Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents
Abstract
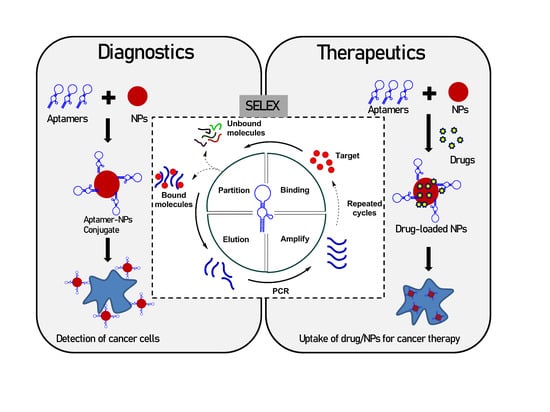
1. Introduction
2. Aptamers in Cancer Diagnosis
3. Aptamers in Infectious Disease Diagnosis
4. Aptamers as Therapeutic Agents
5. Challenges
6. Conclusions
Funding
Conflicts of Interest
References
- Yüce, M.; Ullah, N.; Budak, H. Trends in aptamer selection methods and applications. Analyst 2015, 140, 5379–5399. [Google Scholar] [CrossRef] [PubMed]
- Yüce, M.; Kurt, H.; Hussain, B.; Budak, H. Systematic Evolution of Ligands by Exponential Enrichment for Aptamer Selection. In Biomedical Applications of Functionalized Nanomaterials: Concepts, Development and Clinical Translation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 211–243. ISBN 9780323508797. [Google Scholar]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Sun, H.; Zu, Y. Aptamers and Their Applications in Nanomedicine. Small 2015, 11, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- Kurt, H.; Yüce, M.; Hussain, B.; Budak, H. Dual-excitation upconverting nanoparticle and quantum dot aptasensor for multiplexed food pathogen detection. Biosens. Bioelectron. 2016, 81, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Yüce, M.; Kurt, H.; Hussain, B.; Ow-Yang, C.W.; Budak, H. Exploiting Stokes and anti-Stokes type emission profiles of aptamer-functionalized luminescent nanoprobes for multiplex sensing applications. Chem. Select 2018, 3, 5814–5823. [Google Scholar] [CrossRef]
- Kurt, H.; Eyüpoǧlu, A.E.; Sütlü, T.; Budak, H.; Yüce, M. Plasmonic Selection of ssDNA Aptamers against Fibroblast Growth Factor Receptor. ACS Comb. Sci. 2019, 21, 578–587. [Google Scholar] [CrossRef]
- Hussain, B.; Yüce, M.; Ullah, N.; Budak, H. Bioconjugated nanomaterials for monitoring food contamination. In Nanobiosensors; Elsevier: Amsterdam, The Netherlands, 2017; pp. 93–127. [Google Scholar]
- Bai, C.; Lu, Z.; Jiang, H.; Yang, Z.; Liu, X.; Ding, H.; Li, H.; Dong, J.; Huang, A.; Fang, T.; et al. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 2018, 110, 162–167. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, Y.; Di, Y.; Xiu, C.; He, L.; Liao, S.; Li, D.; Huang, B. DNA aptamers from whole-serum SELEX as new diagnostic agents against gastric cancer. RSC Adv. 2019, 9, 950–957. [Google Scholar] [CrossRef]
- Zamay, G.S.; Ivanchenko, T.I.; Zamay, T.N.; Grigorieva, V.L.; Glazyrin, Y.E.; Kolovskaya, O.S.; Garanzha, I.V.; Barinov, A.A.; Krat, A.V.; Mironov, G.G.; et al. DNA Aptamers for the Characterization of Histological Structure of Lung Adenocarcinoma. Mol. Ther. Nucleic Acids 2017, 6, 150–162. [Google Scholar] [CrossRef]
- Yüce, M.; Kurt, H. How to make nanobiosensors: Surface modification and characterisation of nanomaterials for biosensing applications. RSC Adv. 2017, 7, 49386–49403. [Google Scholar] [CrossRef]
- Le, A.T.H.; Krylova, S.M.; Kanoatov, M.; Desai, S.; Krylov, S.N. Ideal-Filter Capillary Electrophoresis (IFCE) Facilitates the One-Step Selection of Aptamers. Angew. Chem. 2019, 131, 2765–2769. [Google Scholar] [CrossRef]
- Wang, J.; Yu, J.; Yang, Q.; McDermott, J.; Scott, A.; Vukovich, M.; Lagrois, R.; Gong, Q.; Greenleaf, W.; Eisenstein, M.; et al. Multiparameter Particle Display (MPPD): A Quantitative Screening Method for the Discovery of Highly Specific Aptamers. Angew. Chem. Int. Ed. 2017, 56, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Dembowski, S.K.; Bowser, M.T. Microfluidic methods for aptamer selection and characterization. Analyst 2018, 143, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Li, L.; Xu, S.; Yan, H.; Li, X.; Yazd, H.S.; Li, X.; Huang, T.; Cui, C.; Jiang, J. Nucleic Acid Aptamers for Molecular Diagnostics and Therapeutics: Advances and Perspectives. Angew. Chem. Int. Ed. 2020. [Google Scholar] [CrossRef]
- Liu, M.; Yin, Q.; Chang, Y.; Zhang, Q.; Brennan, J.D.; Li, Y. In Vitro Selection of Circular DNA Aptamers for Biosensing Applications. Angew. Chem. Int. Ed. 2019, 58, 8013–8017. [Google Scholar] [CrossRef]
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef]
- Zhong, W.; Pu, Y.; Tan, W.; Liu, J.; Liao, J.; Liu, B.; Chen, K.; Yu, B.; Hu, Y.; Deng, Y.; et al. Identification and Application of an Aptamer Targeting Papillary Thyroid Carcinoma Using Tissue-SELEX. Anal. Chem. 2019, 91, 8289–8297. [Google Scholar] [CrossRef]
- Chen, L.; He, W.; Jiang, H.; Wu, L.; Xiong, W.; Li, B.; Zhou, Z.; Qian, Y. In vivo SELEX of bone targeting aptamer in prostate cancer bone metastasis model. Int. J. Nanomed. 2018, 14, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.G.; Marangoni, K.; Fujimura, P.T.; Alves, P.T.; Silva, M.J.; Bastos, V.A.F.; Goulart, L.R.; Goulart, V.A. 3D Cell-SELEX: Development of RNA aptamers as molecular probes for PC-3 tumor cell line. Exp. Cell Res. 2016, 341, 147–156. [Google Scholar] [CrossRef]
- Ciancio, D.R.; Vargas, M.R.; Thiel, W.H.; Bruno, M.A.; Giangrande, P.H.; Mestre, M.B. Aptamers as diagnostic tools in cancer. Pharmaceuticals 2018, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S.P.; Ohtsu, T.; Nakamura, Y. Selection of RNA aptamers against recombinant transforming growth factor-β type III receptor displayed on cell surface. Biochimie 2006, 88, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Futami, K.; Kimoto, M.; Lim, Y.W.S.; Hirao, I. Genetic Alphabet Expansion Provides Versatile Specificities and Activities of Unnatural-Base DNA Aptamers Targeting Cancer Cells. Mol. Ther. Nucleic Acids 2019, 14, 158–170. [Google Scholar] [CrossRef]
- Matsunaga, K.; Kimoto, M.; Hirao, I. High-Affinity DNA Aptamer Generation Targeting von Willebrand Factor A1-Domain by Genetic Alphabet Expansion for Systematic Evolution of Ligands by Exponential Enrichment Using Two Types of Libraries Composed of Five Different Bases. J. Am. Chem. Soc. 2017, 139, 324–334. [Google Scholar] [CrossRef]
- Klußmann, S.; Nolte, A.; Bald, R.; Erdmann, V.A.; Fürste, J.P. Mirror-image RNA that binds D-adenosine. Nat. Biotechnol. 1996, 14, 1112–1115. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, W.; Liu, L.; Zhu, T.F. A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat. Chem. 2016, 8, 698–704. [Google Scholar] [CrossRef]
- Pech, A.; Achenbach, J.; Jahnz, M.; Schülzchen, S.; Jarosch, F.; Bordusa, F.; Klussmann, S. A thermostable d-polymerase for mirror-image PCR. Nucleic Acids Res. 2017, 45, 3997–4005. [Google Scholar] [CrossRef]
- Mai, J.; Li, X.; Zhang, G.; Huang, Y.; Xu, R.; Shen, Q.; Lokesh, G.L.; Thiviyanathan, V.; Chen, L.; Liu, H.; et al. DNA Thioaptamer with Homing Specificity to Lymphoma Bone Marrow Involvement. Mol. Pharm. 2018, 15, 1814–1825. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, Y.; Kitadume, S.; Morihiro, K.; Kuwahara, M.; Ozaki, H.; Sawai, H.; Imanishi, T.; Obika, S. Effect of 3′-end capping of aptamer with various 2′,4′-bridged nucleotides: Enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorg. Med. Chem. Lett. 2010, 20, 1626–1629. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Clawson, G. The Shorter the Better: Reducing Fixed Primer Regions of Oligonucleotide Libraries for Aptamer Selection. Molecules 2009, 14, 1353–1369. [Google Scholar] [CrossRef]
- Macdonald, J.; Henri, J.; Goodman, L.; Xiang, D.; Duan, W.; Shigdar, S. Development of a Bifunctional Aptamer Targeting the Transferrin Receptor and Epithelial Cell Adhesion Molecule (EpCAM) for the Treatment of Brain Cancer Metastases. ACS Chem. Neurosci. 2017, 8, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Denoyer, D.; Henri, J.; Jamieson, A.; Burvenich, I.J.G.; Pouliot, N.; Shigdar, S. Bifunctional Aptamer–Doxorubicin Conjugate Crosses the Blood–Brain Barrier and Selectively Delivers Its Payload to EpCAM-Positive Tumor Cells. Nucleic Acid Ther. 2020, 30, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Borghei, Y.S.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R.; Sheikhnejad, R. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal. Chim. Acta 2016, 904, 92–97. [Google Scholar] [CrossRef]
- Civit, L.; Theodorou, I.; Frey, F.; Weber, H.; Lingnau, A.; Gröber, C.; Blank, M.; Dambrune, C.; Stunden, J.; Beyer, M.; et al. Targeting hormone refractory prostate cancer by in vivo selected DNA libraries in an orthotopic xenograft mouse model. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Yang, N.; Hu, Z.; Su, J.; Zhong, J.; Yang, Y.; Yu, Y.; Zhu, J.; Xue, D.; Huang, Y.; et al. Aptamer-Functionalized Fluorescent Silica Nanoparticles for Highly Sensitive Detection of Leukemia Cells. Nanoscale Res. Lett. 2016, 11. [Google Scholar] [CrossRef]
- Khoshfetrat, S.M.; Mehrgardi, M.A. Amplified detection of leukemia cancer cells using an aptamer-conjugated gold-coated magnetic nanoparticles on a nitrogen-doped graphene modified electrode. Bioelectrochemistry 2017, 114, 24–32. [Google Scholar] [CrossRef]
- Hu, Z.; Tan, J.; Lai, Z.; Zheng, R.; Zhong, J.; Wang, Y.; Li, X.; Yang, N.; Li, J.; Yang, W.; et al. Aptamer Combined with Fluorescent Silica Nanoparticles for Detection of Hepatoma Cells. Nanoscale Res. Lett. 2017, 12. [Google Scholar] [CrossRef]
- Lee, J.; Kang, H.J.; Jang, H.; Lee, Y.J.; Lee, Y.S.; Ali, B.A.; Al-Khedhairy, A.A.; Kim, S. Simultaneous imaging of two different cancer biomarkers using aptamer-conjugated quantum dots. Sensors 2015, 15, 8595–8604. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, X.; Luo, X.; Wang, L.; Ma, N. DNA-Programmed Quantum Dot Polymerization for Ultrasensitive Molecular Imaging of Cancer Cells. Anal. Chem. 2016, 88, 9355–9358. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, N.; Zhang, X.; Zhou, T.; Tan, Y.; Pi, J.; Pi, L.; Cheng, S.; Zheng, H.; Cheng, Y. Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma. Int. J. Nanomed. 2017, 12, 3899–3911. [Google Scholar] [CrossRef]
- Yuan, B.; Jiang, X.; Chen, Y.; Guo, Q.; Wang, K.; Meng, X.; Huang, Z.; Wen, X. Metastatic cancer cell and tissue-specific fluorescence imaging using a new DNA aptamer developed by Cell-SELEX. Talanta 2017, 170, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Q.; Zhang, H.; Deng, T.; Feng, P.; Hu, B.; Jiang, Y.; Cao, L. Characterization of a DNA aptamer for ovarian cancer clinical tissue recognition and in vivo imaging. Cell. Physiol. Biochem. 2019, 51, 2564–2574. [Google Scholar] [CrossRef]
- Pan, Q.; Law, C.O.K.; Yung, M.M.H.; Han, K.C.; Pon, Y.L.; Lau, T.C.K. Novel RNA aptamers targeting gastrointestinal cancer biomarkers CEA, CA50 and CA72-4 with superior affinity and specificity. PLoS ONE 2018, 13, e0198980. [Google Scholar] [CrossRef]
- Hashkavayi, A.B.; Raoof, J.B.; Ojani, R.; Kavoosian, S. Ultrasensitive electrochemical aptasensor based on sandwich architecture for selective label-free detection of colorectal cancer (CT26) cells. Biosens. Bioelectron. 2017, 92, 630–637. [Google Scholar] [CrossRef]
- Beltrán-Gastélum, M.; Esteban-Fernández de Ávila, B.; Gong, H.; Venugopalan, P.L.; Hianik, T.; Wang, J.; Subjakova, V. Rapid Detection of AIB1 in Breast Cancer Cells Based on Aptamer-Functionalized Nanomotors. Chem. Phys. Chem. 2019, 20, 3177–3180. [Google Scholar] [CrossRef]
- Chen, K.; Georgiev, T.Z.; Sheng, W.; Zheng, X.; Varillas, J.I.; Zhang, J.; Hugh Fan, Z. Tumor cell capture patterns around aptamer-immobilized microposts in microfluidic devices. Biomicrofluidics 2017, 11, 054110. [Google Scholar] [CrossRef]
- Wei, Z.; Wu, Y.; Zhao, Y.; Mi, L.; Wang, J.; Wang, J.; Zhao, J.; Wang, L.; Liu, A.; Li, Y.; et al. Multifunctional nanoprobe for cancer cell targeting and simultaneous fluorescence/magnetic resonance imaging. Anal. Chim. Acta 2016, 938, 156–164. [Google Scholar] [CrossRef]
- Keshtkar, M.; Shahbazi-Gahrouei, D.; Khoshfetrat, S.; Mehrgardi, M.; Aghaei, M. Aptamer-conjugated magnetic nanoparticles as targeted magnetic resonance imaging contrast agent for breast cancer. J. Med. Signals Sens. 2016, 6, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Paiva, A.; Cabral Campello, M.P.; Paulo, A.; Mergny, J.-L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Aptamer-based Targeted Delivery of a G-quadruplex Ligand in Cervical Cancer Cells. Sci. Rep. 2019, 9, 7945. [Google Scholar] [CrossRef] [PubMed]
- Porciani, D.; Cardwell, L.N.; Tawiah, K.D.; Alam, K.K.; Lange, M.J.; Daniels, M.A.; Burke, D.H. Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Liang, C.; Li, F.; Wang, L.; Zhang, Z.K.; Wang, C.; He, B.; Li, J.; Chen, Z.; Shaikh, A.B.; Liu, J.; et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 2017, 147, 68–85. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Hu, Y.; Li, X.; Li, Z.; Duan, J.; Yang, X. Da Selection of a novel DNA aptamer against OFA/iLRP for targeted delivery of doxorubicin to AML cells. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Leitner, M.; Poturnayova, A.; Lamprecht, C.; Weich, S.; Snejdarkova, M.; Karpisova, I.; Hianik, T.; Ebner, A. Characterization of the specific interaction between the DNA aptamer sgc8c and protein tyrosine kinase-7 receptors at the surface of T-cells by biosensing AFM. Anal. Bioanal. Chem. 2017, 409, 2767–2776. [Google Scholar] [CrossRef]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Multistate Point-Prevalence Survey of Health Care–Associated Infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef]
- Suh, S.H.; Choi, S.J.; Dwivedi, H.P.; Moore, M.D.; Escudero-Abarca, B.I.; Jaykus, L.A. Use of DNA aptamer for sandwich type detection of Listeria monocytogenes. Anal. Biochem. 2018, 557, 27–33. [Google Scholar] [CrossRef]
- Pehlivan, Z.S.; Torabfam, M.; Kurt, H.; Ow-Yang, C.; Hildebrandt, N.; Yüce, M. Aptamer and nanomaterial based FRET biosensors: A review on recent advances (2014–2019). Microchim. Acta 2019, 186, 563. [Google Scholar] [CrossRef]
- Jin, B.; Wang, S.; Lin, M.; Jin, Y.; Zhang, S.; Cui, X.; Gong, Y.; Li, A.; Xu, F.; Lu, T.J. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection. Biosens. Bioelectron. 2017, 90, 525–533. [Google Scholar] [CrossRef]
- Chen, H.-L.; Hsiao, W.-H.; Lee, H.-C.; Wu, S.-C.; Cheng, J.-W. Selection and Characterization of DNA Aptamers Targeting All Four Serotypes of Dengue Viruses. PLoS ONE 2015, 10, e0131240. [Google Scholar] [CrossRef]
- Shubham, S.; Hoinka, J.; Banerjee, S.; Swanson, E.; Dillard, J.A.; Lennemann, N.J.; Przytycka, T.M.; Maury, W.; Nilsen-Hamilton, M. A 2′FY-RNA Motif Defines an Aptamer for Ebolavirus Secreted Protein. Sci. Rep. 2018, 8, 12373. [Google Scholar] [CrossRef] [PubMed]
- Saraf, N.; Villegas, M.; Willenberg, B.J.; Seal, S. Multiplex Viral Detection Platform Based on a Aptamers-Integrated Microfluidic Channel. ACS Omega 2019, 4, 2234–2240. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Haßel, S.K.; Mayer, G. Aptamers as Therapeutic Agents: Has the Initial Euphoria Subsided? Mol. Diagn. Ther. 2019, 23, 301–309. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Quirico, L.; Orso, F.; Esposito, C.L.; Bertone, S.; Coppo, R.; Conti, L.; Catuogno, S.; Cavallo, F.; de Franciscis, V.; Taverna, D. Axl-148b chimeric aptamers inhibit breast cancer and melanoma progression. Int. J. Biol. Sci. 2020, 16, 1238–1251. [Google Scholar] [CrossRef]
- Jin, D.; Takai, S.; Nonaka, Y.; Yamazaki, S.; Fujiwara, M.; Nakamura, Y. A Chymase Inhibitory RNA Aptamer Improves Cardiac Function and Survival after Myocardial Infarction. Mol. Ther. Nucleic Acids 2019, 14, 41–51. [Google Scholar] [CrossRef]
- Yoshitomi, T.; Hayashi, M.; Oguro, T.; Kimura, K.; Wayama, F.; Furusho, H.; Yoshimoto, K. Binding and Structural Properties of DNA Aptamers with VEGF-A-Mimic Activity. Mol. Ther. Nucleic Acids 2020, 19, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, H.; Sun, J.; Liu, Y.; Li, J.; Li, J.; Li, J.; Yang, H. Bispecific Aptamer Induced Artificial Protein-Pairing: A Strategy for Selective Inhibition of Receptor Function. J. Am. Chem. Soc. 2019, 141, 12673–12681. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Joyce, G.F. Specific Inhibition of MicroRNA Processing Using L -RNA Aptamers. J. Am. Chem. Soc. 2015, 137, 16032–16037. [Google Scholar] [CrossRef] [PubMed]
- Zboralski, D.; Hoehlig, K.; Eulberg, D.; Frömming, A.; Vater, A. Increasing Tumor-Infiltrating T Cells through Inhibition of CXCL12 with NOX-A12 Synergizes with PD-1 Blockade. Cancer Immunol. Res. 2017, 5, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Sett, A.; Borthakur, B.B.; Bora, U. Selection of DNA aptamers for extra cellular domain of human epidermal growth factor receptor 2 to detect HER2 positive carcinomas. Clin. Transl. Oncol. 2017, 19, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ghamande, S.; Liu, H.; Xue, L.; Zhao, S.; Tan, W.; Zhao, L.; Tang, S.-C.; Wu, D.; Korkaya, H.; et al. Targeting EGFR/HER2/HER3 with a Three-in-One Aptamer-siRNA Chimera Confers Superior Activity against HER2+ Breast Cancer. Mol. Ther. Nucleic Acids 2018, 10, 317–330. [Google Scholar] [CrossRef]
- Gilboa, E.; McNamara, J.; Pastor, F. Use of Oligonucleotide Aptamer Ligands to Modulate the Function of Immune Receptors. Clin. Cancer Res. 2013, 19, 1054–1062. [Google Scholar] [CrossRef]
- Ii, J.O.M.; Sullenger, B.; Gilboa, E.; Ii, J.O.M.; Kolonias, D.; Pastor, F.; Mittler, R.S.; Chen, L.; Giangrande, P.H.; Sullenger, B.; et al. Multivalent 4-1BB binding aptamers costimulate CD8 + T cells and inhibit tumor growth in mice Find the latest version: Technical advance Multivalent 4-1BB binding aptamers costimulate CD8 + T cells and inhibit tumor growth in mice. J. Clin. Investig. 2008, 118, 376–386. [Google Scholar] [CrossRef]
- Pastor, F.; Soldevilla, M.M.; Villanueva, H.; Kolonias, D.; Inoges, S.; de Cerio, A.L.; Kandzia, R.; Klimyuk, V.; Gleba, Y.; Gilboa, E.; et al. CD28 Aptamers as Powerful Immune Response Modulators. Mol. Ther. Nucleic Acids 2013, 2, e98. [Google Scholar] [CrossRef]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Ferrara, N.; Adamis, A.P. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discov. 2016, 15, 385–403. [Google Scholar] [CrossRef]
- Lincoff, A.M.; Mehran, R.; Povsic, T.J.; Zelenkofske, S.L.; Huang, Z.; Armstrong, P.W.; Steg, P.G.; Bode, C.; Cohen, M.G.; Buller, C.; et al. Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): A randomised clinical trial. Lancet 2016, 387, 349–356. [Google Scholar] [CrossRef]
- Verheugt, F.W.A. An anticoagulant too good to be true for revascularisation. Lancet 2016, 387, 314–315. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-based Aptamers to the 165-Amino Acid Form of Vascular Endothelial Growth Factor (VEGF 165 ). J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed]
- Biesecker, G.; Dihel, L.; Enney, K.; Bendele, R. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 1999, 42, 219–230. [Google Scholar] [CrossRef]
- Vater, A.; Sahlmann, J.; Kröger, N.; Zöllner, S.; Lioznov, M.; Maasch, C.; Buchner, K.; Vossmeyer, D.; Schwoebel, F.; Purschke, W.G.; et al. Hematopoietic Stem and Progenitor Cell Mobilization in Mice and Humans by a First-in-Class Mirror-Image Oligonucleotide Inhibitor of CXCL12. Clin. Pharmacol. Ther. 2013, 94, 150–157. [Google Scholar] [CrossRef]
- Green, L.S.; Jellinek, D.; Jenison, R.; Östman, A.; Heldin, C.H.; Janjic, N. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry 1996, 35, 14413–14424. [Google Scholar] [CrossRef]
- Floege, J.; Ostendorf, T.; Janssen, U.; Burg, M.; Radeke, H.H.; Vargeese, C.; Gill, S.C.; Green, L.S.; Janjic, N. Novel Approach to Specific Growth Factor Inhibition in Vivo. Am. J. Pathol. 1999, 154, 169–179. [Google Scholar] [CrossRef]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef]
- Gilbert, J.C.; DeFeo-Fraulini, T.; Hutabarat, R.M.; Horvath, C.J.; Merlino, P.G.; Marsh, H.N.; Healy, J.M.; BouFakhreddine, S.; Holohan, T.V.; Schaub, R.G. First-in-Human Evaluation of Anti–von Willebrand Factor Therapeutic Aptamer ARC1779 in Healthy Volunteers. Circulation 2007, 116, 2678–2686. [Google Scholar] [CrossRef]
- Jilma, B.; Paulinska, P.; Jilma-Stohlawetz, P.; Gilbert, J.; Hutabarat, R.; Knöbl, P. A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb. Haemost. 2010, 104, 563–570. [Google Scholar] [CrossRef]
- Gurbel, P.A.; Tantry, U.S. Acute coronary syndromes. In New Oral Anticoagulants; Future Medicine Ltd.: London, UK, 2012; Volume 8, pp. 34–57. ISBN 9781780840581. [Google Scholar]
- Dyke, C.K.; Steinhubl, S.R.; Kleiman, N.S.; Cannon, R.O.; Aberle, L.G.; Lin, M.; Myles, S.K.; Melloni, C.; Harrington, R.A.; Alexander, J.H.; et al. First-in-Human Experience of an Antidote-Controlled Anticoagulant Using RNA Aptamer Technology. Circulation 2006, 114, 2490–2497. [Google Scholar] [CrossRef]
- D’Amico, D.J. Pegaptanib Sodium for Neovascular Age-Related Macular Degeneration. Ophthalmology 2006, 113, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Staurenghi, G. Clinical experience with pegaptanib sodium. Clin. Ophthalmol. 2008, 2, 485. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maier, K.E.; Levy, M. From selection hits to clinical leads: Progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016, 3, 16014. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhao, G.; Zhang, S.; Nigim, F.; Zhou, G.; Yu, Z.; Song, Y.; Chen, Y.; Li, Y. AS1411-Induced Growth Inhibition of Glioma Cells by Up-Regulation of p53 and Down-Regulation of Bcl-2 and Akt1 via Nucleolin. PLoS ONE 2016, 11, e0167094. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Reyes, E.M.; Bates, P.J. Characterizing Oligonucleotide Uptake in Cultured Cells: A Case Study Using AS1411 Aptamer. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 2036, pp. 173–186. [Google Scholar]
- Schwoebel, F.; van Eijk, L.T.; Zboralski, D.; Sell, S.; Buchner, K.; Maasch, C.; Purschke, W.G.; Humphrey, M.; Zöllner, S.; Eulberg, D.; et al. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood 2013, 121, 2311–2315. [Google Scholar] [CrossRef] [PubMed]
- Maasch, C.; Buchner, K.; Eulberg, D.; Vonhoff, S.; Klussmann, S. Physicochemical Stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for Therapeutic Applications. Nucleic Acids Symp. Ser. 2008, 52, 61–62. [Google Scholar] [CrossRef] [PubMed]
- Menne, J.; Eulberg, D.; Beyer, D.; Baumann, M.; Saudek, F.; Valkusz, Z.; Więcek, A.; Haller, H. C-C motif-ligand 2 inhibition with emapticap pegol (NOX-E36) in type 2 diabetic patients with albuminuria. Nephrol. Dial. Transplant. 2016, 32, 307–315. [Google Scholar] [CrossRef]
- Zavyalova, E.; Legatova, V.; Alieva, R.; Zalevsky, A.; Tashlitsky, V.; Arutyunyan, A.; Kopylov, A. Putative Mechanisms Underlying High Inhibitory Activities of Bimodular DNA Aptamers to Thrombin. Biomolecules 2019, 9, 41. [Google Scholar] [CrossRef]
- Spiel, A.O.; Mayr, F.B.; Ladani, N.; Wagner, P.G.; Schaub, R.G.; Gilbert, J.C.; Jilma, B. The aptamer ARC1779 is a potent and specific inhibitor of von willebrand factor mediated ex vivo platelet function in acute myocardial infarction. Platelets 2009, 20, 334–340. [Google Scholar] [CrossRef]
- Jilma-Stohlawetz, P.; Gorczyca, M.; Jilma, B.; Siller-Matula, J.; Gilbert, J.; Knöbl, P. Inhibition of von Willebrand factor by ARC1779 in patients with acute thrombotic thrombocytopenic purpura. Thromb. Haemost. 2011, 105, 545–552. [Google Scholar] [CrossRef]
- Du, H.; Rosbash, M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 2002, 419, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.G.; Purdy, D.A.; Rossi, J.S.; Grinfeld, L.R.; Myles, S.K.; Aberle, L.H.; Greenbaum, A.B.; Fry, E.; Chan, M.Y.; Tonkens, R.M.; et al. First Clinical Application of an Actively Reversible Direct Factor IXa Inhibitor as an Anticoagulation Strategy in Patients Undergoing Percutaneous Coronary Intervention. Circulation 2010, 122, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Gening, L.V.; Klincheva, S.A.; Reshetnjak, A.; Grollman, A.P.; Miller, H. RNA aptamers selected against DNA polymerase inhibit the polymerase activities of DNA polymerases and. Nucleic Acids Res. 2006, 34, 2579–2586. [Google Scholar] [CrossRef] [PubMed]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, Solutions and Prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef]
- Dhiman, A.; Kalra, P.; Bansal, V.; Bruno, J.G.; Sharma, T.K. Aptamer-based point-of-care diagnostic platforms. Sens. Actuators B Chem. 2017, 246, 535–553. [Google Scholar] [CrossRef]
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef]
- Wong, E.; Goldberg, T. Mipomersen (kynamro): A novel antisense oligonucleotide inhibitor for the management of homozygous familial hypercholesterolemia. Pharm. Ther. 2014, 39, 119–122. [Google Scholar]
- Lim, K.R.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 2017, 11, 533–545. [Google Scholar] [CrossRef]
- Ottesen, E.W. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl. Neurosci. 2017, 8, 1–6. [Google Scholar] [CrossRef]
- Ganson, N.J.; Povsic, T.J.; Sullenger, B.A.; Alexander, J.H.; Zelenkofske, S.L.; Sailstad, J.M.; Rusconi, C.P.; Hershfield, M.S. Pre-existing anti–polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J. Allergy Clin. Immunol. 2016, 137, 1610–1613. [Google Scholar] [CrossRef] [PubMed]
- AlQahtani, A.D.; O’Connor, D.; Domling, A.; Goda, S.K. Strategies for the production of long-acting therapeutics and efficient drug delivery for cancer treatment. Biomed. Pharmacother. 2019, 113, 108750. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, K.D.; Gilbert, J.C.; Jilma, B. Pharmacokinetics, pharmacodynamics and safety of aptamers. Adv. Drug Deliv. Rev. 2018, 134, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.C.; Levy, M. Aptamer-Mediated Delivery and Cell-Targeting Aptamers: Room for Improvement. Nucleic Acid Ther. 2018, 28, 194–199. [Google Scholar] [CrossRef]


| Target | Aptamer Sequence (5’-3’) | SELEX Method | Binding Affinity | Brief Result | Ref |
|---|---|---|---|---|---|
| MCF-7 breast cancer cells | GGTGGTGGTGGTT-GTGGTGGTGGTGG | Cell-SELEX | 30–50 nM | AS1411 Aptamer-AuNP probes for color-based visual detection of MCF-7 breast cancer cells with a detection limit of 10 cells. | [38] |
| Prostate cancer cells | GGAGGCAACGGAG-CGGAGACATTGAC-TGAGTGAACGTGT-AGTG | In vivo SELEX | 2–100 nM | D3-21 aptamer conjugated with PEG was used for in vivo detection of prostate cancer by in vivo SELEX | [39] |
| Blood cancer or leukemia cells | TTTTTTTTTTATCT-AACTGCTGCGCCG-CCGGGAAAATACT-GTACGGTTAGA | Live cell-SELEX | - | Amine-labeled Sgc8 aptamers were conjugated to carboxyl-modified fluorescent silica NPs for highly specific and sensitive detection of leukemia cells. | [40] |
| Leukemia cells | TTTTTTTTTTATCT-AACTGCTGCGCCG-CCGGGAAAATACT-GTACGGTTAGA | Live cell-SELEX | 5.16 nM | Sgc8 aptamers were conjugated to AuNP-coated magnetic Fe3O4 NPs for highly specific and sensitive detection of leukemia cells. | [41,58] |
| HepG2 liver cancer cells | ACAGCATCCCCAT-GTGAACAATCGCA-TTGTGATTGTTAC-GGTTTCCGCCTCA-TGGACGTGCTG | Live cell-SELEX | - | TLS11a aptamer-fluorescent silica NPs conjugates for detection of liver cancer cells, HepG2. | [42] |
| Nucleolin & Tenascin-C cancer biomarkers | TTGGTGGTGGTGG-TTGTGGTGGTGGT-GG & CCTGCACTTGGCT-TGGATTTCAGAAG-GGAGACCC | Cell SELEX | - | AS1411 and TTA-1 aptamer-conjugated with QDs were used for multiplex detection of nucleolin and tenascin-C cancer biomarkers. | [43] |
| Glioma cancer cells | GCAATGGTACGGT-ACTTCCTGAATGT-TGTTTTTTCTCTT-TTCTATAGTACAA-AAGTGCACGCTAC-TTTGCTAA | Cell SELEX | - | A32-aptamer-QD conjugates were used for fluorescence-guided surgery for glioma cancer cells | [45] |
| Ovarian cancer Cells | TCTCTAGTTATTG-AGTTTTCTTTTAT-GGGTGGGTGGGG-GGTTTTT | Cell SELEX | 29.24–158 nM | R13 aptamer showed a high binding affinity with several ovarian cancer cell lines (Caov3, HO8910, A2780, and SKOV3) | [47] |
| Gastric cancer cells | GGATCCGACACGA-CCCTATAGTGAGT-CGTATTA | Cell SELEX | 16.5–156, 52.7–71.2, 30.7–38 nM | Aptamers with high affinity against gastric cancer biomarkers (CEA, CA72-4, and CA50) were selected. | [48] |
| Gastric cancer cells | CCTCGGCACGTTC-TCAGTAGCGCTCG-CTGGTCATCCCAC-A | Whole-serum subtractive SELEX | 128 nM | Highly specific aptamer (Seq-3) for gastric cancer was selected through the whole-serum subtractive SELEX | [12] |
| MCF-7 and 4T1 breast cancer cells | GGTGGTGGTGGTT-GTGGTGGTGGTGG | Cell SELEX | 30–50 nM | Fluorescein-labeled AS1411 Aptamers were integrated with an ultrasound-propelled gold nanowire motors (FAM-AIB1-apt) and MRI machine for qualitative diagnosis of breast cancer cells. | [50,53] |
| Leukemia cells | ATCTAACTGCTGC-GCCGCCGGGAAAA-TACTGTACGGTTA-GATTTTTTTTTT | Cell SELEX | 0.04 Hz | Sgc8 aptamers were integrated into a microfluidic device for rapid detection of leukemia cells. | [51] |
| HepG2 liver cancer cells | ACAGCATCCCCAT-GTGAACAATCGCA-TTGTGATTGTTAC-GGTTTCCGCCTCA-TGGACGTGCTG | Cell SELEX | - | TLS11a aptamer was conjugated with Fe3O4 NPs for rapid and specific detection and bio-imaging of HepG2 liver cancer cells in combination with MRI. | [52] |
| AML cancer cells | TGCGTGTGTAGTG-TGTCTGTTGTTTG-TATTGTTGTCTAT-CCTCTTAGGGATT-TGGGCGG | In vitro SELEX | 101 nM | AB3 aptamer was functionalized to deliver doxorubicin (Dox) drug molecules to the acute myeloid leukemia (AML) cancer cells. | [57] |
| Target | Aptamer Sequence (5’-3’) | SELEX Method | Binding Affinity | Brief Result/Specific Nanoparticles | References |
|---|---|---|---|---|---|
| Candida albicans | - | Cell SELEX | 79.76 nm 103.7 nM | AU1 and AD1 aptamers were used for the detection of the (1→3)-β-d-glucans present in the cell wall of the fungus, Candida albicans with high affinity and specificity. | [38] |
| Escherichia coli ATCC 8739 | GCAATGGTACGGT-ACTTCCCCATGAG-TGTTGTGAAATGT-TGGGACACTAGGT-GGCATAGAGCCGC-AAAAGTGCACGCT-ACTTTGCTAA | Cell SELEX | - | The FRET aptasensor detected E. coli ATCC 8739 with a LOD of 3 CFU/mL | |
| Dengue virus 2 (DENV) | GCACCGGGCAGGA-CGTCCGGGGTCCT-CGGGGGGC | In vitro SELEX | 200 nm | Aptamer S15 with high affinity and specific diagnosis of the envelope protein domain III (ED3) of dengue virus 2 (DENV). | [64] |
| Influenza viruses (H3N2 and H1N1) | - | Subtractive SELEX | 5.56–5.84 nM | A8 and A20 DNA aptamers and their truncated sequences used for detection of type A influenza viruses (H3N2 and H1N1 viruses) with high affinity and selectivity | [11] |
| Ebola virus & Ebola Sudan virus | GGGCGCUCAAUUU-UUUAUUGCAUUUU-UCUUUGAGCGCCC | Cell SELEX | 30 nM & 250 nM | An RNA aptamer, 39SGP1A, functionalized with 2’ fluoropyrimidine (2’FY) for efficient detection of Ebola virus (EBOV) and Ebola Sudan virus (SUDV). | [65] |
| Chikungunya & Zika viruses | - | Cell SELEX | 50 pg/mL | An Aptamer-Au NPs conjugate based device for multiplexed colorimetric diagnosis of chikungunya and Zika viruses with high selectivity in a microfluidic channel. The subsequent introduction of silver reagent and its deposition on the AuNPs surface created a gray contrast in the testing zone. | [66] |
| Aptamer | Modification | Target and Binding KD | Application | Clinical Status | Ref. |
|---|---|---|---|---|---|
| Pegaptanib sodium (Macugen) RNA (27 nt) | 2’fluoropyrimidines 2’-O-methyl purines 3’-inverted dT 40 kDa PEG | VEGF165 50 pM | AMD Diabetic macular edema Diabetic retinopathy | FDA approved drug for the treatment of AMD | [95,96] |
| ARC1905 (Zimura) RNA (38 nt) | 2’fluoropyrimidines 2’-O-methyl purines 3’-inverted T 40 kDa PEG | C5 20-40 nM | Dry AMD IPCV | Phase I completed, Phase II and III recruiting (NCT02686658) Zimura in Combination with Anti-VEGF Therapy in Subjects with IPCV (NCT02397954) | [86] |
| E-10030 (Fovista) DNA (29 nt) | 2’-O-methyl purines 3’-inverted dT 40kDaPEG | PDGF 20 pM | Neovascular AMD | Phase II (NCT02214628) Anti-PDGF Pegylated Aptamer with Lucentis (NCT01089517) for neovascular AMD Fovista in Combination with Lucentis as compared to Lucentis monotherapy (NCT01940900) | [97] |
| NOX-A12 RNA (45 nt) | PEGlyated L-RNA (Spiegelmer) | CXCL12 200 pM | CCL Multiple myeloma Colorectal cancer Pancreatic cancer | Phase II (NCT01486797) NOX-A12 in Combination with Bortezomib and Dexamethasone Phase II (NCT 01521533) | [75] |
| AS1411 DNA (26 nt) | G-rich quartets, PEGlyated | Nucleolin 55 nM | AML MRCC | Phase II (NCT01034410) Phase II (NCT00740441) Phase I (NCT00881244) | [98,99] |
| NOX-H94 (lexaptepid pegol) RNA (44 nt) | L-RNA 5’ with 40 kDa PEG | Human Hepcidin 0.65 ± 0.06 nmol/L | Anemia of chronic disease End-Stage Renal Disease | Phase I and II (NCT02079896) | [100] |
| 68Ga-Sgc8 DNA (41 nt) | Bifunctional agent 1,4,7-triazacyclononane-1,4,7-triacetic acid (NOTA) Radioisotope Ga68 | PTK7/ CCk-4 NA | Colorectal cancer | Early Phase 1 (NCT03385148) | |
| NOX-E36 RNA (40 nt) | L-RNA, PEGlyated | MCP-1 1.32 nM | Chronic Inflammatory Diseases Type 2 Diabetes Mellitus Systemic Lupus Erythematosus | Phase I (NCT00976729) | [101,102] |
| NU172 DNA (26 nt) | G-quadruplex structure and unmodified | Thrombin 0.3–0.5 nM | Heart disease | Phase II (NCT00808964) | [103] |
| ARC1779 DNA (39 nt) | 3′-inverted dT 2’-O-methyl group 20 kDa PEGlyated | Von Willebrand factor (A1 domain) 2 nM | Purpura, Thrombotic Thrombocytopenic von Willebrand Disease Type-2b Acute Myocardial Infarction | Phase II (NCT00632242) Phase II (NCT00507338) | [104,105] |
| REG1 anticoagulation system (RB006 and RB007) RNA (37 nt) (RB006) Antidote (RB007) | 2’-ribo purine or 2’fluoropyrimidine 40kDaPEG | Coagulation factor IXa NA | Acute coronary syndrome Coronary artery disease Percutaneous coronary intervention | Phase I and II completed (NCT00113997, NCT00932100, NCT01872572) | [106,107] |
| Drug Candidates | Targeted Disease | Clinical Phase | Result | Company |
|---|---|---|---|---|
| Fovista® (anti-PDGF BB) plus anti-VEGF | Age-related Macular Degeneration | Phase II (NCT02214628) | Terminated | Ophthotech Corporation |
| Drug: E10030 Drug: ranibizumab Drug: E10030 sham intravitreal injection | Age-related Macular Degeneration | Phase III (NCT01944839) | Terminated | Ophthotech Corporation |
| Drug: E10030 Drug: bevacizumab or aflibercept Drug: E10030 sham intravitreal injection | Age-related Macular Degeneration | Phase III (NCT01940887) | Terminated | Ophthotech Corporation |
| Drug: Pegaptanib sodium | Macular Degeneration | Phase IV (NCT00312351) | Terminated | Eyetech Pharmaceuticals |
| Drug: ARC1779 | Von Willebrand Disease | Phase II (NCT00694785) | Withdrawn | Archemix Corp |
| Drug: placebo control Drug: ARC19499 | Hemophilia | Phase I (NCT01191372) | Terminated | Baxalta Inc |
| Drug: AS1411Drug: Cytarabine | Acute Myeloid Leukemia | Phase II (NCT01034410) | Terminated | Antisoma Research |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. https://doi.org/10.3390/pharmaceutics12070646
Kumar Kulabhusan P, Hussain B, Yüce M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics. 2020; 12(7):646. https://doi.org/10.3390/pharmaceutics12070646
Chicago/Turabian StyleKumar Kulabhusan, Prabir, Babar Hussain, and Meral Yüce. 2020. "Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents" Pharmaceutics 12, no. 7: 646. https://doi.org/10.3390/pharmaceutics12070646
APA StyleKumar Kulabhusan, P., Hussain, B., & Yüce, M. (2020). Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics, 12(7), 646. https://doi.org/10.3390/pharmaceutics12070646