmacol. Exp. Ther. 2006, 318, 1178–1186. [Google Scholar] [CrossRef] [Green Version]
- Tye-Din, J.A.; Anderson, R.P.; Ffrench, R.A.; Brown, G.J.; Hodsman, P.; Siegel, M.; Botwick, W.; Shreeniwas, R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin. Immunol. 2010, 134, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Siegel, M.; Garber, M.E.; Spencer, A.G.; Botwick, W.; Kumar, P.; Williams, R.N.; Kozuka, K.; Shreeniwas, R.; Pratha, V.; Adelman, D.C. Safety, tolerability, and activity of ALV003: Results from two phase 1 single, escalating-dose clinical trials. Dig. Dis. Sci. 2012, 57, 440–450. [Google Scholar] [CrossRef]
- Savvateeva, L.V.; Zamyatnin, A.A. Prospects of developing medicinal therapeutic strategies and pharmaceutical design for effective gluten intolerance treatment. Curr. Pharm. Des. 2016, 22, 2439–2449. [Google Scholar] [CrossRef] [Green Version]
- Gorokhovets, N.V.; Makarov, V.A.; Petushkova, A.I.; Prokopets, O.S.; Rubtsov, M.A.; Savvateeva, L.V.; Zernii, E.Y.; Zamyatnin, A.A. Rational design of recombinant papain-like cysteine protease: Optimal domain structure and expression conditions for wheat-derived enzyme triticain-α. Int. J. Mol. Sci. 2017, 18, 1395. [Google Scholar] [CrossRef] [PubMed]
- Cornell, H.J.; Doherty, W.; Stelmasiak, T. Papaya latex enzymes capable of detoxification of gliadin. Amino Acids 2010, 38, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Buddrick, O.; Cornell, H.J.; Small, D.M. Reduction of toxic gliadin content of whole grain bread by the enzyme caricain. Food Chem. 2015, 170, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Cornell, H.J.; Stelmasiak, T.; Small, D.M.; Buddrick, O. Application of the rat liver lysosome assay to determining the reduction of toxic gliadin content during breadmaking. Food Chem. 2016, 192, 924–927. [Google Scholar] [CrossRef] [PubMed]
- Pultz, I.S.; Hill, M.; Vitanza, J.M.; Wolf, C.; Saaby, L.; Liu, T.; Winkle, P.; Leffler, D.A. Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology 2021, 161, 81–93. [Google Scholar] [CrossRef]
- Takeda Pharmaceutical Company Limited. Available online: https://www.takeda.com (accessed on 20 February 2021).
- Amyra Biotech. Available online: https://www.amyra.com (accessed on 20 February 2021).
- NEMYSIS LIMITED. Available online: https://nemysisltd.com (accessed on 20 February 2021).
- Janssen, G.; Christis, C.; Kooy-Winkelaar, Y.; Edens, L.; Smith, D.; van Veelen, P.; Koning, F. Ineffective degradation of immunogenic gluten epitopes by currently available digestive enzyme supplements. PLoS ONE 2015, 10, e0128065. [Google Scholar] [CrossRef] [Green Version]
- König, J.; Brummer, R.J. Is an enzyme supplement for celiac disease finally on the cards? Expert Rev. Gastroenterol. Hepatol. 2018, 12, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Sapone, A.; Lammers, K.M.; Mazzarella, G.; Mikhailenko, I.; Cartenì, M.; Casolaro, V.; Fasano, A. Differential mucosal IL-17 expression in two gliadin-induced disorders: Gluten sensitivity and the autoimmune enteropathy celiac disease. Int. Arch. Allergy Immunol. 2010, 152, 75–80. [Google Scholar] [CrossRef] [Green Version]
- Uhde, M.; Ajamian, M.; Caio, G.; De Giorgio, R.; Indart, A.; Green, P.H.; Verna, E.C.; Volta, U.; Alaedini, A. Intestinal cell damage and systemic immune activation in individuals reporting sensitivity to wheat in the absence of coeliac disease. Gut 2016, 65, 1930–1937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biesiekierski, J.R.; Newnham, E.D.; Irving, P.M.; Barrett, J.S.; Haines, M.; Doecke, J.D.; Shepherd, S.J.; Muir, J.G.; Gibson, P.R. Gluten causes gastrointestinal symptoms in subjects without celiac disease: A double-blind randomized placebo-controlled trial. Am. J. Gastroenterol. 2011, 106, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Di Sabatino, A.; Volta, U.; Salvatore, C.; Biancheri, P.; Caio, G.; De Giorgio, R.; Di Stefano, M.; Corazza, G.R. Small amounts of gluten in subjects with suspected nonceliac gluten sensitivity: A randomized, double-blind, placebo-controlled, cross-over trial. Clin. Gastroenterol. Hepatol. 2015, 13, 1604–1612. [Google Scholar] [CrossRef]
- Rees, D.; Holtrop, G.; Chope, G.; Moar, K.M.; Cruickshank, M.; Hoggard, N. A randomised, double-blind, cross-over trial to evaluate bread, in which gluten has been pre-digested by prolyl endoprotease treatment, in subjects self-reporting benefits of adopting a gluten-free or low-gluten diet. Br. J. Nutr. 2018, 119, 496–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biesiekierski, J.R.; Peters, S.L.; Newnham, E.D.; Rosella, O.; Muir, J.G.; Gibson, P.R. No effects of gluten in patients with self-reported non-celiac gluten sensitivity after dietary reduction of fermentable, poorly absorbed, short-chain carbohydrates. Gastroenterology 2013, 145, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Skodje, G.I.; Sarna, V.K.; Minelle, I.H.; Rolfsen, K.L.; Muir, J.G.; Gibson, P.R.; Veierød, M.B.; Henriksen, C.; Lundin, K.E. Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology 2018, 154, 529–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
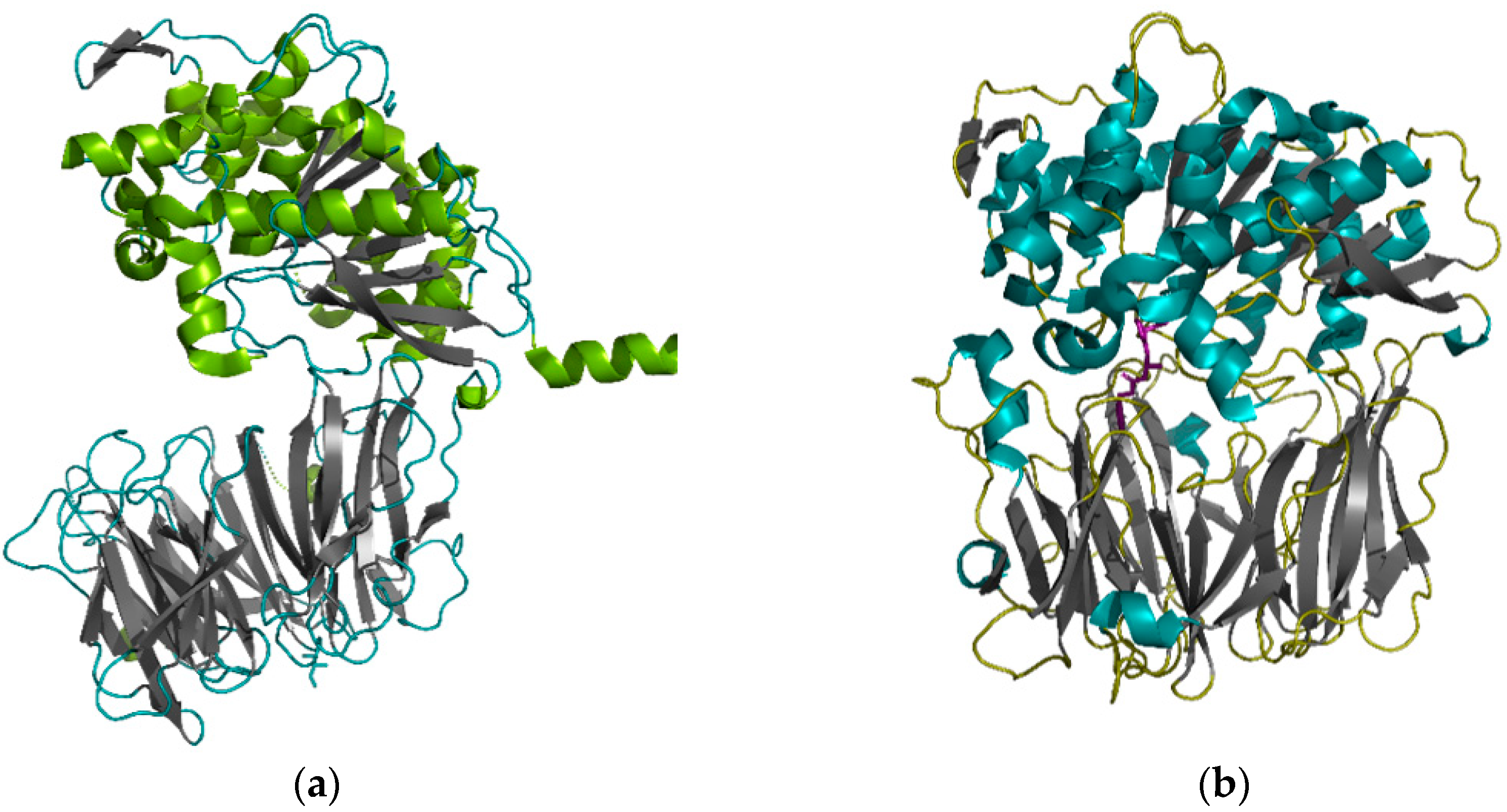
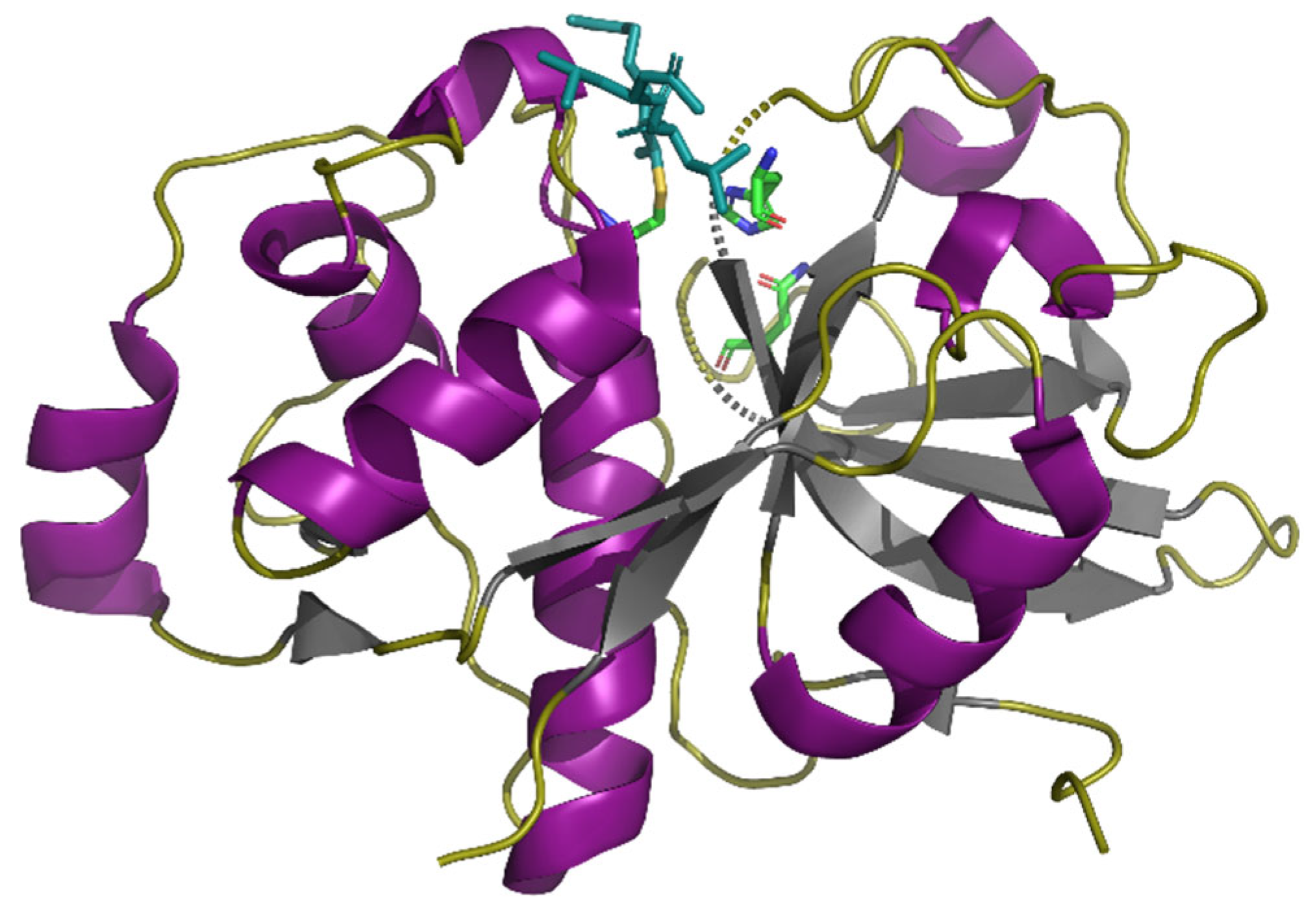
| Size | Peptide Sequence | Origin | Position | Composition (Pro, Gln), % |
|---|---|---|---|---|
| 33-mer | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | α-2 gliadin | 56–88 | Pro 40, Gln 30 |
| 26-mer | FLQPQQPFPQQPQQPYPQQPQQPFPQ | γ-5 gliadin | 26–51 | Pro 35, Gln 46 |
| 20-mer | LQPQQPFPQQPQQPYPQQPQ | γ-5 gliadin | 60–79 | Pro 35, Gln 50 |
| 20-mer | QQQQPPFSQQQQSPFSQQQQ | glutenin | Pro 15, Gln 60 | |
| 19-mer | LGQQQPFPPQQPYPQPQPF | α-gliadin | 31–49 | Pro 37, Gln 37 |
| 17-mer | QLQPFPQPELPYPQPQS | α-gliadin | 57–73 | Pro 35, Gln 29 |
| 15-mer | VQGQGIIQPQQPAQL | γ-gliadin | Pro 13, Gln 40 | |
| 15-mer | QQPPFSQQQQQPLPQ | glutenin | Pro 27, Gln 53 | |
| 14-mer | PQPQLPYPQPQLPY | α-2 gliadin | 62–75 | Pro 43, Gln 29 |
| 13-mer | LGQQQPFPPQQPY | α-gliadin | 31–43 | Pro 31, Gln 38 |
| 12-mer | FSQPQQQFPQPQ | γ-5 gliadin | 102–113 | Pro 25, Gln 50 |
| 12-mer | QLQPFPQPQLPY | α-9 gliadin | 57–68 | Pro 33, Gln 33 |
| 10-mer | QPQQSFPQQQ | γ-gliadin | Pro 20, Gln 60 |
| Number | Peptidase Class | Enzymes | Substrates 1 |
|---|---|---|---|
| 1 | Serine peptidases | Prolyloligopeptidase (POP), prolylendopeptidase (РЕР), fibroblast activation protein (FAP) | (Xaa)n-Xbb-Pro↓Xbb-(Xaa)n, n = 1–13 (the length of the peptide is approximately 30 amino acid residues) |
| 2 | Dipeptidylpeptidases (DPP) 2, DPP 4, DPP 8, DPP 9, FAP | Xbb-Pro↓Xbb-(Xaa)n, n = 2–12 | |
| 3 | Prolylcarboxypeptidase (PRCP) | (Xaa)n-Xbb-Pro↓Xbb, n—any number | |
| 4 | Metallopeptidases | Aminopeptidases P (APP) 1, APP2, APP3 | Xbb↓Pro(Xaa)n, n = 1–9 |
| 5 | Prolidase | Xbb↓Pro |
| Product | Company | Base of the Drug (Origin) | Clinical Trial Phase | References |
|---|---|---|---|---|
| KumaMax, Kuma030, Kuma062, TAK-062 | Takeda Pharmaceutical Company Limited, Tokyo, Japan | Kumamolysine-As Alicyclobacillus sendaiensis | 1 | [66,105,106] |
| Latiglutenase ALV003 | Alvine Pharmaceuticals Inc., San Carlos, CA, USA | Prolyl oligopeptidase (POP) Sphingomonas capsule + cysteine peptidase from barley (EP-B2) | 2 | [67,68,69,71,73] |
| Tolerase G | DSM Nutritional Products, Kaiseraugst, Switzerland | Prolylendopeptidase of the mold fungus Aspergillus niger (AN-PEP) | Dietary supplement | [77,81] |
| AMYRA’s enzymes, AMY01 | AMYRA Biotech AGBasel, Switzerland | Combination of fungal exopeptidases | Dietary supplement | [107] |
| AMYRA’s enzymes, AMY02 | AMYRA Biotech AGBasel, Switzerland | Combination of fungal exopeptidases | Pre-clinical | [107] |
| Nemysis E40 | Nemysis Ltd., Dublin, Ireland | Endopeptidase soil Actinoallomurus strain | Pre-clinical | [108] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunaevsky, Y.E.; Tereshchenkova, V.F.; Belozersky, M.A.; Filippova, I.Y.; Oppert, B.; Elpidina, E.N. Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity. Pharmaceutics 2021, 13, 1603. https://doi.org/10.3390/pharmaceutics13101603
Dunaevsky YE, Tereshchenkova VF, Belozersky MA, Filippova IY, Oppert B, Elpidina EN. Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity. Pharmaceutics. 2021; 13(10):1603. https://doi.org/10.3390/pharmaceutics13101603
Chicago/Turabian StyleDunaevsky, Yakov E., Valeriia F. Tereshchenkova, Mikhail A. Belozersky, Irina Y. Filippova, Brenda Oppert, and Elena N. Elpidina. 2021. "Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity" Pharmaceutics 13, no. 10: 1603. https://doi.org/10.3390/pharmaceutics13101603
APA StyleDunaevsky, Y. E., Tereshchenkova, V. F., Belozersky, M. A., Filippova, I. Y., Oppert, B., & Elpidina, E. N. (2021). Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity. Pharmaceutics, 13(10), 1603. https://doi.org/10.3390/pharmaceutics13101603