Pulmonary Metabolism of Substrates for Key Drug-Metabolizing Enzymes by Human Alveolar Type II Cells, Human and Rat Lung Microsomes, and the Isolated Perfused Rat Lung Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Compound Selection
2.3. Human Hepatocytes
2.4. Patients and Human Lung Tissue Preparation
2.5. Isolation of ATII Cells from Human Lungs
2.6. Incubation of Drug-Metabolizing Enzyme Substrates with Rat and Human Lung Microsomes
2.7. Incubation of Drug-Metabolizing Enzyme Substrates with Isolated Human ATII Cells and Human Hepatocytes
2.8. Assessment of Drug Metabolism in the Ex Vivo Isolated Perfused Rat Lung (IPRL)
2.9. Bioanalysis
3. Results
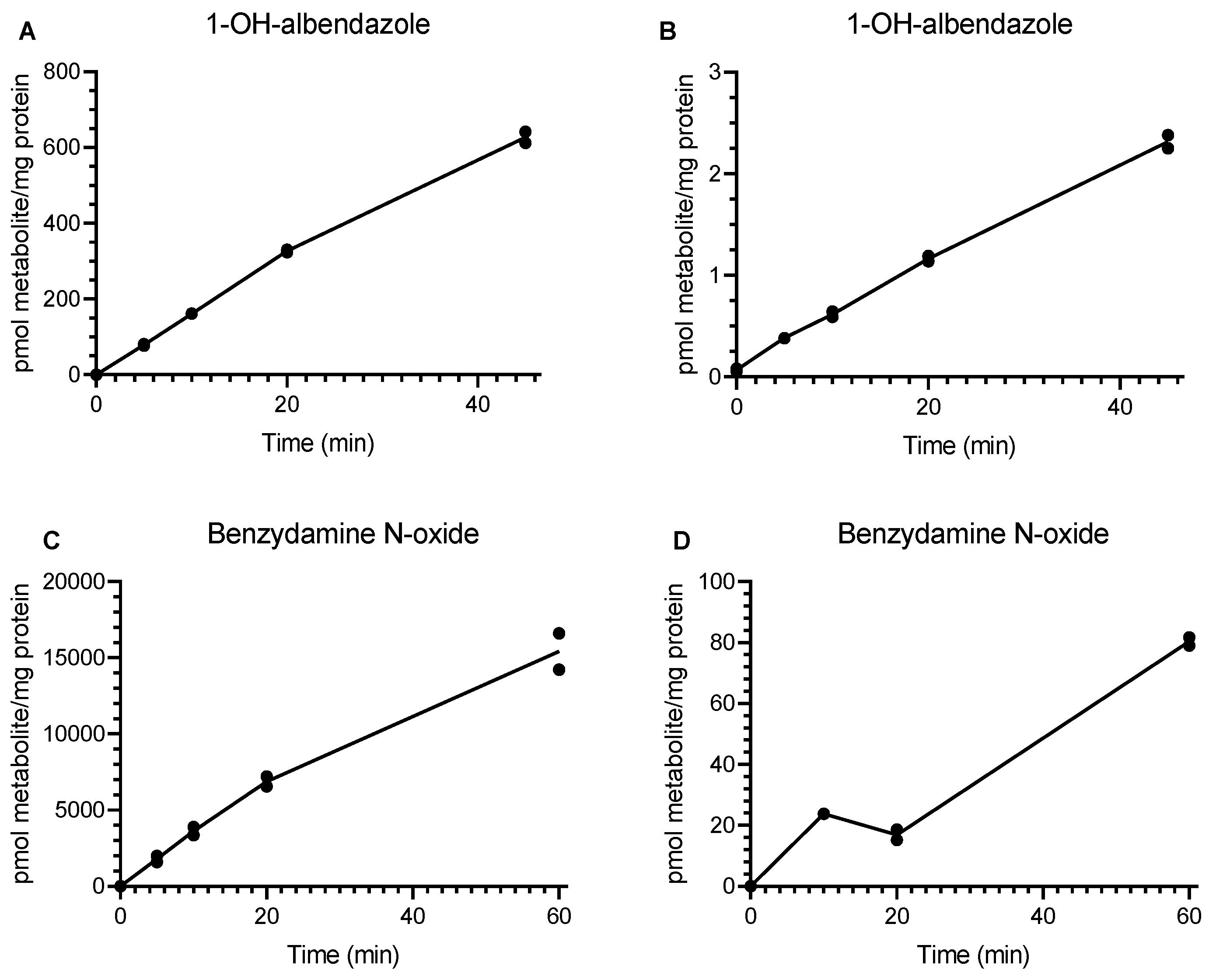
3.1. Incubation of Drug-Metabolizing Enzyme Substrates with Rat and Human Lung Microsomes
3.2. Analysis of Drug-Metabolizing Enzyme Activity in Isolated Human ATII Cells and Human Hepatocytes
3.3. IPRL
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATI | alveolar type I |
| ATI | alveolar type I |
| ATII | alveolar type II |
| CLint | intrinsic clearance |
| CYP | cytochrome P450 |
| BLOQ | below limit of quantification |
| EH | epoxide hydrolase |
| FMO | flavin-containing monooxygenases |
| IPRL | isolated perfused rat lung |
| MOA | monoamine oxidase |
| MS/MS | tandem mass spectrometry |
| PAH | polycyclic aromatic hydrocarbons |
| QToF | quadropole time of flight |
| UPLC | Ultra-performance liquid chromatography |
| XO/AO | xanthine oxidase/aldehyde oxidase |
References
- Patton, J.S.; Byron, P.R. Inhaling medicines: Delivering drugs to the body through the lungs. Nat. Rev. Drug Discov. 2007, 6, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, A. Biotransformation of xenobiotics. In Casarett & Doull’s Toxicology: The Basic Science of Poisons; Klaassen, C.D., Ed.; McGraw-Hill, Inc.: New York, NY, USA, 2001; pp. 161–304. [Google Scholar]
- Guengerich, F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Jhajra, S.; Ramesh Varkhede, N.; Suresh Ahire, D.; Vidyasagar Naik, B.; Prasad, B.; Paliwal, J.; Singh, S. Extrahepatic drug-metabolizing enzymes and their significance. In Encyclopedia of Drug Metabolism and Interactions; Lyubimov, A.V., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 1–99. [Google Scholar]
- Zhang, J.Y.; Wang, Y.; Prakash, C. Xenobiotic-metabolizing enzymes in human lung. Curr. Drug Metab. 2006, 7, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Raunio, H.; Hakkola, J.; Hukkanen, J.; Lassila, A.; Paivarinta, K.; Pelkonen, O.; Anttila, S.; Piipari, R.; Boobis, A.; Edwards, R.J. Expression of xenobiotic-metabolizing CYPs in human pulmonary tissue. Exp. Toxicol. Pathol. 1999, 51, 412–417. [Google Scholar] [CrossRef]
- Berg, T.; Hegelund Myrback, T.; Olsson, M.; Seidegard, J.; Werkstrom, V.; Zhou, X.H.; Grunewald, J.; Gustavsson, L.; Nord, M. Gene expression analysis of membrane transporters and drug-metabolizing enzymes in the lung of healthy and COPD subjects. Pharmacol. Res. Perspect. 2014, 2, e00054. [Google Scholar] [CrossRef]
- Somers, G.I.; Lindsay, N.; Lowdon, B.M.; Jones, A.E.; Freathy, C.; Ho, S.; Woodrooffe, A.J.; Bayliss, M.K.; Manchee, G.R. A comparison of the expression and metabolizing activities of phase I and II enzymes in freshly isolated human lung parenchymal cells and cryopreserved human hepatocytes. Drug Metab. Dispos. 2007, 35, 1797–1805. [Google Scholar] [CrossRef]
- Stone, K.C.; Mercer, R.R.; Gehr, P.; Stockstill, B.; Crapo, J.D. Allometric relationships of cell numbers and size in the mammalian lung. Am. J. Respir. Cell Mol. Biol. 1992, 6, 235–243. [Google Scholar] [CrossRef]
- Backstrom, E.; Hamm, G.; Nilsson, A.; Fihn, B.M.; Strittmatter, N.; Andren, P.; Goodwin, R.J.A.; Friden, M. Uncovering the regional localization of inhaled salmeterol retention in the lung. Drug Deliv. 2018, 25, 838–845. [Google Scholar] [CrossRef]
- Chiba, M.; Ishii, Y.; Sugiyama, Y. Prediction of hepatic clearance in human from in vitro data for successful drug development. AAPS J. 2009, 11, 262–276. [Google Scholar] [CrossRef]
- Kamata, S.; Fujino, N.; Yamada, M.; Grime, K.; Suzuki, S.; Ota, C.; Tando, Y.; Okada, Y.; Sakurada, A.; Noda, M.; et al. Expression of cytochrome P450 mRNAs in Type II alveolar cells from subjects with chronic obstructive pulmonary disease. Pharmacol. Res. Perspect. 2018, 6, e00405. [Google Scholar] [CrossRef]
- Weaver, R.; Graham, K.S.; Beattie, I.G.; Riley, R.J. Cytochrome P450 inhibition using recombinant proteins and mass spectrometry/multiple reaction monitoring technology in a cassette incubation. Drug Metab. Dispos. 2003, 31, 955–966. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, C.J.; Grime, K.; Courtney, P.; Slee, D.; Riley, R.J. The development of a cocktail CYP2B6, CYP2C8, and CYP3A5 inhibition assay and a preliminary assessment of utility in a drug discovery setting. Drug Metab. Dispos. 2007, 35, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Stormer, E.; Roots, I.; Brockmoller, J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br. J. Clin. Pharm. 2000, 50, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lee, D.; Joo, J.; Shin, J.H.; Kang, W.; Oh, S.; Lee, D.Y.; Lee, S.J.; Yea, S.S.; Lee, H.S.; et al. CYP2J2 and CYP2C19 are the major enzymes responsible for metabolism of albendazole and fenbendazole in human liver microsomes and recombinant P450 assay systems. Antimicrob. Agents Chemother. 2013, 57, 5448–5456. [Google Scholar] [CrossRef]
- Li, X.Q.; Bjorkman, A.; Andersson, T.B.; Gustafsson, L.L.; Masimirembwa, C.M. Identification of human cytochrome P(450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur. J. Clin. Pharmacol. 2003, 59, 429–442. [Google Scholar] [CrossRef]
- Fujino, N.; Kubo, H.; Ota, C.; Suzuki, T.; Suzuki, S.; Yamada, M.; Takahashi, T.; He, M.; Suzuki, T.; Kondo, T.; et al. A novel method for isolating individual cellular components from the adult human distal lung. Am. J. Respir. Cell Mol. Biol. 2012, 46, 422–430. [Google Scholar] [CrossRef]
- Ewing, P.; Eirefelt, S.J.; Andersson, P.; Blomgren, A.; Ryrfeldt, A.; Gerde, P. Short inhalation exposures of the isolated and perfused rat lung to respirable dry particle aerosols; the detailed pharmacokinetics of budesonide, formoterol, and terbutaline. J. Aerosol. Med. Pulm. Drug Deliv. 2008, 21, 169–180. [Google Scholar] [CrossRef]
- Olsson, B.; Bondesson, E.; Borgström, L.; Edsbäcker, S.; Eirefelt, S.; Ekelund, K.; Gustavsson, L.; Hegelund-Myrbäck, T. Pulmonary Drug Metabolism, Clearance, and Absorption. In Controlled Pulmonary Drug Delivery; Smyth, D.C.H., Hickey, J.A., Eds.; Springer: New York, NY, USA, 2011; pp. 21–50. [Google Scholar]
- Cooper, A.E.; Ferguson, D.; Grime, K. Optimisation of DMPK by the inhaled route: Challenges and approaches. Curr. Drug Metab. 2012, 13, 457–473. [Google Scholar] [CrossRef]
- Delaney, S.; Biffen, M.; Maltby, J.; Bell, J.; Asimus, S.; Aggarwal, A.; Kraan, M.; Keeling, D. Tolerability in man following inhalation dosing of the selective TLR7 agonist, AZD8848. BMJ Open. Respir. Res. 2016, 3, e000113. [Google Scholar] [CrossRef]
- Chovan, J.P.; Ring, S.C.; Yu, E.; Baldino, J.P. Cytochrome P450 probe substrate metabolism kinetics in Sprague Dawley rats. Xenobiotica Fate Foreign Compd. Biol. Syst. 2007, 37, 459–473. [Google Scholar] [CrossRef]
- Kawaji, A.; Ohara, K.; Takabatake, E. An assay of flavin-containing monooxygenase activity with benzydamine N-oxidation. Anal. Biochem. 1993, 214, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, Y.; Williams, G.; Walles, M.; Manevski, N.; Krahenbuhl, S.; Camenisch, G. Comparison of Rat and Human Pulmonary Metabolism Using Precision-cut Lung Slices (PCLS). Drug Metab. Lett. 2019, 13, 53–63. [Google Scholar] [CrossRef] [PubMed]

| Substrate | Test System | Enzyme | Metabolites |
|---|---|---|---|
| Albendazole | Microsomes, ATII, hepatocytes, IPRL | CYP2J2 | 1-OH-albendazole |
| Phenacetin | Microsomes | CYP1A2 | Acetaminophen |
| Benzydamine | Microsomes, ATII, hepatocytes, IPRL | FMO | Benzydamine N-oxide |
| Bufuralol | Microsomes, ATII, hepatocytes | CYP2D6 | 1-OH-bufuralol |
| Bupropion | Microsomes, ATII, hepatocytes | CYP2B6 | 2-OH-Bupropion |
| P-nitrophenol | Microsomes | CYP2E1 | 4-nitrocatechol |
| Flutamide | Microsomes, ATII, hepatocytes | CYP1B1 | 2-OH-flutamide |
| Midazolam | Microsomes, ATII, hepatocytes | CYP3A4/5 | 1-OH-midazolam 4-OH-midazolam |
| Paclitaxel | Microsomes | CYP2C8 | 1-OH-paclitaxel |
| Diclofenac | Microsomes | CYP2C9 | 4-OH-diclofenac |
| S-mephenytoin | Microsomes | CYP2C19 | 4-OH-mephenytoin |
| Metabolite | Transition | CE (volts) | Rt (min) | LLOQ (nM) |
|---|---|---|---|---|
| Acetaminophen | 152.1 > 110.0 | 23 | 0.73 | 3.7 |
| 2-OH-flutamide | 291.1 > 205.1 | 18 | 1.5 | 0.07 |
| 2-OH-bupropion | 256.1 > 238.0 | 14 | 2.6 | 1.5 |
| 6α-OH-paclitaxel | 870.0 > 104.9 | 80 | 1.5 | 3.3 |
| 4-OH-diclofenac | 312.0 > 230.0 | 42 | 1.4 | 0.05 |
| 4-OH-mephenytoin | 235.1 > 149.9 | 22 | 1.0 | 4.4 |
| 1-OH-bufuralol | 278.2 > 185.9 | 18 | 0.88 | 0.06 |
| 1-OH-albendazole | 282.1 > 250.1 | 18 | 2.4 | 0.03 |
| 4-nitrocatechol | 154.0 > 124.0 | 14 | 1.0 | 0.2 |
| 1-OH-midazolam | 342.1 > 203.1 | 34 | 1.6 | 0.4 |
| 4-OH-midazolam | 342.1 > 325.1 | 26 | 1.5 | 0.08 |
| Benzydamine N-oxide | 326.2 > 84.0 | 38 | 1.2 | 4.6 |
| Metabolite Formed | Rat Lung Microsomes (pmol/min/mg Protein) | Human Lung Microsomes (pmol/min/mg Protein) |
|---|---|---|
| OH-albendazole | 16.5 | 0.053 |
| 16.2 | 0.055 | |
| Benzydamine N-oxide | 358 | 1.36 |
| 329 | 1.39 | |
| 4-nitrocatechol | 14.9 | n/a |
| 14.6 | ||
| 2-OH-bupropion | 19.7 | n/a |
| 10.8 | ||
| 1-OH-bufuralol | 0.089 | n/a |
| 0.102 | ||
| 4-OH-midazolam | 1.79 | n/a |
| 1.26 | ||
| 1-OH-midazolam | 0.447 | 0.103 |
| 0.363 | 0.089 | |
| 2-OH-flutamide | 0.933 | 0.099 |
| 0.897 | 0.123 | |
| 4-OH-diclofenac * | 0.102 | 0.033 |
| Acetaminophen | 5.20 | n/a |
| 5.53 |
| Sample ID | Time (h) | 1-OH-albendazole | Benzydamine N-oxide | 1-OH-bufuralol | 2-OH-bupropion | 2-OH-flutamide | 4-OH midazolam |
|---|---|---|---|---|---|---|---|
| ATII cells | |||||||
| 15-017 | 2 | 78 | 75 | n/a | n/d | n/a | n/a |
| 15-017 | 4 | 93 | 120 | n/a | n/d | n/a | n/a |
| 15-018 | 2 | 87 | 93 | n/a | n/d | n/a | n/a |
| 15-018 | 4 | 123 | 138 | n/a | n/d | n/a | n/a |
| 15-019 | 2 | 102 | 75 | n/a | n/d | n/a | n/a |
| 15-019 | 4 | 177 | 135 | n/a | n/d | n/a | n/a |
| 15-020 | 2 | 75 | 114 | 9 | n/d | 9 | 6 |
| 15-020 | 4 | 90 | 159 | 9 | n/d | 18 | 9 |
| 15-022 | 2 | 75 | 129 | n/a | n/d | n/a | 9 |
| 15-022 | 4 | 90 | 141 | n/a | n/d | n/a | n/a |
| Hepatocyte | |||||||
| S1193T-1 | 2 | 766 | 3024 | 1697 | 1318 | 3769 | 300 |
| S1193T-1 | 4 | 931 | 4817 | 2477 | 1649 | 4721 | 336 |
| S1193T-2 | 2 | 724 | 2694 | 1339 | 1111 | 3360 | 270 |
| S1193T-2 | 4 | 826 | 4084 | 1799 | 1300 | 3928 | 282 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubin, K.; Ewing, P.; Bäckström, E.; Abrahamsson, A.; Bonn, B.; Kamata, S.; Grime, K. Pulmonary Metabolism of Substrates for Key Drug-Metabolizing Enzymes by Human Alveolar Type II Cells, Human and Rat Lung Microsomes, and the Isolated Perfused Rat Lung Model. Pharmaceutics 2020, 12, 117. https://doi.org/10.3390/pharmaceutics12020117
Rubin K, Ewing P, Bäckström E, Abrahamsson A, Bonn B, Kamata S, Grime K. Pulmonary Metabolism of Substrates for Key Drug-Metabolizing Enzymes by Human Alveolar Type II Cells, Human and Rat Lung Microsomes, and the Isolated Perfused Rat Lung Model. Pharmaceutics. 2020; 12(2):117. https://doi.org/10.3390/pharmaceutics12020117
Chicago/Turabian StyleRubin, Katarina, Pär Ewing, Erica Bäckström, Anna Abrahamsson, Britta Bonn, Satoshi Kamata, and Ken Grime. 2020. "Pulmonary Metabolism of Substrates for Key Drug-Metabolizing Enzymes by Human Alveolar Type II Cells, Human and Rat Lung Microsomes, and the Isolated Perfused Rat Lung Model" Pharmaceutics 12, no. 2: 117. https://doi.org/10.3390/pharmaceutics12020117
APA StyleRubin, K., Ewing, P., Bäckström, E., Abrahamsson, A., Bonn, B., Kamata, S., & Grime, K. (2020). Pulmonary Metabolism of Substrates for Key Drug-Metabolizing Enzymes by Human Alveolar Type II Cells, Human and Rat Lung Microsomes, and the Isolated Perfused Rat Lung Model. Pharmaceutics, 12(2), 117. https://doi.org/10.3390/pharmaceutics12020117




