Surface-Initiated Atom Transfer Polymerized Anionic Corona on Gold Nanoparticles for Anti-Cancer Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
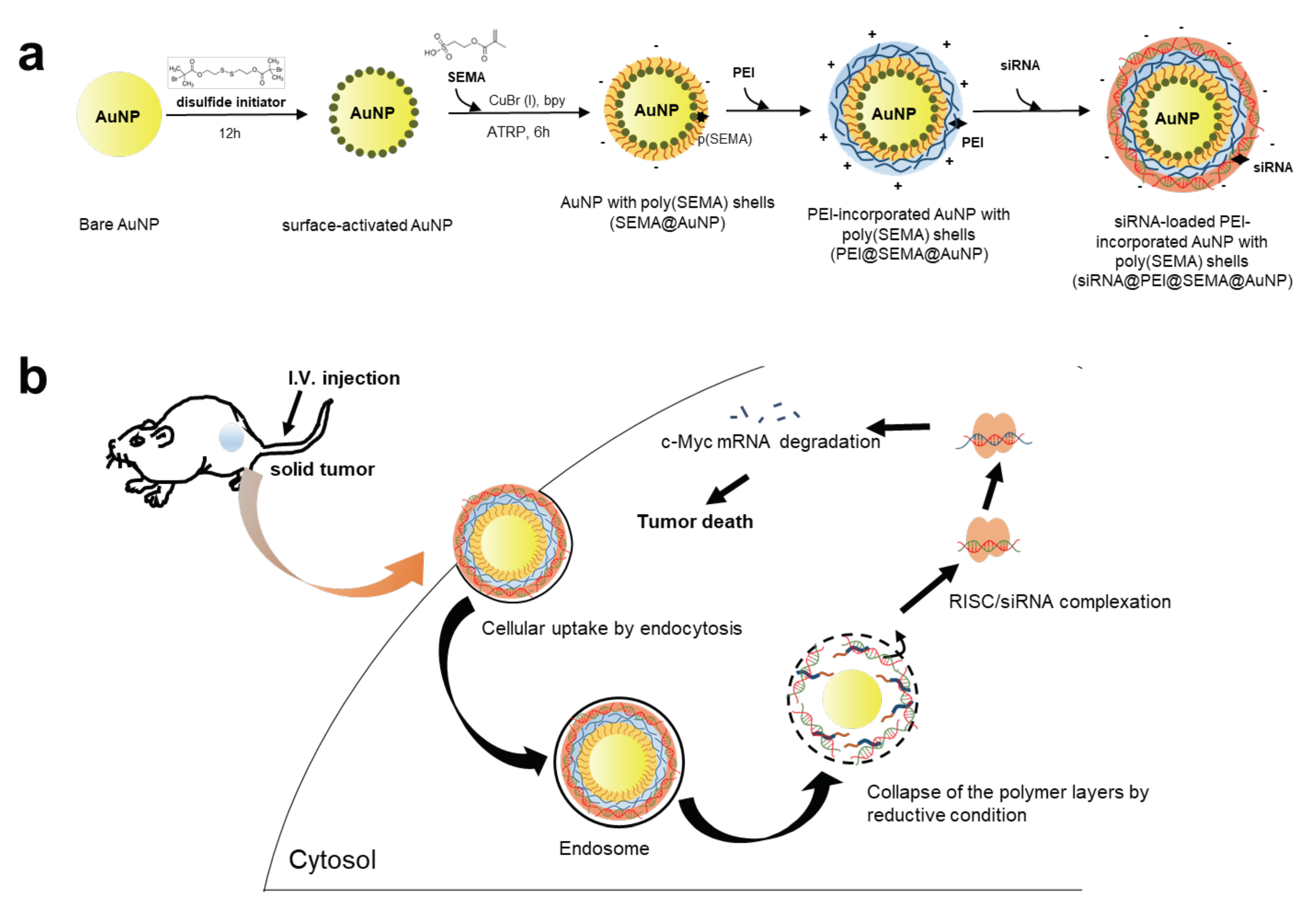
2.2. Synthesis of Gold Nanoparticles (AuNPs) and Surface Decoration by ATRP
2.3. PEI Decoration on SEMA@AuNPs
2.4. Incorporation of siRNA on PEI@SEMA@AuNPs
2.5. In Vitro Anti-Cancer Effect
2.6. siRNA Silencing of c-Myc Gene
2.7. Apoptotic Cell Death
2.8. Tumor Model and siRNA Treatment for Anti-Cancer Activity and Imaging
2.9. c-Myc Expression in the Tumor Tissue
2.10. Immunohistochemical Staining from the Tumor Tissue
2.11. Statistical Analysis
3. Results and Discussion
3.1. Preparation of SEMA@AuNP, PEI@SEMA@AuNP, and siRNA@PEI@SEMA@AuNP
3.2. Characterization of SEMA@AuNP, PEI@SEMA@AuNP and siRNA@PEI@SEMA@AuNP
3.2.1. Size and Zeta Potential Measurement of AuNP, SEMA@AuNP, and PEI@SEMA@AuNP
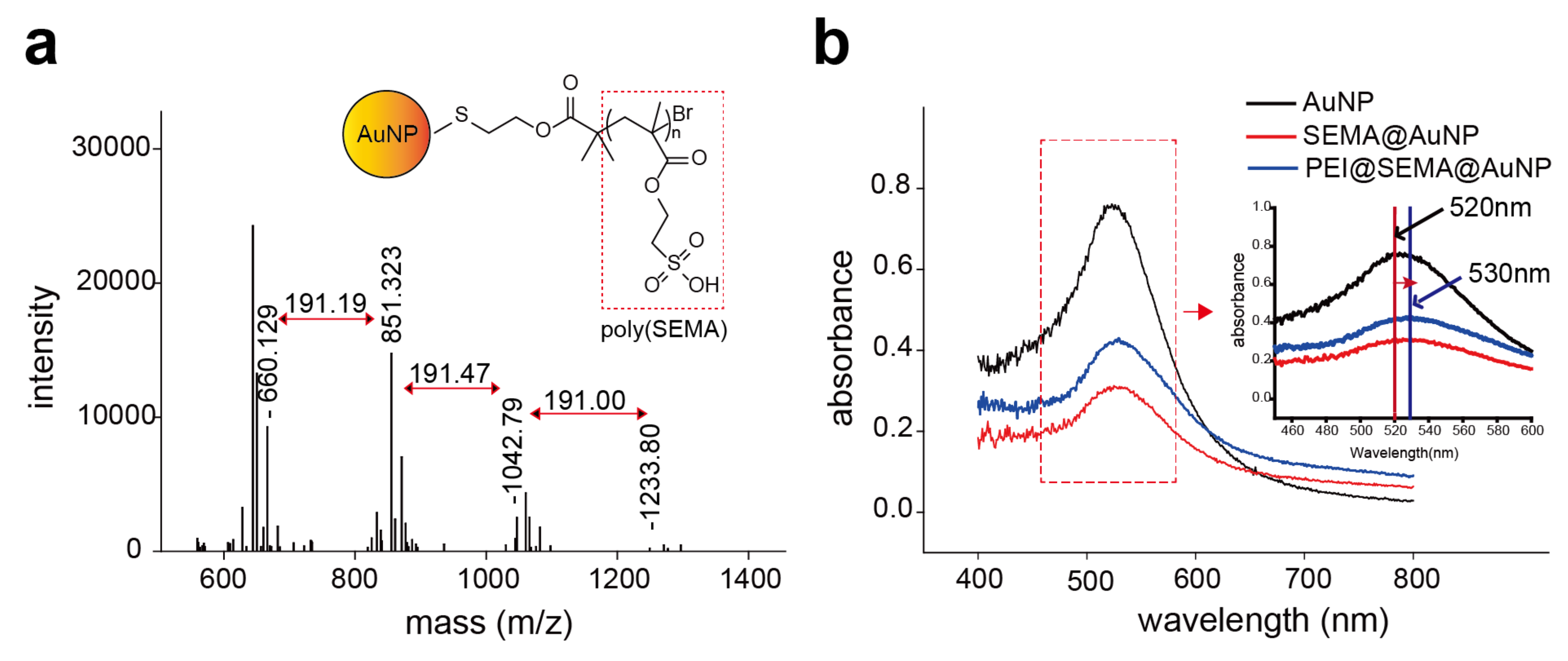
3.2.2. Characterization of pSEMA Presence and PEI Release from AuNPs
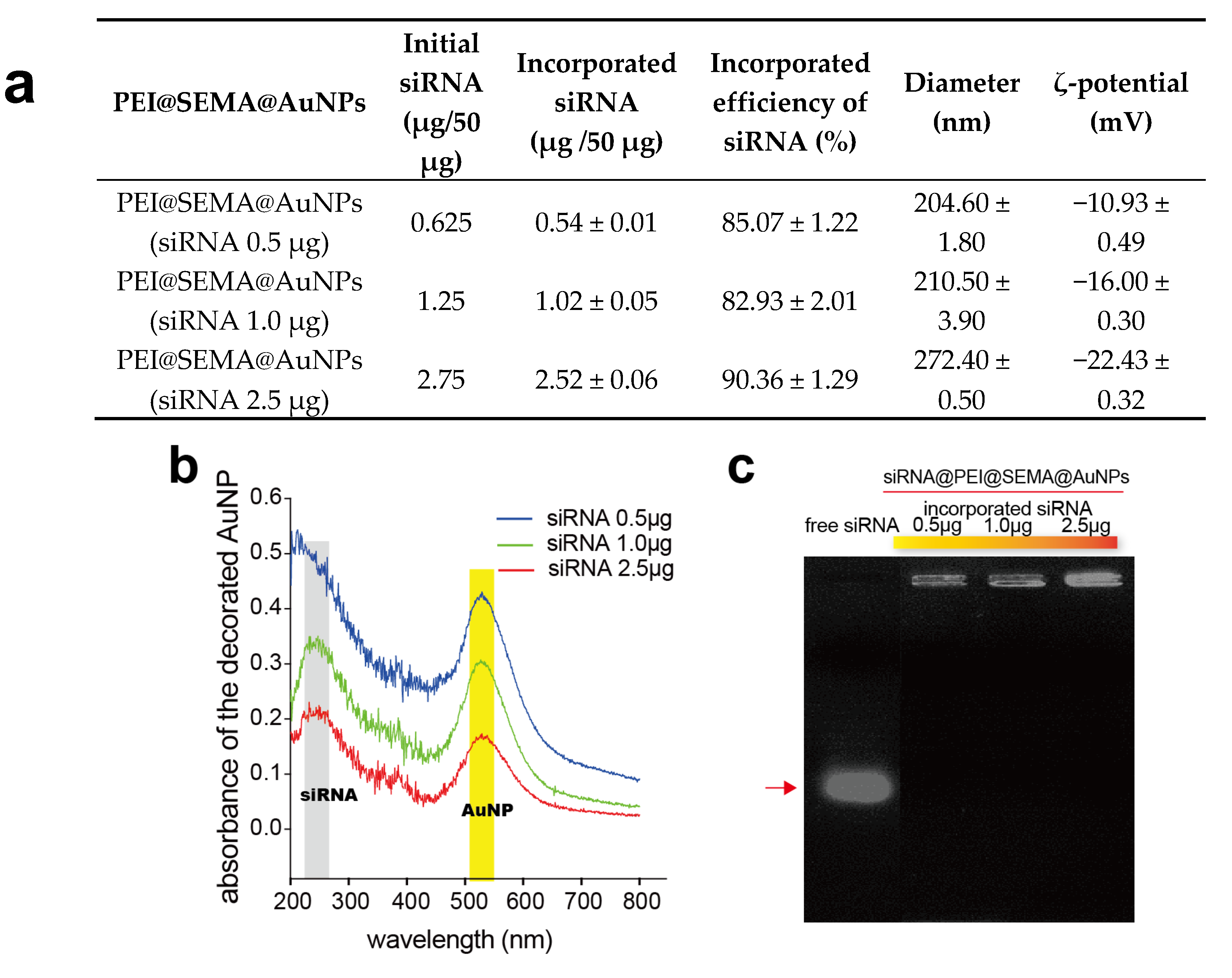
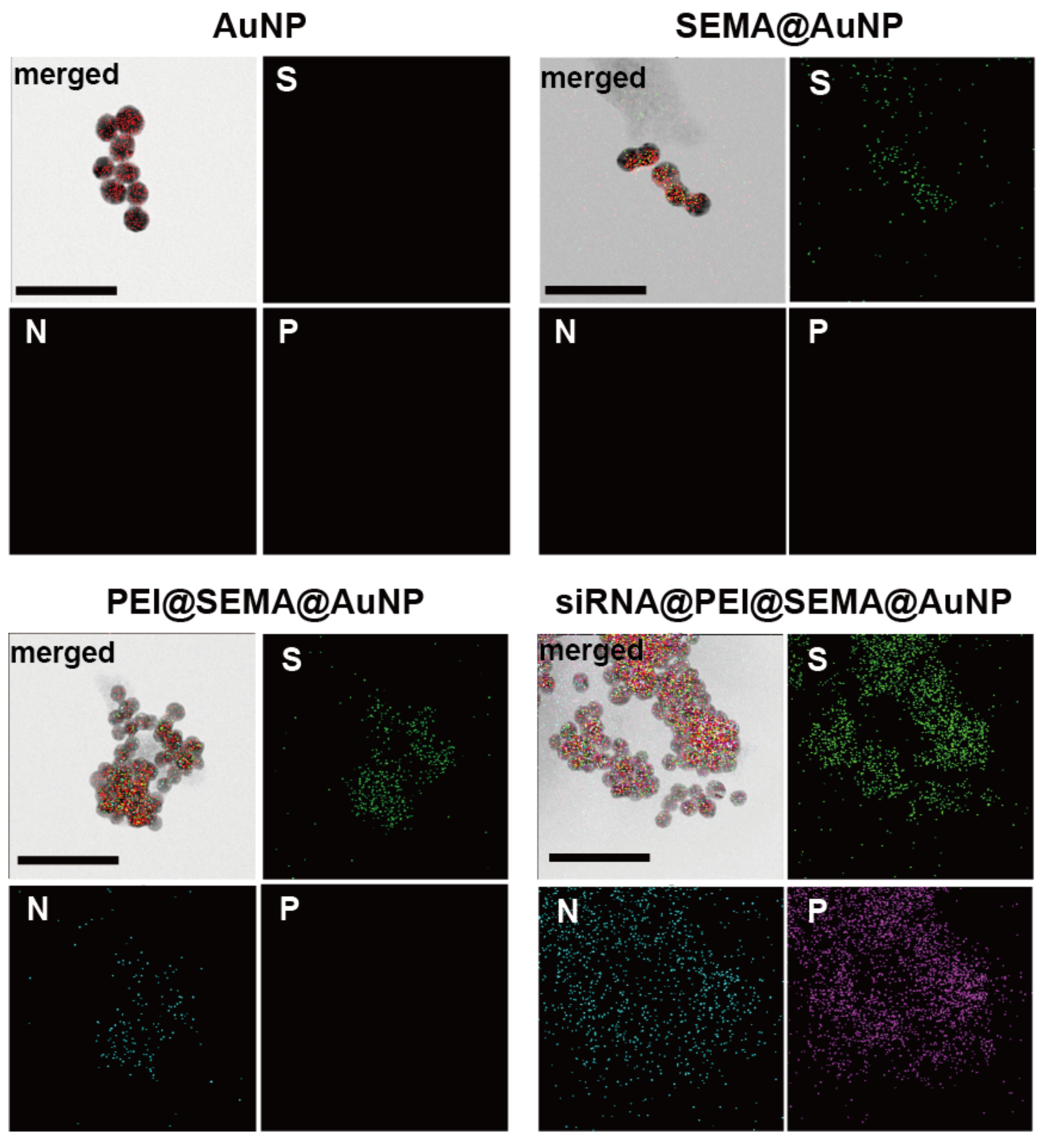
3.2.3. Confirmation of siRNA Incorporation on PEI@SEMA@AuNPs
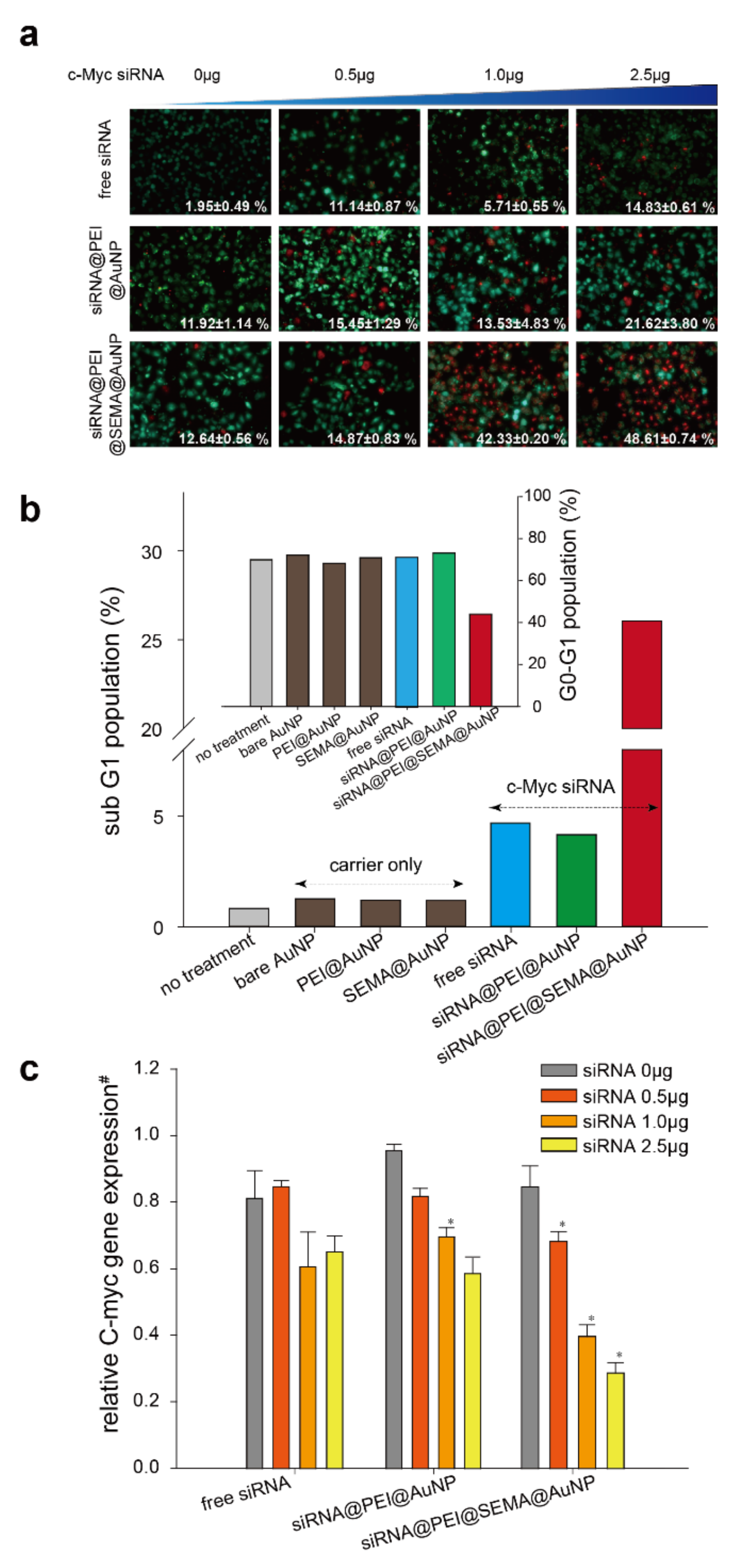
3.3. In Vitro Gene-Silencing Efficincy Evaluation of Free siRNA, siRNA@PEI@AuNP, and siRNA@PEI@SEMA@AuNP
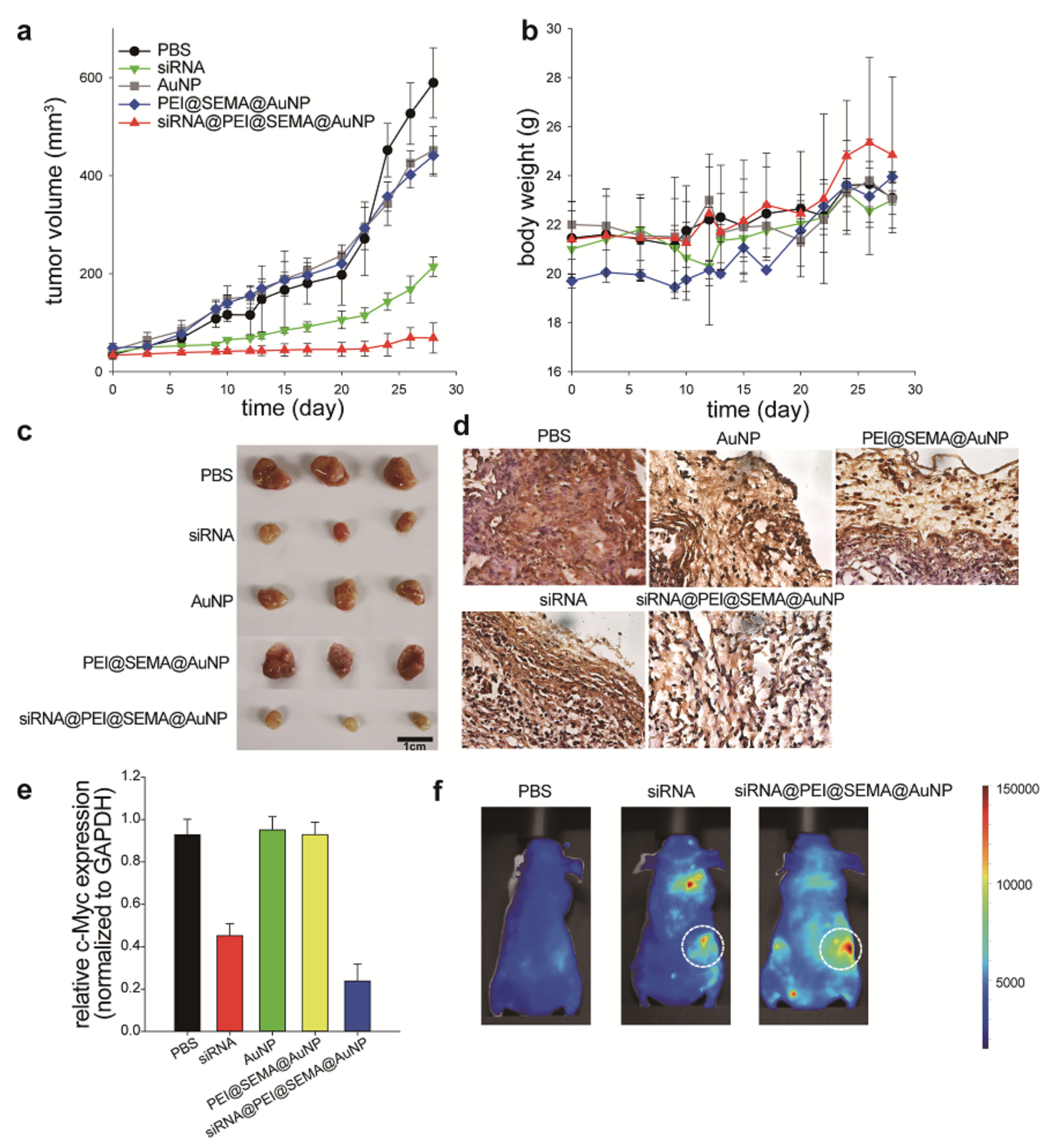
3.4. In Vivo Anti-Cancer Efficincy Evaluation of Free siRNA and siRNA@PEI@SEMA@AuNP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Glinel, K.; Jonas, A.M.; Jouenne, T.; Leprince, J.; Galas, L.; Huck, W.T.S. Antibacterial and Antifouling Polymer Brushes Incorporating Antimicrobial Peptide. Bioconjug. Chem. 2009, 20, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Matyjaszewski, K.; Xia, J. Atom Transfer Radical Polymerization. Chem. Rev. 2001, 101, 2921–2990. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Edmondson, S. Polydopamine-melanin initiators for Surface-initiated ATRP. Polymer (Guildf) 2011, 52, 2141–2149. [Google Scholar] [CrossRef]
- Pyun, J.; Kowalewski, T.; Matyjaszewski, K. Synthesis of Polymer Brushes Using Atom Transfer Radical Polymerization. Macromol. Rapid Commun. 2003, 24, 1043–1059. [Google Scholar] [CrossRef]
- Sun, H.; Yang, L.; Thompson, M.P.; Schara, S.; Cao, W.; Choi, W.; Hu, Z.; Zang, N.; Tan, W.; Gianneschi, N.C. Recent Advances in Amphiphilic Polymer–Oligonucleotide Nanomaterials via Living/Controlled Polymerization Technologies. Bioconjug. Chem. 2019, 30, 1889–1904. [Google Scholar] [CrossRef]
- Sun, H.; Choi, W.; Zang, N.; Battistella, C.; Thompson, M.P.; Cao, W.; Zhou, X.; Forman, C.; Gianneschi, N.C. Bioactive Peptide Brush Polymers via Photoinduced Reversible-Deactivation Radical Polymerization. Angew. Chem. Int. Ed. 2019, 58, 17359–17364. [Google Scholar] [CrossRef]
- Lee, S.Y.; Huh, M.S.; Lee, S.; Lee, S.J.; Chung, H.; Park, J.H.; Oh, Y.K.; Choi, K.; Kim, K.; Kwon, I.C. Stability and cellular uptake of polymerized siRNA (poly-siRNA)/polyethylenimine (PEI) complexes for efficient gene silencing. J. Control. Release 2010, 141, 339–346. [Google Scholar] [CrossRef]
- Lu, S.; Morris, V.B.; Labhasetwar, V. Effectiveness of siRNA Delivery via Arginine-Rich PEI-based Polyplex in Metastatic and Doxorubicin Resistant Breast Cancer Cells. J. Pharmacol. Exp. Ther. 2019. [Google Scholar] [CrossRef]
- Kim, H.S.; Son, Y.J.; Mao, W.; Leong, K.W.; Yoo, H.S. Atom Transfer Radical Polymerization of Multishelled Cationic Corona for the Systemic Delivery of siRNA. Nano Lett. 2018, 18, 314–325. [Google Scholar] [CrossRef]
- Yoon, K.R.; Lee, S.M.; Ramaraj, B.; Kim, D.P. Surface initiated-atom transfer radical polymerization of a sugar methacrylate on gold nanoparticles. Surf. Interface Anal. 2008, 40, 1139–1143. [Google Scholar] [CrossRef]
- Kang, S.M.; Lee, K.B.; Kim, D.J.; Choi, I.S. Biomimetic approach to the formation of gold nanoparticle/silica core/shell structures and subsequent bioconjugation. Nanotechnology 2006, 17, 4719–4725. [Google Scholar] [CrossRef] [PubMed]
- Yenice, Z.; Schön, S.; Bildirir, H.; Genzer, J.; von Klitzing, R. Thermoresponsive PDMAEMA Brushes: Effect of Gold Nanoparticle Deposition. J. Phys. Chem. B 2015, 119, 10348–10358. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Qiu, J.; Sun, W.; Yang, J.; Shen, M.; Wang, L.; Shi, X. Multifunctional PEI-entrapped gold nanoparticles enable efficient delivery of therapeutic siRNA into glioblastoma cells. Biomater. Sci. 2017, 5, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, R.; Asik, E.; Kahraman, N.; Turk, M.; Ozpolat, B.; Ulubayram, K. Modified gold-based siRNA nanotherapeutics for targeted therapy of triple-negative breast cancer. Nanomedicine 2017, 12, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; O’Driscoll, C.M.; Holmes, J.D.; Rahme, K. Bioconjugated gold nanoparticles enhance cellular uptake: A proof of concept study for siRNA delivery in prostate cancer cells. Int. J. Pharm. 2016, 509, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Byeon, H.J.; Thao, L.Q.; Lee, S.; Min, S.Y.; Lee, E.S.; Shin, B.S.; Choi, H.G.; Youn, Y.S. Doxorubicin-loaded nanoparticles consisted of cationic- and mannose-modified-albumins for dual-targeting in brain tumors. J. Control. Release 2016, 225, 301–313. [Google Scholar] [CrossRef]
- Ishii, T.; Kira, N.; Yoshida, T.; Narahara, H. Cucurbitacin D induces growth inhibition, cell cycle arrest, and apoptosis in human endometrial and ovarian cancer cells. Tumor Biol. 2013, 34, 285–291. [Google Scholar] [CrossRef]
- Bettadapur, A.; Suh, G.C.; Geisse, N.A.; Wang, E.R.; Hua, C.; Huber, H.A.; Viscio, A.A.; Kim, J.Y.; Strickland, J.B.; McCain, M.L. Prolonged Culture of Aligned Skeletal Myotubes on Micromolded Gelatin Hydrogels. Sci. Rep. 2016, 6, 28855. [Google Scholar] [CrossRef]
- Zhao, B.; Brittain, W.J. Polymer brushes: Surface-immobilized macromolecules. Prog. Polym. Sci. 2000, 25, 677–710. [Google Scholar] [CrossRef]
- Richards Grayson, A.C.; Doody, A.M.; Putnam, D. Biophysical and Structural Characterization of Polyethylenimine-Mediated siRNA Delivery in Vitro. Pharm. Res. 2006, 23, 1868–1876. [Google Scholar] [CrossRef]
- Ma, H.; Hyun, J.; Stiller, P.; Chilkoti, A. “Non-Fouling” Oligo(ethylene glycol)-Functionalized Polymer Brushes Synthesized by Surface-Initiated Atom Transfer Radical Polymerization. Adv. Mater. 2004, 16, 338–341. [Google Scholar] [CrossRef]
- Morandi, G.; Heath, L.; Thielemans, W. Cellulose Nanocrystals Grafted with Polystyrene Chains through Surface-Initiated Atom Transfer Radical Polymerization (SI-ATRP). Langmuir 2009, 25, 8280–8286. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, W.J.; Yoo, H.S. Dual-responsive breakdown of nanostructures with high doxorubicin payload for apoptotic anticancer therapy. Small 2013, 9, 284–293. [Google Scholar] [CrossRef]
- Kafil, V.; Omidi, Y. Cytotoxic impacts of linear and branched polyethylenimine nanostructures in a431 cells. Bioimpacts 2011, 1, 23–30. [Google Scholar] [PubMed]
- Dang, C. V c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef]
- Ghosh, P.; Han, G.; De, M.; Kim, C.K.; Rotello, V.M. Gold Nanoparticles in Delivery Applications. Adv. Drug Deliv. Rev. 2008, 60, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Crnkovic-Mertens, I.; Hoppe-Seyler, F.; Butz, K. Induction of apoptosis in tumor cells by siRNA-mediated silencing of the livin/ML-IAP/KIAP gene. Oncogene 2003, 22, 8330–8336. [Google Scholar] [CrossRef][Green Version]
- Ormerod, M.G.; Sun, X.M.; Brown, D.; Snowden, R.T.; Cohen, G.M. Quantification of Apoptosis and Necrosis by Flow Cytometry. Acta Oncol. 1993, 32, 417–424. [Google Scholar] [CrossRef]
- Gary, D.J.; Puri, N.; Won, Y.Y. Polymer-based siRNA delivery: Perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J. Control. Release 2007, 121, 64–73. [Google Scholar] [CrossRef]
- Jain, P.K.; Qian, W.; El-Sayed, M.A. Ultrafast Cooling of Photoexcited Electrons in Gold Nanoparticle−Thiolated DNA Conjugates Involves the Dissociation of the Gold−Thiol Bond. J. Am. Chem. Soc. 2006, 128, 2426–2433. [Google Scholar] [CrossRef]
- Austin, C.D.; Wen, X.; Gazzard, L.; Nelson, C.; Scheller, R.H.; Scales, S.J. Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody–drug conjugates. Proc. Natl. Acad. Sci. USA 2005, 102, 17987–17992. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Little, H.C.; Tiambeng, T.N.; Williams, G.A.; Guan, Z. Multifunctional Dendronized Peptide Polymer Platform for Safe and Effective siRNA Delivery. J. Am. Chem. Soc. 2013, 135, 4962–4965. [Google Scholar] [CrossRef] [PubMed]
- Tai, Z.; Wang, X.; Tian, J.; Gao, Y.; Zhang, L.; Yao, C.; Wu, X.; Zhang, W.; Zhu, Q.; Gao, S. Biodegradable Stearylated Peptide with Internal Disulfide Bonds for Efficient Delivery of siRNA In Vitro and In Vivo. Biomacromolecules 2015, 16, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Won, Y.W.; Yoon, S.M.; Lee, K.M.; Kim, Y.H. Poly(oligo-D-arginine) With Internal Disulfide Linkages as a Cytoplasm-sensitive Carrier for siRNA Delivery. Mol. Ther. 2011, 19, 372–380. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, Y.L.; Tian, R.H. The knockdown of c-myc expression by RNAi inhibits cell proliferation in human colon cancer HT-29 cells in vitro and in vivo. Cell. Mol. Biol. Lett. 2009, 14, 305–318. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, J.J.; Huang, L. Nanoparticles Targeted With NGR Motif Deliver c-myc siRNA and Doxorubicin for Anticancer Therapy. Mol. Ther. 2010, 18, 828–834. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Zhang, G.; Zhou, C.; Zhu, H.; Zhou, X.; Quan, L.; Bai, J.; Xu, N. Knockdown of c-Myc expression by RNAi inhibits MCF-7 breast tumor cells growth in vitro and in vivo. Breast Cancer Res. 2004, 7, R220. [Google Scholar] [CrossRef]
- Conde, J.; Tian, F.; Hernández, Y.; Bao, C.; Cui, D.; Janssen, K.P.; Ibarra, M.R.; Baptista, P.V.; Stoeger, T.; de la Fuente, J.M. In vivo tumor targeting via nanoparticle-mediated therapeutic siRNA coupled to inflammatory response in lung cancer mouse models. Biomaterials 2013, 34, 7744–7753. [Google Scholar] [CrossRef]
- Calabrese, C.M.; Merkel, T.J.; Briley, W.E.; Randeria, P.S.; Narayan, S.P.; Rouge, J.L.; Walker, D.A.; Scott, A.W.; Mirkin, C.A. Biocompatible Infinite-Coordination-Polymer Nanoparticle–Nucleic-Acid Conjugates for Antisense Gene Regulation. Angew. Chem. Int. Ed. 2015, 54, 476–480. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Rahme, K.; Guo, J.; Holmes, J.D.; O’Driscoll, C.M. Anisamide-targeted gold nanoparticles for siRNA delivery in prostate cancer–synthesis, physicochemical characterisation and in vitro evaluation. J. Mater. Chem. B 2016, 4, 2242–2252. [Google Scholar] [CrossRef]
- Wang, R.; Degirmenci, V.; Xin, H.; Li, Y.; Wang, L.; Chen, J.; Hu, X.; Zhang, D. PEI-Coated Fe3O4 Nanoparticles Enable Efficient Delivery of Therapeutic siRNA Targeting REST into Glioblastoma Cells. Int. J. Mol. Sci. 2018, 19, 2230. [Google Scholar] [CrossRef] [PubMed]
| Carriers | Diameter (nm) | ζ-Potential (mV) |
|---|---|---|
| bare AuNPs | 25.55 ± 0.49 | −23.87 ± 0.50 |
| SEMA@AuNPs | 222.20 ± 4.75 | −26.55 ± 1.91 |
| PEI@SEMA@AuNPs | 161.08 ± 1.79 | 36.93 ± 0.97 |
| PEI@AuNPs | 180.63 ± 1.94 | 14.37 ± 1.94 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, W.; Lee, S.; Shin, J.U.; Yoo, H.S. Surface-Initiated Atom Transfer Polymerized Anionic Corona on Gold Nanoparticles for Anti-Cancer Therapy. Pharmaceutics 2020, 12, 261. https://doi.org/10.3390/pharmaceutics12030261
Mao W, Lee S, Shin JU, Yoo HS. Surface-Initiated Atom Transfer Polymerized Anionic Corona on Gold Nanoparticles for Anti-Cancer Therapy. Pharmaceutics. 2020; 12(3):261. https://doi.org/10.3390/pharmaceutics12030261
Chicago/Turabian StyleMao, Wei, Sol Lee, Ji Un Shin, and Hyuk Sang Yoo. 2020. "Surface-Initiated Atom Transfer Polymerized Anionic Corona on Gold Nanoparticles for Anti-Cancer Therapy" Pharmaceutics 12, no. 3: 261. https://doi.org/10.3390/pharmaceutics12030261
APA StyleMao, W., Lee, S., Shin, J. U., & Yoo, H. S. (2020). Surface-Initiated Atom Transfer Polymerized Anionic Corona on Gold Nanoparticles for Anti-Cancer Therapy. Pharmaceutics, 12(3), 261. https://doi.org/10.3390/pharmaceutics12030261







