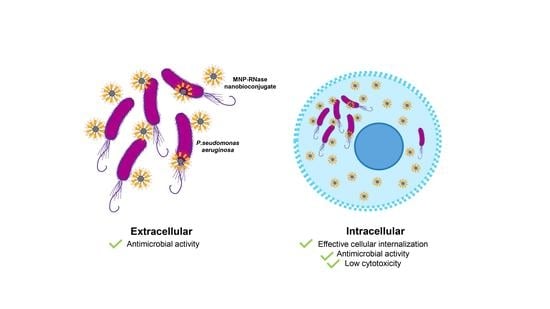
Magnetite Nanoparticles Functionalized with RNases against Intracellular Infection of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Magnetic Nanoparticles
2.3. Surface Functionalization
2.4. Ribonuclease A and Ribonuclease 3/1 Immobilization
2.5. Labeling of MNPs-RNase Nanobioconjugates with Rhodamine B
2.6. Hemolysis Assay
2.7. Platelet Aggregation Test
2.8. LDH Cytotoxicity Assay
2.9. Catalytic Evaluation of MNPs-RNases by Activity Staining Plates
2.10. Bacterial Cell Viability Luminescent Assay
2.11. THP-1 Cell Culture and Macrophage Induction
2.12. Macrophage Infection
2.13. Cell Translocation of Nanobioconjugates
2.14. Internalization Percent Determination
2.15. Infection of Macrophages with Stained P. aeruginosa
2.16. Statistical Analysis
3. Results
3.1. Ribonuclease A and Ribonuclease 3/1 Immobilization on Magnetite Nanoparticles (MNPs)
3.2. Catalytic Activity Evaluation of MNPs-RNases
3.3. Biocompatibility Assessment
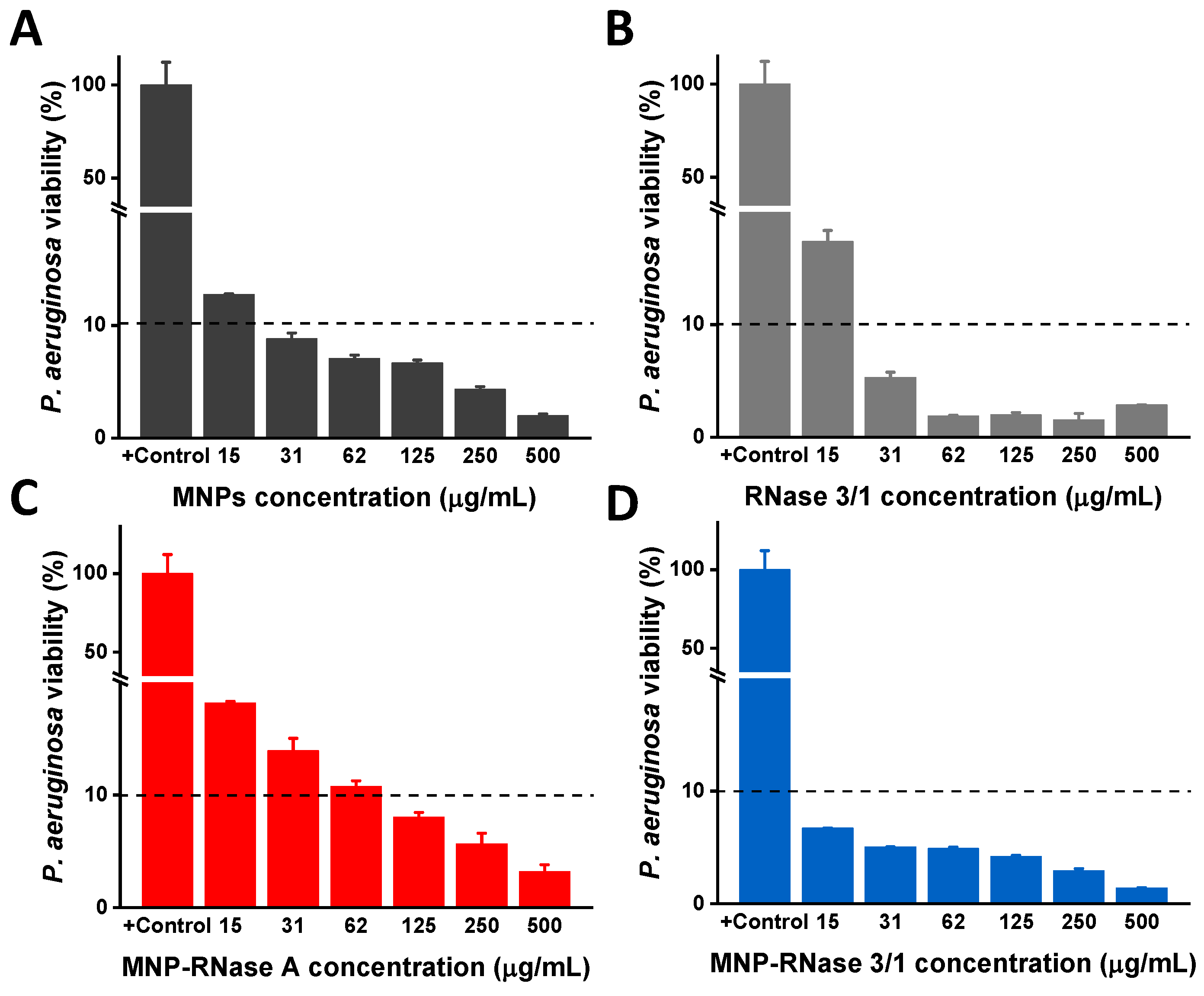
3.4. Antibacterial Activity of MNPs-RNases Toward Extracellular and Intracellular P. aeruginosa
3.5. Translocation of Nanobioconjugates and Intracellular Colocalization
3.6. Analysis of Infected Macrophages with P. aeruginosa and Exposed to MNPs-RNase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A Review of Antimicrobial Peptides and Their Therapeutic Potential as Anti-Infective Drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef]
- Auvynet, C.; Rosenstein, Y. Multifunctional host defense peptides: Antimicrobial peptides, the small yet big players in innate and adaptive immunity. FEBS J. 2009, 276, 6497–6508. [Google Scholar] [CrossRef]
- Koczera, P.; Martin, L.; Marx, G.; Schuerholz, T. The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity. Int. J. Mol. Sci. 2016, 17, 1278. [Google Scholar] [CrossRef]
- Sorrentino, S. The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010, 584, 2194–2200. [Google Scholar] [CrossRef]
- Lu, L.; Li, J.; Moussaoui, M.; Boix, E. Immune Modulation by Human Secreted RNases at the Extracellular Space. Front. Immunol. 2018, 9, 1012. [Google Scholar] [CrossRef]
- Sorrentino, S.; Naddeo, M.; Russo, A.; D’Alessio, G. Degradation of Double-Stranded RNA by Human Pancreatic Ribonuclease: Crucial Role of Noncatalytic Basic Amino Acid Residues. Biochemistry 2003, 42, 10182–10190. [Google Scholar] [CrossRef]
- Landré, J.B.; Hewett, P.W.; Olivot, J.-M.; Friedl, P.; Ko, Y.D.; Sachinidis, A.; Moenner, M. Human endothelial cells selectively express large amounts of pancreatic-type ribonuclease (RNase 1). J. Cell. Biochem. 2002, 86, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Pulido-Gomez, D.; Prats-Ejarque, G.; Villalba, C.; Albacar, M.; Gonzalez-Lopez, J.J.; Burgas, M.T.; Moussaoui, M.; Boix, E. A Novel RNase 3/ECP Peptide for Pseudomonas aeruginosa Biofilm Eradication That Combines Antimicrobial, Lipopolysaccharide Binding, and Cell-Agglutinating Activities. Antimicrob. Agents Chemother. 2016, 60, 6313–6325. [Google Scholar] [CrossRef] [PubMed]
- Pulido-Gomez, D.; Prats-Ejarque, G.; Villalba, C.; Albacar, M.; Moussaoui, M.; Andreu, D.; Volkmer, R.; Burgas, M.T.; Boix, E. Positional scanning library applied to the human eosinophil cationic protein/RNase3 N-terminus reveals novel and potent anti-biofilm peptides. Eur. J. Med. Chem. 2018, 152, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Burgas, M.T.; Badia, M.; Moussaoui, M.; Sánchez, D.; Nogués, M.V.; Boix, E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010, 277, 1713–1725. [Google Scholar] [CrossRef]
- Prats-Ejarque, G.; Li, J.; Ait-Ichou, F.; Lorente, H.; Boix, E. Testing a Human Antimicrobial RNase Chimera Against Bacterial Resistance. Front. Microbiol. 2019, 10, 1357. [Google Scholar] [CrossRef]
- Lu, L.; Arranz-Trullén, J.; Prats-Ejarque, G.; Pulido, D.; Bhakta, S.; Boix, E. Human Antimicrobial RNases Inhibit Intracellular Bacterial Growth and Induce Autophagy in Mycobacteria-Infected Macrophages. Front. Immunol. 2019, 10, 1500. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Barbier, F.; Andremont, A.; Wolff, M.; Bouadma, L. Hospital-acquired pneumonia and ventilator-associated pneumonia. Curr. Opin. Pulm. Med. 2013, 19, 216–228. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017; Volume 7, pp. 318–327. [Google Scholar]
- Moussouni, M.; Nogaret, P.; Garai, P.; Ize, B.; Vivès, E.; Blanc-Potard, A.-B. Activity of a Synthetic Peptide Targeting MgtC on Pseudomonas aeruginosa Intramacrophage Survival and Biofilm Formation. Front. Microbiol. 2019, 9, 84. [Google Scholar] [CrossRef]
- Garai, P.; Berry, L.; Moussouni, M.; Bleves, S.; Blanc-Potard, A.-B. Killing from the inside: Intracellular role of T3SS in the fate of Pseudomonas aeruginosa within macrophages revealed by mgtC and oprF mutants. PLoS Pathog. 2019, 15, e1007812. [Google Scholar] [CrossRef]
- Deng, Q.; Wang, Y.; Zhang, Y.; Li, M.; Li, D.; Huang, X.; Wu, Y.; Pu, J.; Wu, M. Pseudomonas aeruginosa Triggers Macrophage Autophagy To Escape Intracellular Killing by Activation of the NLRP3 Inflammasome. Infect. Immun. 2015, 84, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Bastaert, F.; Kheir, S.; Saint-Criq, V.; Villeret, B.; Dang, P.M.-C.; El-Benna, J.; Sirard, J.-C.; Voulhoux, R.; Sallenave, J.-M. Pseudomonas aeruginosa LasB Subverts Alveolar Macrophage Activity by Interfering With Bacterial Killing Through Downregulation of Innate Immune Defense, Reactive Oxygen Species Generation, and Complement Activation. Front. Immunol. 2018, 9, 1675. [Google Scholar] [CrossRef] [PubMed]
- Kroken, A.; Chen, C.K.; Evans, D.; Yahr, T.L.; Fleiszig, S.M.J. The Impact of ExoS on Pseudomonas aeruginosa Internalization by Epithelial Cells is Independent of fleQ and Correlates with Bistability of Type Three Secretion System Gene Expression. MBio 2018, 9, e00668. [Google Scholar] [CrossRef] [PubMed]
- Buyck, J.M.; Tulkens, P.M.; Van Bambeke, F. Pharmacodynamic Evaluation of the Intracellular Activity of Antibiotics towards Pseudomonas aeruginosa PAO1 in a Model of THP-1 Human Monocytes. Antimicrob. Agents Chemother. 2013, 57, 2310–2318. [Google Scholar] [CrossRef] [PubMed]
- Cabot, G.; Zamorano, L.; Moyà, B.; Juan, C.; Navas, A.; Blázquez, J.; Oliver, A. Evolution of Pseudomonas aeruginosa Antimicrobial Resistance and Fitness under Low and High Mutation Rates. Antimicrob. Agents Chemother. 2016, 60, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, P.; Heidenreich, N.; Podeyn, B.; Bürck, J.; Ulrich, A.S. Antibiotic gold: Tethering of antimicrobial peptides to gold nanoparticles maintains conformational flexibility of peptides and improves trypsin susceptibility. Biomater. Sci. 2017, 5, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Skwarecki, A.; Milewski, S.; Schielmann, M.; Milewska, M.J. Antimicrobial molecular nanocarrier—drug conjugates. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 2215–2240. [Google Scholar] [CrossRef] [PubMed]
- Sadat, S.M.A.; Jahan, S.T.; Haddadi, A. Effects of Size and Surface Charge of Polymeric Nanoparticles on in Vitro and in Vivo Applications. J. Biomater. Nanotechnol. 2016, 7, 91–108. [Google Scholar] [CrossRef]
- Niemirowicz, K.; Durnaś, B.; Tokajuk, G.; Piktel, E.; Michalak, G.; Gu, X.; Kułakowska, A.; Savage, P.B.; Bucki, R. Formulation and candidacidal activity of magnetic nanoparticles coated with cathelicidin LL-37 and ceragenin CSA-13. Sci. Rep. 2017, 7, 4610. [Google Scholar] [CrossRef]
- Morales-Avila, E.; Ferro-Flores, G.; Ocampo-García, B.E.; López-Téllez, G.; López-Ortega, J.; Rogel-Ayala, D.G.; Sánchez-Padilla, D. Antibacterial Efficacy of Gold and Silver Nanoparticles Functionalized with the Ubiquicidin (29–41) Antimicrobial Peptide. J. Nanomater. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Findlay, F.; Pohl, J.; Svoboda, P.; Shakamuri, P.; McLean, K.; Inglis, N.F.; Proudfoot, L.; Barlow, P.G. Carbon Nanoparticles Inhibit the Antimicrobial Activities of the Human Cathelicidin LL-37 through Structural Alteration. J. Immunol. 2017, 199, 2483–2490. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Cifuentes, J.; Cuellar, M.; Suarez-Arnedo, A.; Cruz, J.; Muñoz-Camargo, C.; Rueda, J. Cell-Penetrating And Antibacterial BUF-II Nanobioconjugates: Enhanced Potency Via Immobilization On Polyetheramine-Modified Magnetite Nanoparticles. Int. J. Nanomed. 2019, 14, 8483–8497. [Google Scholar] [CrossRef] [PubMed]
- Cuellar, M.; Cifuentes, J.; Pérez, J.; Suarez-Arnedo, A.; Serna, J.A.; Groot, H.; Muñoz-Camargo, C.; Cruz, J. Novel BUF2-magnetite nanobioconjugates with cell-penetrating abilities. Int. J. Nanomed. 2018, 13, 8087–8094. [Google Scholar] [CrossRef]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial Peptides and Nanotechnology, Recent Advances and Challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef] [PubMed]
- Israel, L.L.; Galstyan, A.; Holler, E.; Ljubimova, J.Y. Magnetic iron oxide nanoparticles for imaging, targeting and treatment of primary and metastatic tumors of the brain. J. Control. Release 2020, 320, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Sun, J.; Wang, Y.; Sheng, J.; Wang, F.; Sun, M. Application of Iron Magnetic Nanoparticles in Protein Immobilization. Molecules 2014, 19, 11465–11486. [Google Scholar] [CrossRef]
- Gao, R.; Kong, X.; Wang, X.; Hea, X.; Chen, L.; Zhangad, Y. Preparation and characterization of uniformly sized molecularly imprinted polymers functionalized with core–shell magnetic nanoparticles for the recognition and enrichment of protein. J. Mater. Chem. 2011, 21, 17863–17871. [Google Scholar] [CrossRef]
- Wang, W.; Guo, N.; Huang, W.; Zhang, Z.; Mao, X. Immobilization of Chitosanases onto Magnetic Nanoparticles to Enhance Enzyme Performance. Catalysts 2018, 8, 401. [Google Scholar] [CrossRef]
- Salehi, Z.; Ghahfarokhi, H.H.; Kodadadi, A.A.; Rahimnia, R. Thiol and urea functionalized magnetic nanoparticles with highly enhanced loading capacity and thermal stability for lipase in transesterification. J. Ind. Eng. Chem. 2016, 35, 224–230. [Google Scholar] [CrossRef]
- Defaei, M.; Taheri-Kafrani, A.; Miroliaei, M.; Yaghmaei, P. Improvement of stability and reusability of α-amylase immobilized on naringin functionalized magnetic nanoparticles: A robust nanobiocatalyst. Int. J. Biol. Macromol. 2018, 113, 354–360. [Google Scholar] [CrossRef]
- Song, J.; Su, P.; Yang, Y.; Yang, Y. Efficient immobilization of enzymes onto magnetic nanoparticles by DNA strand displacement: A stable and high-performance biocatalyst. New J. Chem. 2017, 41, 6089–6097. [Google Scholar] [CrossRef]
- Sahu, A.; Badhe, P.S.; Adivarekar, R.; Ladole, M.R.; Pandit, A. Synthesis of glycinamides using protease immobilized magnetic nanoparticles. Biotechnol. Rep. 2016, 12, 13–25. [Google Scholar] [CrossRef]
- Muljajew, I.; Erlebach, A.; Weber, C.; Buchheim, J.R.; Sierka, M.; Schubert, U.S. A polyesteramide library from dicarboxylic acids and 2,2′-bis(2-oxazoline): Synthesis, characterization, nanoparticle formulation and molecular dynamics simulations. Polym. Chem. 2020, 11, 112–124. [Google Scholar] [CrossRef]
- Jendelova, P.; Rittich, B.; Safar, J.; Španová, A.; Lenfeld, J.; Beneš, M.J. Properties of RNase A Immobilized on Magnetic Poly(2-hydroxyethyl methacrylate) Microspheres. Biotechnol. Prog. 2001, 17, 447–452. [Google Scholar] [CrossRef]
- Kordalivand, N.; Li, D.; Beztsinna, N.; Toraño, J.S.; Mastrobattista, E.; Van Nostrum, C.F.; Hennink, W.E.; Vermonden, T. Polyethyleneimine coated nanogels for the intracellular delivery of RNase A for cancer therapy. Chem. Eng. J. 2018, 340, 32–41. [Google Scholar] [CrossRef]
- Niu, Y.; Yu, M.; Meka, A.; Liu, Y.; Zhang, J.; Yang, Y.; Yu, C. Understanding the contribution of surface roughness and hydrophobic modification of silica nanoparticles to enhanced therapeutic protein delivery. J. Mater. Chem. B 2015, 4, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Li, Q.; Neufeld, C.I.; Pouli, D.; Sun, S.; Yang, L.; Deng, P.; Wang, M.; Georgakoudi, I.; et al. Hyaluronic acid modification of RNase A and its intracellular delivery using lipid-like nanoparticles. J. Control. Release 2017, 263, 39–45. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, S.; Wen, K.; Li, Q. Nano-Scaled Zeolitic Imidazole Framework-8 as an Efficient Carrier for the Intracellular Delivery of RNase A in Cancer Treatment. Int. J. Nanomed. 2019, 14, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- McVey, C.; Huang, F.; Elliott, C.; Cao, C. Endonuclease controlled aggregation of gold nanoparticles for the ultrasensitive detection of pathogenic bacterial DNA. Biosens. Bioelectron. 2017, 92, 502–508. [Google Scholar] [CrossRef]
- Otieno, B.A.; Krause, C.E.; Rusling, J.F. Bioconjugation of Antibodies and Enzyme Labels onto Magnetic Beads. In Methods in Enzymology; Academic Press Inc.: New York, NY, USA, 2016; Volume 571, pp. 135–150. [Google Scholar]
- Bucior, I.; Tran, C.; Engel, J. Assessing Pseudomonas virulence using host cells. Breast Cancer 2014, 1149, 741–755. [Google Scholar] [CrossRef]
- Atwal, S.; Giengkam, S.; VanNieuwenhze, M.S.; Salje, J. Live imaging of the genetically intractable obligate intracellular bacteria Orientia tsutsugamushi using a panel of fluorescent dyes. J. Microbiol. Methods 2016, 130, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, W.M.; Gonzalez, E.E. Study of agglomeration and magnetic sedimentation of Glutathione@Fe3O4 nanoparticles in water medium. Dyna 2018, 85, 19–26. [Google Scholar] [CrossRef]
- Stoia, M.; Istratie, R.; Păcurariu, C. Investigation of magnetite nanoparticles stability in air by thermal analysis and FTIR spectroscopy. J. Therm. Anal. Calorim. 2016, 125, 1185–1198. [Google Scholar] [CrossRef]
- Xue, Y.; Patel, A.; Sant, V.; Sant, S. Semiquantitative FTIR Analysis of the Crosslinking Density of Poly(ester amide)-Based Thermoset Elastomers. Macromol. Mater. Eng. 2015, 301, 296–305. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Cheng, H.; Yan, Y.; Iqbal, H.M. Multi-point enzyme immobilization, surface chemistry, and novel platforms: A paradigm shift in biocatalyst design. Crit. Rev. Biotechnol. 2018, 39, 202–219. [Google Scholar] [CrossRef]
- Radomski, A.; Jurasz, P.; Alonso-Escolano, D.; Drews, M.; Morandi, M.; Malinski, T.; Radomski, M.W. Nanoparticle-induced platelet aggregation and vascular thrombosis. Br. J. Pharmacol. 2005, 146, 882–893. [Google Scholar] [CrossRef]
- Agotegaray, M.; Campelo, A.E.; Zysler, R.D.; Gumilar, F.; Bras, C.; Gandini, A.; Minetti, A.; Massheimer, V.L.; Lassalle, V.L. Magnetic nanoparticles for drug targeting: From design to insights into systemic toxicity. Preclinical evaluation of hematological, vascular and neurobehavioral toxicology. Biomater. Sci. 2017, 5, 772–783. [Google Scholar] [CrossRef]
- Kumar, R.; Kanwar, S.S. Biotechnological Production and Applications of Ribonucleases; Elsevier BV: Amsterdam, The Netherlands, 2020; pp. 363–389. ISBN 9780444643230. [Google Scholar]
- Perryman, A.L.; Patel, J.S.; Russo, R.; Singleton, E.; Connell, N.; Ekins, S.; Freundlich, J.S. Naïve Bayesian Models for Vero Cell Cytotoxicity. Pharm. Res. 2018, 35, 170. [Google Scholar] [CrossRef]
- Otake, S.; Kobayashi, M.; Narumi, K.; Sasaki, S.; Kikutani, Y.; Furugen, A.; Watanabe, M.; Takahashi, N.; Ogura, J.; Yamaguchi, H.; et al. Regulation of the Expression and Activity of Glucose and Lactic Acid Metabolism-Related Genes by Protein Kinase C in Skeletal Muscle Cells. Biol. Pharm. Bull. 2013, 36, 1435–1439. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, Y.; Huang, J.; Chen, K.; Huang, J.; Xiao, K. Uptake, distribution, clearance, and toxicity of iron oxide nanoparticles with different sizes and coatings. Sci. Rep. 2018, 8, 2082. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Duan, F.; Liu, S.; Wu, T.; Shang, Y.; Tian, R.; Liu, J.; Wang, Z.-G.; Jiang, Q.; Ding, B. Efficient Intracellular Delivery of RNase A Using DNA Origami Carriers. ACS Appl. Mater. Interfaces 2019, 11, 11112–11118. [Google Scholar] [CrossRef] [PubMed]
- De Toledo, L.d.A.S.; Rosseto, H.C.; Bruschi, M.L. Iron oxide magnetic nanoparticles as antimicrobials for therapeutics. Pharm. Dev. Technol. 2017, 23, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F. Vertebrate Secretory (RNase A) Ribonucleases and Host Defense; Springer: Berlin/Heidelberg, Germany, 2011; pp. 35–53. [Google Scholar]
- Zhang, J. Human RNase 7: A new cationic ribonuclease of the RNase A superfamily. Nucleic Acids Res. 2003, 31, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.; Cohen, A.; Thomas, J.; Spencer, J.D. The Immunomodulatory and Antimicrobial Properties of the Vertebrate Ribonuclease A Superfamily. Vaccines 2018, 6, 76. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, Y.T.; Rao, K.V.; Kumari, B.S.; Kumar, V.S.S.; Pavani, T. Synthesis of Fe3O4 nanoparticles and its antibacterial application. Int. Nano Lett. 2015, 5, 85–92. [Google Scholar] [CrossRef]
- Taimoory, S.M.; Rahdar, A.; Aliahmad, M.; Sadeghfar, F.; Hajinezhad, M.R.; Jahantigh, M.; Shahbazi, P.; Trant, J.F. The synthesis and characterization of a magnetite nanoparticle with potent antibacterial activity and low mammalian toxicity. J. Mol. Liq. 2018, 265, 96–104. [Google Scholar] [CrossRef]
- Caamaño, M.A.; Olivares-Trejo, J.D.J. Iron Oxide Nanoparticle Improve the Antibacterial Activity of Erythromycin. J. Bacteriol. Parasitol. 2016, 7. [Google Scholar] [CrossRef]
- Luciani, N.; Gazeau, F.; Wilhelm, C. Reactivity of the monocyte/macrophage system to superparamagnetic anionic nanoparticles. J. Mater. Chem. 2009, 19, 6373–6380. [Google Scholar] [CrossRef]
- García-Mayoral, M.F.; Canales, A.; Díaz, D.; López-Prados, J.; Moussaoui, M.; De Paz, J.L.; Angulo, J.; Nieto, P.M.M.; Jiménez-Barbero, J.; Boix, E.; et al. Insights into the Glycosaminoglycan-Mediated Cytotoxic Mechanism of Eosinophil Cationic Protein Revealed by NMR. ACS Chem. Biol. 2012, 8, 144–151. [Google Scholar] [CrossRef]
- Burgas, M.T.; Nogués, M.V.; Boix, E. Eosinophil cationic protein (ECP) can bind heparin and other glycosaminoglycans through its RNase active site. J. Mol. Recognit. 2010, 24, 90–100. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.-Y.; Raines, R.T. Mechanism of Ribonuclease A Endocytosis: Analogies to Cell-Penetrating Peptides. Biochemistry 2011, 50, 8374–8382. [Google Scholar] [CrossRef] [PubMed]
- Persson, T.; Calafat, J.; Janssen, H.; Karawajczyk, M.; Carlsson, S.R.; Egesten, A. Specific Granules of Human Eosinophils Have Lysosomal Characteristics: Presence of Lysosome-Associated Membrane Proteins and Acidification upon Cellular Activation. Biochem. Biophys. Res. Commun. 2002, 291, 844–854. [Google Scholar] [CrossRef]
- Lopez-Barbosa, N.; Garcia, J.G.; Cifuentes, J.; Castro, L.M.; Vargas, F.; Ostos, C.; Cardona-Gomez, G.P.; Hernandez, A.M.; Cruz, J.C. Multifunctional magnetite nanoparticles to enable delivery of siRNA for the potential treatment of Alzheimer’s. Drug Deliv. 2020, 27, 864–875. [Google Scholar] [CrossRef]
- Walrant, A.; Bechara, C.; Alves, I.D.; Sagan, S. Molecular partners for interaction and cell internalization of cell-penetrating peptides: How identical are they? Nanomedicine 2012, 7, 133–143. [Google Scholar] [CrossRef]
- Beloqui, A.; Rieux, A.D.; Preat, V. Mechanisms of transport of polymeric and lipidic nanoparticles across the intestinal barrier. Adv. Drug Deliv. Rev. 2016, 106, 242–255. [Google Scholar] [CrossRef]
- Pacheco, P.; White, D.; Sulchek, T. Effects of Microparticle Size and Fc Density on Macrophage Phagocytosis. PLoS ONE 2013, 8, e60989. [Google Scholar] [CrossRef]
- Bernut, A.; Belon, C.; Soscia, C.; Bleves, S.; Blanc-Potard, A.-B. Intracellular phase for an extracellular bacterial pathogen: MgtC shows the way. Microb. Cell 2015, 2, 353–355. [Google Scholar] [CrossRef]
- Kamaruzzaman, N.F.; Kendall, S.; Good, L. Targeting the hard to reach: Challenges and novel strategies in the treatment of intracellular bacterial infections. Br. J. Pharmacol. 2016, 174, 2225–2236. [Google Scholar] [CrossRef]
- Foroozandeh, P.; Azlan, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef]
- Salazar, V.A.; Arranz-Trullén, J.; Navarro, S.; Blanco, J.A.; Sanchez, D.; Moussaoui, M.; Boix, E. Exploring the mechanisms of action of human secretory RNase 3 and RNase 7 againstCandida albicans. Microbiologyopen 2016, 5, 830–845. [Google Scholar] [CrossRef] [PubMed]
- Arakha, M.; Pal, S.; Samantarrai, D.; Panigrahi, T.K.; Mallick, B.C.; Pramanik, K.; Mallick, B.; Jha, S. Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle-bacteria interface. Sci. Rep. 2015, 5, 14813. [Google Scholar] [CrossRef]
- Gupta, S.; Haigh, B.J.; Griffin, F.; Wheeler, T.T. The mammalian secreted RNases: Mechanisms of action in host defence. Innate Immun. 2012, 19, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Salazar, V.A.; Arranz-Trullén, J.; Prats-Ejarque, G.; Burgas, M.T.; Andreu, D.; Pulido, D.; Boix, E. Insight into the Antifungal Mechanism of Action of Human RNase N-terminus Derived Peptides. Int. J. Mol. Sci. 2019, 20, 4558. [Google Scholar] [CrossRef] [PubMed]
- Burgas, M.T.; Pulido, D.; Valle, J.; Nogués, M.V.; Andreu, D.; Boix, E.; Pulido-Gomez, D. Ribonucleases as a host-defence family: Evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem. J. 2013, 456, 99–108. [Google Scholar] [CrossRef]
- Sierra, H.; Cordova, M.; Chen, C.-S.J.; Rajadhyaksha, M. Confocal imaging-guided laser ablation of basal cell carcinomas: An ex vivo study. J. Investig. Dermatol. 2014, 135, 612–615. [Google Scholar] [CrossRef]
- Leland, P.A.; Schultz, L.W.; Kim, B.-M.; Raines, R.T. Ribonuclease A variants with potent cytotoxic activity. Proc. Natl. Acad. Sci. USA 1998, 95, 10407–10412. [Google Scholar] [CrossRef]
- Dickson, K.A.; Haigis, M.C.; Raines, R.T. Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 349–374. [Google Scholar] [CrossRef]
- Lomax, J.E.; Eller, C.H.; Raines, R.T. Rational Design and Evaluation of Mammalian Ribonuclease Cytotoxins. Methods Enzym. 2012, 502, 273–290. [Google Scholar] [CrossRef]
- Suzuki, M.; Saxena, S.K.; Boix, E.; Prill, R.J.; Vasandani, V.M.; Ladner, J.E.; Sung, C.; Youle, R.J. Engineering receptor-mediated cytotoxicity into human ribonucleases by steric blockade of inhibitor interaction. Nat. Biotechnol. 1999, 17, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Scaletti, F.; Hardie, J.; Lee, Y.-W.; Luther, D.; Ray, M.; Rotello, V. Protein delivery into cells using inorganic nanoparticle–protein supramolecular assemblies. Chem. Soc. Rev. 2018, 47, 3421–3432. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Barbosa, N.; Suarez-Arnedo, A.; Cifuentes, J.; Barrios, A.F.G.; Batista, C.A.S.; Osma, J.F.; Muñoz-Camargo, C.; Cruz, J. Magnetite–OmpA Nanobioconjugates as Cell-Penetrating Vehicles with Endosomal Escape Abilities. ACS Biomater. Sci. Eng. 2019, 6, 415–424. [Google Scholar] [CrossRef]
- Gustafson, H.H.; Holt-Casper, D.; Grainger, D.W.; Ghandehari, H. Nanoparticle uptake: The phagocyte problem. Nano Today 2015, 10, 487–510. [Google Scholar] [CrossRef] [PubMed]
- A Makarov, A.; Ilinskaya, O.N. Cytotoxic ribonucleases: Molecular weapons and their targets. FEBS Lett. 2003, 540, 15–20. [Google Scholar] [CrossRef]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangel-Muñoz, N.; Suarez-Arnedo, A.; Anguita, R.; Prats-Ejarque, G.; Osma, J.F.; Muñoz-Camargo, C.; Boix, E.; Cruz, J.C.; Salazar, V.A. Magnetite Nanoparticles Functionalized with RNases against Intracellular Infection of Pseudomonas aeruginosa. Pharmaceutics 2020, 12, 631. https://doi.org/10.3390/pharmaceutics12070631
Rangel-Muñoz N, Suarez-Arnedo A, Anguita R, Prats-Ejarque G, Osma JF, Muñoz-Camargo C, Boix E, Cruz JC, Salazar VA. Magnetite Nanoparticles Functionalized with RNases against Intracellular Infection of Pseudomonas aeruginosa. Pharmaceutics. 2020; 12(7):631. https://doi.org/10.3390/pharmaceutics12070631
Chicago/Turabian StyleRangel-Muñoz, Nathaly, Alejandra Suarez-Arnedo, Raúl Anguita, Guillem Prats-Ejarque, Johann F. Osma, Carolina Muñoz-Camargo, Ester Boix, Juan C. Cruz, and Vivian A. Salazar. 2020. "Magnetite Nanoparticles Functionalized with RNases against Intracellular Infection of Pseudomonas aeruginosa" Pharmaceutics 12, no. 7: 631. https://doi.org/10.3390/pharmaceutics12070631
APA StyleRangel-Muñoz, N., Suarez-Arnedo, A., Anguita, R., Prats-Ejarque, G., Osma, J. F., Muñoz-Camargo, C., Boix, E., Cruz, J. C., & Salazar, V. A. (2020). Magnetite Nanoparticles Functionalized with RNases against Intracellular Infection of Pseudomonas aeruginosa. Pharmaceutics, 12(7), 631. https://doi.org/10.3390/pharmaceutics12070631