Pharmacogenetic Testing: A Tool for Personalized Drug Therapy Optimization
Abstract
:1. Introduction
2. Pharmacogenetic Studies of Drugs
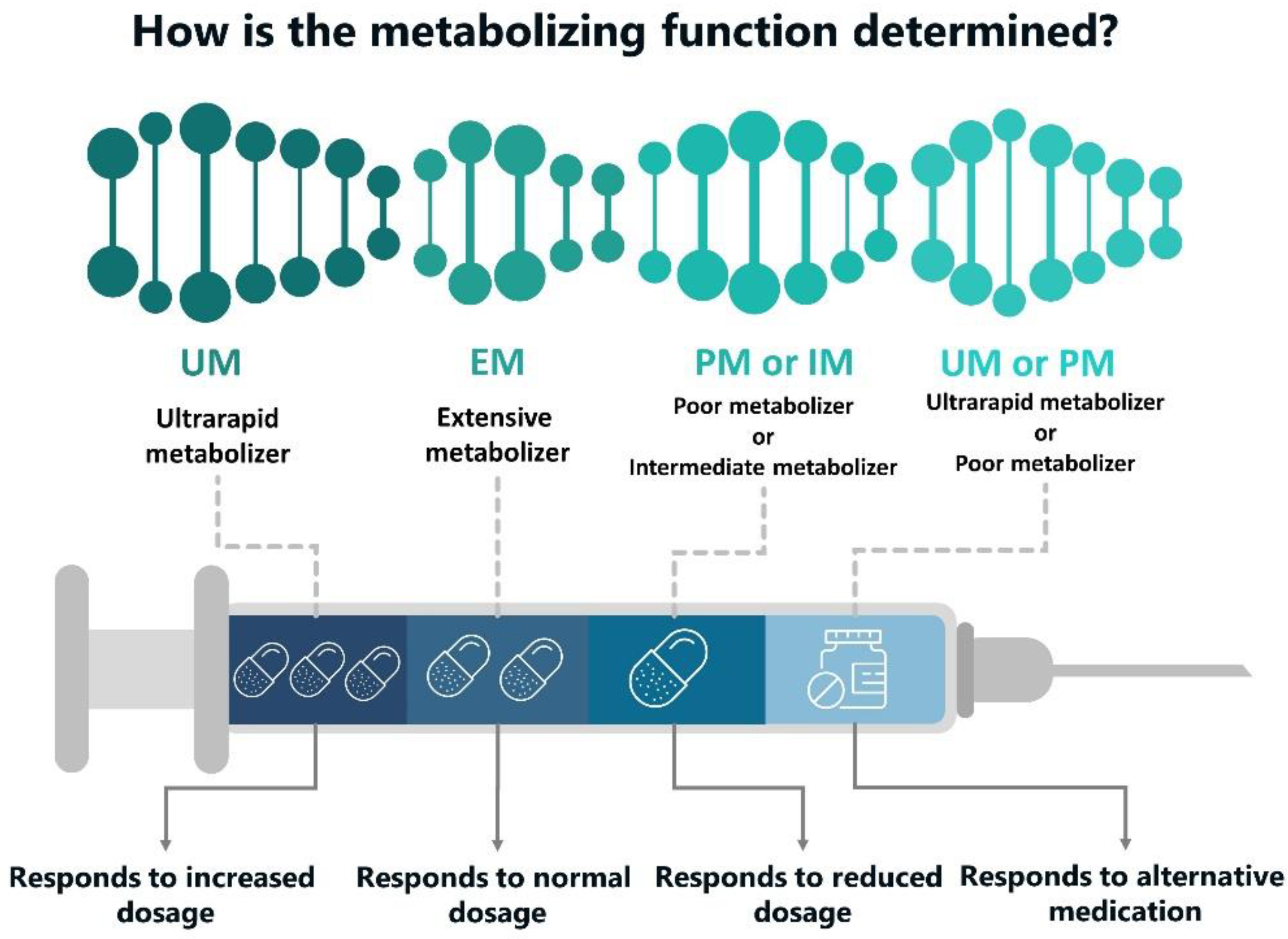
- poor metabolism (PM): a type of alleles that carry the mutated gene(s) encoding important metabolizing enzyme that participates in drug transformation and exhibition of drug activity. Such mutations cause the synthesis of either an insufficient amount of enzyme or produce its inactive gene product which entails decreasing of enzymatic activity and even complete loss of activity. They are much slower to eliminate various drugs metabolized by the same enzyme. Therefore, the patient runs a risk to reach a high plasma concentration of the drug, causing dose-dependent adverse effects. In this regard, slow metabolizers require a careful drug dose selection:
- Extensive metabolizers (EM): they provide a regular rate of drug biotransformation. They usually have two active allelic genes or one functional and one partially active allele;
- Intermediate metabolizers (IM): heterozygous carriers of the mutation (with an autosomal recessive inheritance). To achieve an optimal therapeutic effect, they may require a lower pharmacological dosage than the usual one;
- Ultra-fast metabolizers (UM): they are characterized with an increased gene expression—owing to the presence of three or more functional alleles following the duplication or multiple duplications of a functional allele (e.g., duplication of the CYP2D6 gene). Ultra-fast metabolizers may require a higher drug dose for an optimal effect (Figure 1).
2.1. Comparative Cohort Study with Posterior Analysis
2.2. Comparative Cohort Study with Genotyping of Participants before Inclusion
- Posterior analysis, in which genotyping is carried out before inclusion in the study to form subgroups of equal number with the “wild” genotype and polymorphic variant and does not affect the appointment of pharmacotherapy;
2.3. Comparative Study of the Pharmacogenetic Approach
3. Prospects for the Introduction of a Pharmacogenetic Test into the Clinic
3.1. Audience for PGx Testing
3.2. Resources in the Pharmacogenetic Sector
3.3. Choice of PGx Testing
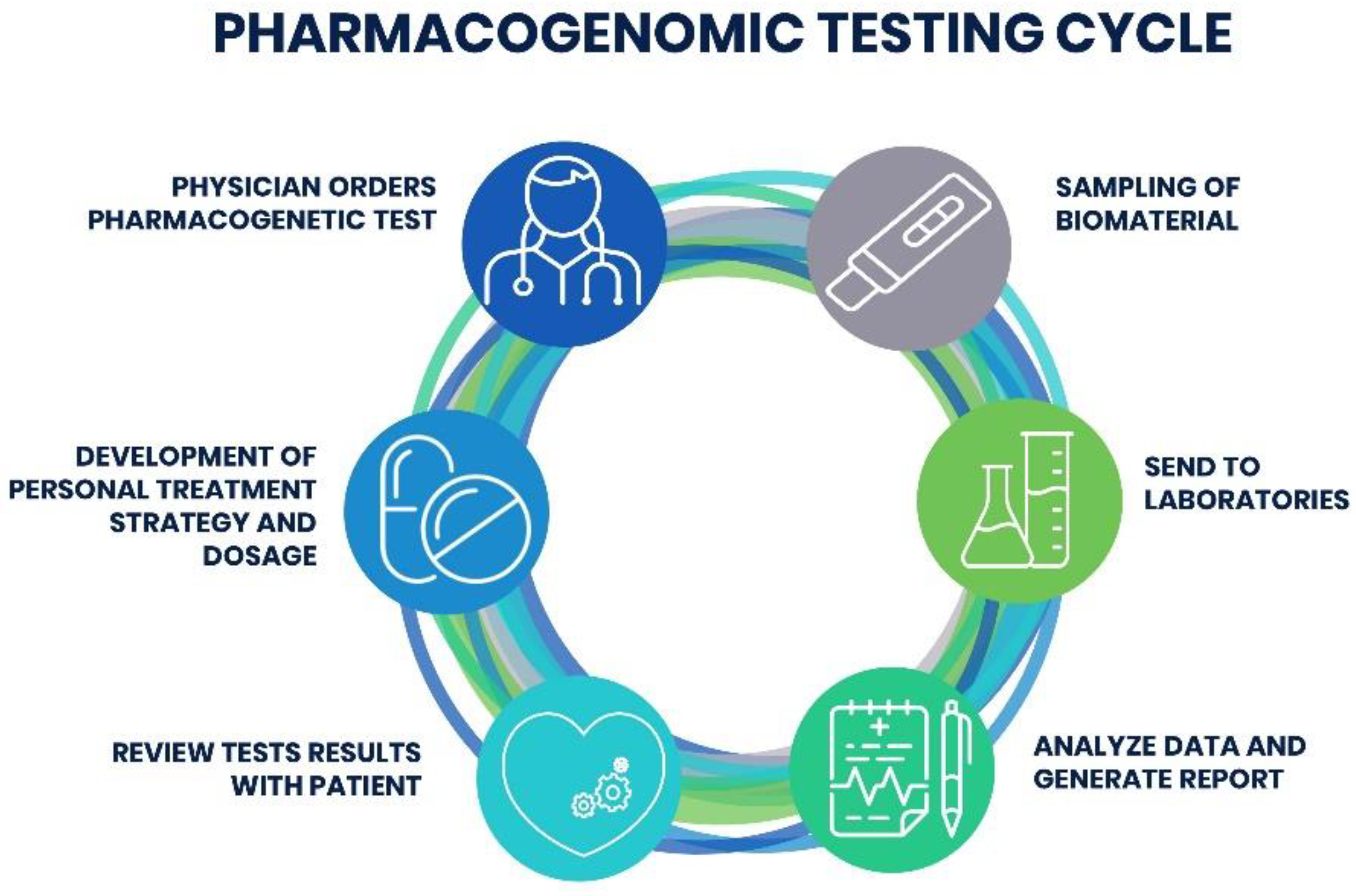
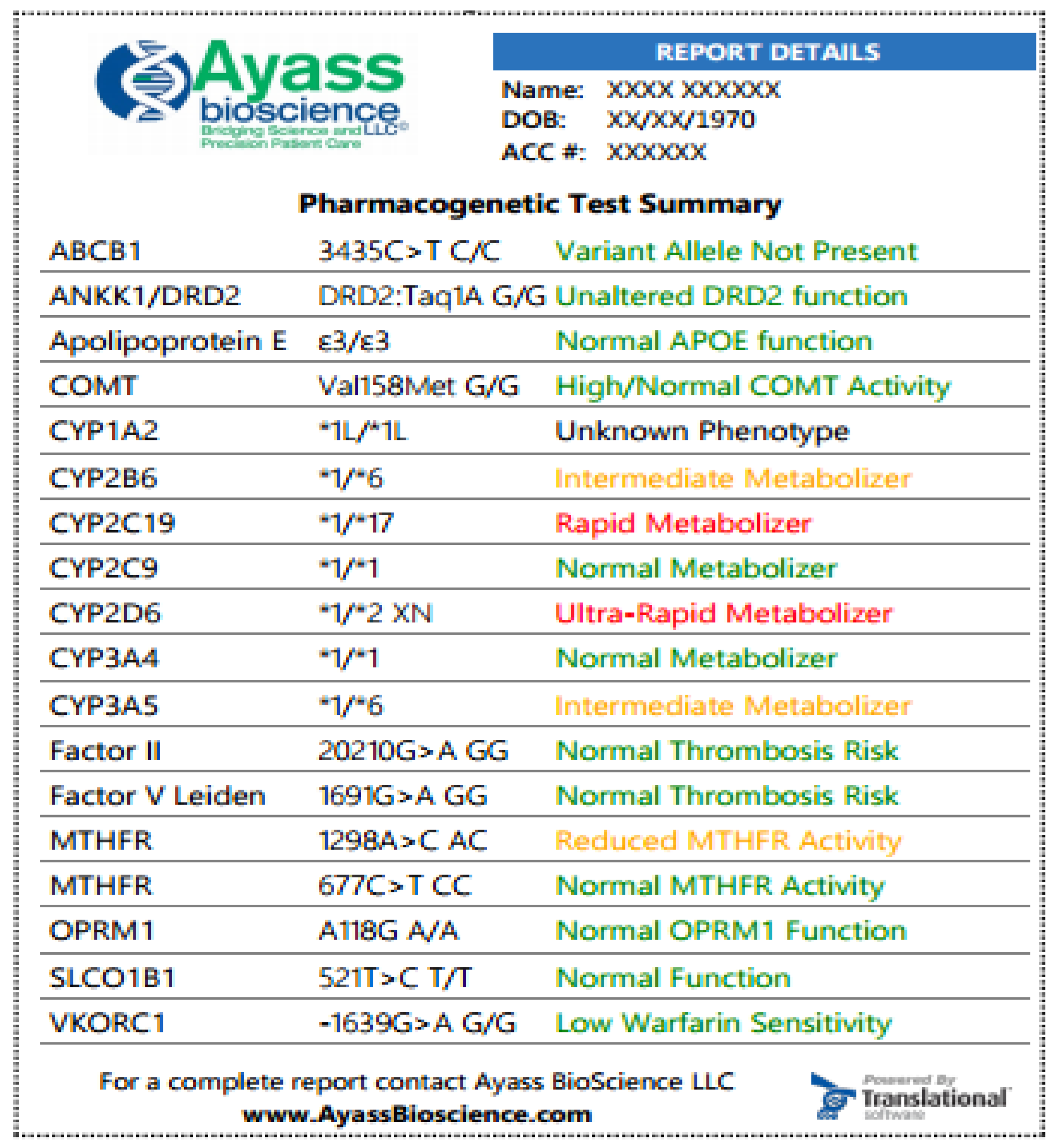
3.4. Interpreting PGx Test Results
3.5. Automation Tools for Integrating PGx Testing into the Clinic
4. Side Effects of Drugs and Safety
5. Obstacles on the Way to the Introduction of Pharmacogenetic Tests into Clinical Practice
- Instructions for the medical use of a medicinal product (FDA and European Medicines Agency (EMA)), recommendations of international and national professional scientific public organizations (Recommendations of the experts of the European Science Foundation (ESF), discussed and approved by the participants European Conference on Pharmacogenetics and Pharmacogenomics in Barcelona in June 2010 (published in March 2011) [113],
- Expert recommendations of the Pharmacogenetics Working Group of the Royal Dutch Pharmaceutical Association (published in March 2011) [50],
- Expert guidance of the CPIC, beginning of publication—January 2011) [104].
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Alonso, S.G.; de la Torre Díez, I.; Zapiraín, B.G. Predictive, Personalized, Preventive and Participatory (4P) Medicine Applied to Telemedicine and EHealth in the Literature. J. Med. Syst. 2019, 43, 140. [Google Scholar] [CrossRef] [PubMed]
- Vogel, F. Moderne Probleme der Humangenetik. In Ergebnisse der Inneren Medizin und Kinderheilkunde; Heilmeyer, L., Schoen, R., de Rudder, B., Eds.; Springer: Berlin/Heidelberg, Germany, 1959; pp. 52–125. [Google Scholar] [CrossRef]
- Meyer, U.A. Pharmacogenetics—Five Decades of Therapeutic Lessons from Genetic Diversity. Nat. Rev. Genet. 2004, 5, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J. Drug Metabolism and Pharmacogenetics: The British Contribution to Fields of International Significance. Br. J. Pharmacol. 2006, 147 (Suppl. 1), S89–S99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, R.; Allali, I.; Agamah, F.E.; Elsheikh, S.S.M.; Thomford, N.E.; Dandara, C.; Chimusa, E.R. Drug Response in Association with Pharmacogenomics and Pharmacomicrobiomics: Towards a Better Personalized Medicine. Brief. Bioinform. 2020, bbaa292. [Google Scholar] [CrossRef] [PubMed]
- Testa, R.; Bonfigli, A.R.; Sirolla, C.; Boemi, M.; Manfrini, S.; Mari, D.; Testa, I.; Sacchi, E.; Franceschi, C. Effect of 4G/5G PAI-1 Polymorphism on the Response of PAI-1 Activity to Vitamin E Supplementation in Type 2 Diabetic Patients. Diabetes Nutr. Metab. 2004, 17, 217–221. [Google Scholar] [PubMed]
- He, H.-Y.; Liu, M.-Z.; Zhang, Y.-L.; Zhang, W. Vitamin Pharmacogenomics: New Insight into Individual Differences in Diseases and Drug Responses. Genom. Proteom. Bioinform. 2017, 15, 94–100. [Google Scholar] [CrossRef]
- Awh, C.C.; Lane, A.-M.; Hawken, S.; Zanke, B.; Kim, I.K. CFH and ARMS2 Genetic Polymorphisms Predict Response to Antioxidants and Zinc in Patients with Age-Related Macular Degeneration. Ophthalmology 2013, 120, 2317–2323. [Google Scholar] [CrossRef]
- Mosolov, S.N. (Ed.) Biological Methods of Therapy for Mental Disorders; Sociopolitical thought (Socialno-politicheskaya mysl): Moscow, Russia, 2012. (In Russian) [Google Scholar]
- Fabbri, C.; Zohar, J.; Serretti, A. Pharmacogenetic Tests to Guide Drug Treatment in Depression: Comparison of the Available Testing Kits and Clinical Trials. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 86, 36–44. [Google Scholar] [CrossRef]
- Carter, C.A.; Frischmeyer-Guerrerio, P.A. The Genetics of Food Allergy. Curr. Allergy Asthma Rep. 2018, 18, 2. [Google Scholar] [CrossRef]
- Noble J., A.; Valdes A., M. Genetics of the HLA Region in the Prediction of Type 1 Diabetes. Curr. Diabetes Rep. 2011, 11, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Schaeffeler, E.; Schwab, M.; Eichelbaum, M.; Zanger, U.M. CYP2D6 Genotyping Strategy Based on Gene Copy Number Determination by TaqMan Real-Time PCR. Hum. Mutat. 2003, 22, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.W.F. Scientific Challenges and Implementation Barriers to Translation of Pharmacogenomics in Clinical Practice. ISRN Pharmacol. 2013, 2013, 641089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stingl (formerly Kirchheiner), J.; Brockmöller, J. Study Designs in Clinical Pharmacogenetic and Pharmacogenomic Research. In Pharmacogenomics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 309–341. [Google Scholar] [CrossRef]
- Matsui, S. Genomic Biomarkers for Personalized Medicine: Development and Validation in Clinical Studies. Comput. Math. Methods Med. 2013, 2013, 865980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roden, D.M.; Wilke, R.A.; Kroemer, H.K.; Stein, C.M. Pharmacogenomics: The Genetics of Variable Drug Responses. Circulation 2011, 123, 1661–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardon, L.R.; Harris, T. Precision Medicine, Genomics and Drug Discovery. Hum. Mol. Genet. 2016, 25, R166–R172. [Google Scholar] [CrossRef] [Green Version]
- Marx, V. The DNA of a Nation. Nature 2015, 524, 503–505. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Lewis, C.M.; Traylor, M. Pharmacogenetic testing through the direct-to-consumer genetic testing company 23andMe. BMC Med. Genomics 2017, 10, 47. [Google Scholar] [CrossRef] [Green Version]
- Crews, K.R.; Hicks, J.K.; Pui, C.-H.; Relling, M.V.; Evans, W.E. Pharmacogenomics and Individualized Medicine: Translating Science into Practice. Clin. Pharmacol. Ther. 2012, 92, 467–475. [Google Scholar] [CrossRef] [Green Version]
- Veenstra, D.L. The Value of Routine Pharmacogenomic Screening—Are We There yet? A Perspective on the Costs and Benefits of Routine Screening—Shouldn’t Everyone Have This Done? Clin. Pharmacol. Ther. 2016, 99, 164–166. [Google Scholar] [CrossRef]
- Haga, S.B.; Kantor, A. Horizon Scan of Clinical Laboratories Offering Pharmacogenetic Testing. Health Aff. 2018, 37, 717–723. [Google Scholar] [CrossRef]
- de Leon, J.; Susce, M.T.; Murray-Carmichael, E. The AmpliChip CYP450 Genotyping Test: Integrating a New Clinical Tool. Mol. Diagn. Ther. 2006, 10, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Burmester, J.K.; Sedova, M.; Shapero, M.H.; Mansfield, E. DMET Microarray Technology for Pharmacogenomics-Based Personalized Medicine. Methods Mol. Biol. 2010, 632, 99–124. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Burkley, B.M.; Langaee, T.Y.; Clare-Salzler, M.J.; Klein, T.E.; Altman, R.B. Implementing Personalized Medicine: Development of a Cost-Effective Customized Pharmacogenetics Genotyping Array. Clin. Pharmacol. Ther. 2012, 92, 437–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielinski, S.J.; Olson, J.E.; Pathak, J.; Weinshilboum, R.M.; Wang, L.; Lyke, K.J.; Ryu, E.; Targonski, P.V.; Van Norstrand, M.D.; Hathcock, M.A.; et al. Preemptive Genotyping for Personalized Medicine: Design of the Right Drug, Right Dose, Right Time-Using Genomic Data to Individualize Treatment Protocol. Mayo Clin. Proc. 2014, 89, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, C.; Jansen, L.A.; Dhamija, R. Review of Commercially Available Epilepsy Genetic Panels. J. Genet. Couns. 2016, 25, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Platt, J.; Cox, R.; Enns, G.M. Points to Consider in the Clinical Use of NGS Panels for Mitochondrial Disease: An Analysis of Gene Inclusion and Consent Forms. J. Genet. Couns. 2014, 23, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Bush, W.S.; Crosslin, D.R.; Owusu-Obeng, A.; Wallace, J.; Almoguera, B.; Basford, M.A.; Bielinski, S.J.; Carrell, D.S.; Connolly, J.J.; Crawford, D.; et al. Genetic Variation among 82 Pharmacogenes: The PGRNseq Data from the EMERGE Network. Clin. Pharmacol. Ther. 2016, 100, 160–169. [Google Scholar] [CrossRef]
- Janssens, A.C.J.W.; Deverka, P.A. Useless Until Proven Effective: The Clinical Utility of Preemptive Pharmacogenetic Testing. Clin. Pharmacol. Ther. 2014, 96, 652–654. [Google Scholar] [CrossRef]
- Lazaridis, K.N. Improving Therapeutic Odyssey: Preemptive Pharmacogenomics Utility in Patient Care. Clin. Pharmacol. Ther. 2017, 101, 39–41. [Google Scholar] [CrossRef]
- Dunnenberger, H.M.; Crews, K.R.; Hoffman, J.M.; Caudle, K.E.; Broeckel, U.; Howard, S.C.; Hunkler, R.J.; Klein, T.E.; Evans, W.E.; Relling, M.V. Preemptive Clinical Pharmacogenetics Implementation: Current Programs in Five US Medical Centers. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 89–106. [Google Scholar] [CrossRef] [Green Version]
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Müller, D.J.; Shimoda, K.; Bishop, J.R.; Kharasch, E.D.; Skaar, T.C.; Gaedigk, A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants: 2016 Update. Clin. Pharmacol. Ther. 2017, 102, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, J.K.; Bishop, J.R.; Sangkuhl, K.; Müller, D.J.; Ji, Y.; Leckband, S.G.; Leeder, J.S.; Graham, R.L.; Chiulli, D.L.; LLerena, A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 2015, 98, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Vilches, S.; Tuson, M.; Vieta, E.; Álvarez, E.; Espadaler, J. Effectiveness of a Pharmacogenetic Tool at Improving Treatment Efficacy in Major Depressive Disorder: A Meta-Analysis of Three Clinical Studies. Pharmaceutics 2019, 11, 453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazer, A.; Benmansour, S. Delayed Pharmacological Effects of Antidepressants. Mol. Psychiatry 2002, 7 (Suppl. 1), S23–S28. [Google Scholar] [CrossRef] [PubMed]
- GlaxoSmithKline Inc. ZIAGEN (GlaxoSmithKline Inc): FDA Package Insert. Available online: https://druginserts.com/lib/rx/meds/ziagen-6/ (accessed on 24 August 2020).
- Cho, S.-M.; Lee, K.-Y.; Choi, J.R.; Lee, K.-A. Development and Comparison of Warfarin Dosing Algorithms in Stroke Patients. Yonsei Med. J. 2016, 57, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, L.E.; Rieder, M.; van den Anker, J.; Malkin, B.; Ross, C.; Neely, M.N.; Carleton, B.; Hayden, M.R.; Madadi, P.; Koren, G. More Codeine Fatalities after Tonsillectomy in North American Children. Pediatrics 2012, 129, e1343–e1347. [Google Scholar] [CrossRef] [Green Version]
- Thorn, C.F.; Klein, T.E.; Altman, R.B. Codeine and Morphine Pathway. Pharm. Genom. 2009, 19, 556–558. [Google Scholar] [CrossRef]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Klein, T.E.; Shen, D.D.; Callaghan, J.T.; Kharasch, E.D.; Skaar, T.C. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for Codeine Therapy in the Context of Cytochrome P450 2D6 (CYP2D6) Genotype. Clin. Pharmacol. Ther. 2012, 91, 321–326. [Google Scholar] [CrossRef]
- Lemke, A.A.; Hulick, P.J.; Wake, D.T.; Wang, C.; Sereika, A.W.; Yu, K.D.; Glaser, N.S.; Dunnenberger, H.M. Patient Perspectives Following Pharmacogenomics Results Disclosure in an Integrated Health System. Pharmacogenomics 2018, 19, 321–331. [Google Scholar] [CrossRef]
- Patel, H.N.; Ursan, I.D.; Zueger, P.M.; Cavallari, L.H.; Pickard, A.S. Stakeholder Views on Pharmacogenomic Testing. Pharmacotherapy 2014, 34, 151–165. [Google Scholar] [CrossRef]
- Haga, S.B.; Mills, R.; Moaddeb, J.; Allen Lapointe, N.; Cho, A.; Ginsburg, G.S. Patient Experiences with Pharmacogenetic Testing in a Primary Care Setting. Pharmacogenomics 2016, 17, 1629–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielinski, S.J.; St Sauver, J.L.; Olson, J.E.; Wieland, M.L.; Vitek, C.R.; Bell, E.J.; Mc Gree, M.E.; Jacobson, D.J.; McCormick, J.B.; Takahashi, P.Y.; et al. Are Patients Willing to Incur Out-of-Pocket Costs for Pharmacogenomic Testing? Pharm. J. 2017, 17, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haga, S.B.; O’Daniel, J.M.; Tindall, G.M.; Lipkus, I.R.; Agans, R. Survey of US Public Attitudes toward Pharmacogenetic Testing. Pharm. J. 2012, 12, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Dunnenberger, H.M.; Biszewski, M.; Bell, G.C.; Sereika, A.; May, H.; Johnson, S.G.; Hulick, P.J.; Khandekar, J. Implementation of a Multidisciplinary Pharmacogenomics Clinic in a Community Health System. Am. J. Health Syst. Pharm. 2016, 73, 1956–1966. [Google Scholar] [CrossRef]
- Wake, D.T.; Ilbawi, N.; Dunnenberger, H.M.; Hulick, P.J. Pharmacogenomics. Med. Clin. N. Am. 2019, 103, 977–990. [Google Scholar] [CrossRef]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Rongen, G.a.P.J.M.; van Schaik, R.H.N.; Schalekamp, T.; Touw, D.J.; et al. Pharmacogenetics: From Bench to Byte--An Update of Guidelines. Clin. Pharmacol. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef]
- Caudle, K.E.; Klein, T.E.; Hoffman, J.M.; Muller, D.J.; Whirl-Carrillo, M.; Gong, L.; McDonagh, E.M.; Sangkuhl, K.; Thorn, C.F.; Schwab, M.; et al. Incorporation of Pharmacogenomics into Routine Clinical Practice: The Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. Curr. Drug Metab. 2014, 15, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Rubinstein, W.S.; Maglott, D.R.; Lee, J.M.; Kattman, B.L.; Malheiro, A.J.; Ovetsky, M.; Hem, V.; Gorelenkov, V.; Song, G.; Wallin, C.; et al. The NIH Genetic Testing Registry: A New, Centralized Database of Genetic Tests to Enable Access to Comprehensive Information and Improve Transparency. Nucleic Acids Res. 2013, 41, D925–D935. [Google Scholar] [CrossRef] [Green Version]
- Sangkuhl, K.; Berlin, D.S.; Altman, R.B.; Klein, T.E. PharmGKB: Understanding the Effects of Individual Genetic Variants. Drug Metab. Rev. 2008, 40, 539–551. [Google Scholar] [CrossRef]
- Volpi, S.; Bult, C.J.; Chisholm, R.L.; Deverka, P.A.; Ginsburg, G.S.; Jacob, H.J.; Kasapi, M.; McLeod, H.L.; Roden, D.M.; Williams, M.S.; et al. Research Directions in the Clinical Implementation of Pharmacogenomics: An Overview of US Programs and Projects. Clin. Pharmacol. Ther. 2018, 103, 778–786. [Google Scholar] [CrossRef]
- Caraballo, P.J.; Hodge, L.S.; Bielinski, S.J.; Stewart, A.K.; Farrugia, G.; Schultz, C.G.; Rohrer-Vitek, C.R.; Olson, J.E.; St Sauver, J.L.; Roger, V.L.; et al. Multidisciplinary Model to Implement Pharmacogenomics at the Point of Care. Genet. Med. 2017, 19, 421–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Wouden, C.H.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.C.; Dávila-Fajardo, C.L.; Deneer, V.H.; Dolžan, V.; Ingelman-Sundberg, M.; Jönsson, S.; Karlsson, M.O.; et al. Ubiquitous Pharmacogenomics Consortium. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2017, 101, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Luzum, J.A.; Pakyz, R.E.; Elsey, A.R.; Haidar, C.E.; Peterson, J.F.; Whirl-Carrillo, M.; Handelman, S.K.; Palmer, K.; Pulley, J.M.; Beller, M.; et al. Pharmacogenomics Research Network Translational Pharmacogenetics Program. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clin. Pharmacol. Ther. 2017, 102, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Sangkuhl, K.; Stein, C.M.; Hulot, J.-S.; Mega, J.L.; Roden, D.M.; Klein, T.E.; Sabatine, M.S.; Johnson, J.A.; Shuldiner, A.R. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and Clopidogrel Therapy: 2013 Update. Clin. Pharmacol. Ther. 2013, 94, 317–323. [Google Scholar] [CrossRef]
- Vo, T.T.; Bell, G.C.; Obeng, A.O.; Hicks, J.K.; Dunnenberger, H.M. Pharmacogenomics Implementation: Considerations for Selecting a Reference Laboratory. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017, 37, 1014–1022. [Google Scholar] [CrossRef]
- Bousman, C.; Maruf, A.A.; Müller, D.J. Towards the Integration of Pharmacogenetics in Psychiatry: A Minimum, Evidence-Based Genetic Testing Panel. Curr. Opin. Psychiatry 2019, 32, 7–15. [Google Scholar] [CrossRef]
- Phillips, E.J.; Sukasem, C.; Whirl-Carrillo, M.; Müller, D.J.; Dunnenberger, H.M.; Chantratita, W.; Goldspiel, B.; Chen, Y.-T.; Carleton, B.C.; George, A.L.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 574–581. [Google Scholar] [CrossRef] [Green Version]
- Hosono, N.; Kato, M.; Kiyotani, K.; Mushiroda, T.; Takata, S.; Sato, H.; Amitani, H.; Tsuchiya, Y.; Yamazaki, K.; Tsunoda, T.; et al. CYP2D6 Genotyping for Functional-Gene Dosage Analysis by Allele Copy Number Detection. Clin. Chem. 2009, 55, 1546–1554. [Google Scholar] [CrossRef] [Green Version]
- Caudle, K.E.; Dunnenberger, H.M.; Freimuth, R.R.; Peterson, J.F.; Burlison, J.D.; Whirl-Carrillo, M.; Scott, S.A.; Rehm, H.L.; Williams, M.S.; Klein, T.E.; et al. Standardizing Terms for Clinical Pharmacogenetic Test Results: Consensus Terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 2017, 19, 215–223. [Google Scholar] [CrossRef]
- Hershfield, M.S.; Callaghan, J.T.; Tassaneeyakul, W.; Mushiroda, T.; Thorn, C.F.; Klein, T.E.; Lee, M.T.M. Clinical Pharmacogenetics Implementation Consortium Guidelines for Human Leukocyte Antigen-B Genotype and Allopurinol Dosing. Clin. Pharmacol. Ther. 2013, 93, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Leckband, S.G.; Kelsoe, J.R.; Dunnenberger, H.M.; George, A.L.; Tran, E.; Berger, R.; Müller, D.J.; Whirl-Carrillo, M.; Caudle, K.E.; Pirmohamed, M. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Carbamazepine Dosing. Clin. Pharmacol. Ther. 2013, 94, 324–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinderer, M.; Boeker, M.; Wagner, S.A.; Lablans, M.; Newe, S.; Hülsemann, J.L.; Neumaier, M.; Binder, H.; Renz, H.; Acker, T.; et al. Integrating Clinical Decision Support Systems for Pharmacogenomic Testing into Clinical Routine—A Scoping Review of Designs of User-System Interactions in Recent System Development. BMC Med. Inform. Decis. Mak. 2017, 17, 81. [Google Scholar] [CrossRef] [Green Version]
- Blagec, K.; Koopmann, R.; Crommentuijn-van Rhenen, M.; Holsappel, I.; van der Wouden, C.H.; Konta, L.; Xu, H.; Steinberger, D.; Just, E.; Swen, J.J.; et al. Implementing Pharmacogenomics Decision Support across Seven European Countries: The Ubiquitous Pharmacogenomics (U-PGx) Project. J. Am. Med. Inform. Assoc 2018, 25, 893–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krebs, K.; Milani, L. Translating Pharmacogenomics into Clinical Decisions: Do Not Let the Perfect Be the Enemy of the Good. Hum. Genom. 2019, 13, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plumpton, C.O.; Roberts, D.; Pirmohamed, M.; Hughes, D.A. A Systematic Review of Economic Evaluations of Pharmacogenetic Testing for Prevention of Adverse Drug Reactions. Pharmacoeconomics 2016, 34, 771–793. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Cacabelos, N.; Carril, J.C. The Role of Pharmacogenomics in Adverse Drug Reactions. Expert Rev. Clin. Pharmacol. 2019, 12, 407–442. [Google Scholar] [CrossRef]
- Shepherd, G.; Mohorn, P.; Yacoub, K.; May, D.W. Adverse Drug Reaction Deaths Reported in United States Vital Statistics, 1999–2006. Ann. Pharmacother. 2012, 46, 169–175. [Google Scholar] [CrossRef]
- Ekhart, C.; van Hunsel, F.; Scholl, J.; de Vries, S.; van Puijenbroek, E. Sex Differences in Reported Adverse Drug Reactions of Selective Serotonin Reuptake Inhibitors. Drug Saf. 2018, 41, 677–683. [Google Scholar] [CrossRef]
- Routledge, P.A.; O’Mahony, M.S.; Woodhouse, K.W. Adverse Drug Reactions in Elderly Patients. Br. J. Clin. Pharmacol. 2004, 57, 121–126. [Google Scholar] [CrossRef] [Green Version]
- Wilke, R.A.; Lin, D.W.; Roden, D.M.; Watkins, P.B.; Flockhart, D.; Zineh, I.; Giacomini, K.M.; Krauss, R.M. Identifying Genetic Risk Factors for Serious Adverse Drug Reactions: Current Progress and Challenges. Nat. Rev. Drug Discov. 2007, 6, 904–916. [Google Scholar] [CrossRef]
- Chyka, P.A. How many deaths occur annually from adverse drug reactions in the United States? Am. J. Med. 2000, 109, 122–130. [Google Scholar] [CrossRef]
- Shrestha, S.; Shakya, R.; Shrestha, S.; Shakya, S. Adverse Drug Reaction due to Cancer Chemotherapy and its Financial Burden in Different Hospitals of Nepal. Int. J. Pharmacovigil. 2017, 2, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Watson, S.; Caster, O.; Rochon, P.A.; den Ruijter, H. Reported Adverse Drug Reactions in Women and Men: Aggregated Evidence from Globally Collected Individual Case Reports during Half a Century. EClinicalMedicine 2019, 17, 100188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, K. Sex-Related Differences in Drug Disposition in Man. Clin. Pharmacokinet. 1984, 9, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Mallik, S.; Palaian, S.; Ojha, P.; Mishra, P. Pattern of Adverse Drug Reactions Due to Cancer Chemotherapy in a Tertiary Care Teaching Hospital in Nepal. Pak. J. Pharm. Sci. 2007, 20, 214–218. [Google Scholar] [PubMed]
- Jose, J.; Rao, P.G.M. Pattern of Adverse Drug Reactions Notified by Spontaneous Reporting in an Indian Tertiary Care Teaching Hospital. Pharmacol. Res. 2006, 54, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-W.; Liu, W.; Zhou, S.-F. Pharmacogenomics-Guided Approaches to Avoiding Adverse Drug Reactions. Clin. Pharmacol. Biopharm. 2012, 1. [Google Scholar] [CrossRef]
- Cargnin, S.; Jommi, C.; Canonico, P.L.; Genazzani, A.A.; Terrazzino, S. Diagnostic Accuracy of HLA-B*57:01 Screening for the Prediction of Abacavir Hypersensitivity and Clinical Utility of the Test.: A Meta-Analytic Review. Pharmacogenomics 2014, 15, 963–976. [Google Scholar]
- Mallal, S.; Phillips, E.; Carosi, G.; Molina, J.-M.; Workman, C.; Tomažič, J.; Jägel-Guedes, E.; Rugina, S.; Kozyrev, O.; Cid, J.F.; et al. HLA-B*5701 Screening for Hypersensitivity to Abacavir. N. Engl. J. Med. 2008, 358, 568–579. [Google Scholar] [CrossRef] [Green Version]
- Schackman, B.R.; Scott, C.A.; Walensky, R.P.; Losina, E.; Freedberg, K.A.; Sax, P.E. The Cost-Effectiveness of HLA-B*5701 Genetic Screening to Guide Initial Antiretroviral Therapy for HIV. AIDS 2008, 22, 2025–2033. [Google Scholar] [CrossRef] [Green Version]
- Hippman, C.; Nislow, C. Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges. J. Pers. Med. 2019, 9, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, G.P.; Fonseca, E.; Scott, R.; Fagerness, J. Pharmacogenomic and Pharmacogenetic-Guided Therapy as a Tool in Precision Medicine: Current State and Factors Impacting Acceptance by Stakeholders. Genet. Res. 2015, 97, e13. [Google Scholar] [CrossRef] [PubMed]
- Botkin, J.R.; Teutsch, S.M.; Kaye, C.I.; Hayes, M.; Haddow, J.E.; Bradley, L.A.; Szegda, K.; Dotson, W.D. Outcomes of Interest in Evidence-Based Evaluations of Genetic Tests. Genet. Med. 2010, 12, 228–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomaki, G.E.; Bradley, L.A.; Douglas, M.P.; Kolor, K.; Dotson, W.D. Can UGT1A1 Genotyping Reduce Morbidity and Mortality in Patients with Metastatic Colorectal Cancer Treated with Irinotecan? An Evidence-Based Review. Genet. Med. 2009, 11, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. The EGAPP initiative: Lessons learned. Genet. Med. 2014, 16, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Relling, M.V.; Evans, W.E. Pharmacogenomics in the Clinic. Nature 2015, 526, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Sorich, M.J.; Coory, M. Interpreting the Clinical Utility of a Pharmacogenomic Marker Based on Observational Association Studies. Pharm. J. 2014, 14, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Dias, M.M.; Sorich, M.J.; Rowland, A.; Wiese, M.D.; McKinnon, R.A. The Routine Clinical Use of Pharmacogenetic Tests: What It Will Require? Pharm. Res. 2017, 34, 1544–1550. [Google Scholar] [CrossRef]
- Teutsch, S.M.; Bradley, L.A.; Palomaki, G.E.; Haddow, J.E.; Piper, M.; Calonge, N.; Dotson, W.D.; Douglas, M.P.; Berg, A.O.; EGAPP Working Group. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: Methods of the EGAPP Working Group. Genet. Med. 2009, 11, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.A.; Caudle, K.E.; Gong, L.; Whirl-Carrillo, M.; Stein, C.M.; Scott, S.A.; Lee, M.T.M.; Gage, B.F.; Kimmel, S.E.; Perera, M.A. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin dosing: 2017 Update. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Borenstein, M. (Ed.) Introduction to Meta-Analysis; John Wiley & Sons: Chichester, UK, 2009. [Google Scholar]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. PLoS Med. 2009, 6, e1000100. [Google Scholar] [CrossRef] [PubMed]
- Sorich, M.J.; Polasek, T.M.; Wiese, M.D. Challenges and Limitations in the Interpretation of Systematic Reviews: Making Sense of Clopidogrel and CYP2C19 Pharmacogenetics. Clin. Pharmacol. Ther. 2013, 94, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Genomic Epidemiology of Complex Disease: The Need for an Electronic Evidence-Based Approach to Research Synthesis. Available online: https://www.pubfacts.com/detail/16014778/Genomic-epidemiology-of-complex-disease-the-need-for-an-electronic-evidence-based-approach-to-resear (accessed on 25 August 2020).
- Amstutz, U.; Carleton, B. Pharmacogenetic Testing: Time for Clinical Practice Guidelines. Clin. Pharmacol. Ther. 2011. [CrossRef] [PubMed]
- Sorich, M.J.; Wiese, M.D.; Pekarsky, B. Cost-Effectiveness of Genotyping to Guide Treatment. Pharmacogenomics 2014, 15, 727–729. [Google Scholar] [CrossRef]
- Tansey, K.E.; Guipponi, M.; Hu, X.; Domenici, E.; Lewis, G.; Malafosse, A.; Wendland, J.R.; Lewis, C.M.; McGuffin, P.; Uher, R. Contribution of Common Genetic Variants to Antidepressant Response. Biol. Psychiatry 2013, 73, 679–682. [Google Scholar] [CrossRef]
- Niitsu, T.; Fabbri, C.; Bentini, F.; Serretti, A. Pharmacogenetics in Major Depression: A Comprehensive Meta-Analysis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 45, 183–194. [Google Scholar] [CrossRef]
- Peterson, K.; Dieperink, E.; Anderson, J.; Boundy, E.; Ferguson, L.; Helfand, M. Rapid Evidence Review of the Comparative Effectiveness, Harms, and Cost-Effectiveness of Pharmacogenomics-Guided Antidepressant Treatment versus Usual Care for Major Depressive Disorder. Psychopharmacology 2017, 234, 1649–1661. [Google Scholar] [CrossRef]
- Rosenblat, J.D.; Lee, Y.; McIntyre, R.S. Does Pharmacogenomic Testing Improve Clinical Outcomes for Major Depressive Disorder? A Systematic Review of Clinical Trials and Cost-Effectiveness Studies. J. Clin. Psychiatry 2017, 78, 720–729. [Google Scholar] [CrossRef]
- Bousman, C.A.; Hopwood, M. Commercial Pharmacogenetic-Based Decision-Support Tools in Psychiatry. Lancet Psychiatry 2016, 3, 585–590. [Google Scholar] [CrossRef]
- Gillis, N.K.; Innocenti, F. Evidence Required to Demonstrate Clinical Utility of Pharmacogenetic Testing: The Debate Continues. Clin. Pharmacol. Ther. 2014, 96, 655–657. [Google Scholar] [CrossRef]
- Stanek, E.J.; Sanders, C.L.; Taber, K.A.J.; Khalid, M.; Patel, A.; Verbrugge, R.R.; Agatep, B.C.; Aubert, R.E.; Epstein, R.S.; Frueh, F.W. Adoption of Pharmacogenomic Testing by US Physicians: Results of a Nationwide Survey. Clin. Pharmacol. Ther. 2012, 91, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Conti, R.; Veenstra, D.L.; Armstrong, K.; Lesko, L.J.; Grosse, S.D. Personalized Medicine and Genomics: Challenges and Opportunities in Assessing Effectiveness, Cost-Effectiveness, and Future Research Priorities. Med. Decis. Mak. 2010, 30, 328–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freedman, K.J.; Bastian, A.R.; Chaiken, I.; Kim, M.J. Solid-State Nanopore Detection of Protein Complexes: Applications in Healthcare and Protein Kinetics. Small 2013, 9, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Faruki, H.; Lai-Goldman, M. Application of a Pharmacogenetic Test Adoption Model to Six Oncology Biomarkers. Per. Med. 2010, 7, 441–450. [Google Scholar] [CrossRef]
- Dias, M.M.; Ward, H.M.; Sorich, M.J.; McKinnon, R.A. Exploration of the Perceptions, Barriers and Drivers of Pharmacogenomics Practice among Hospital Pharmacists in Adelaide, South Australia. Pharm. J. 2014, 14, 235–240. [Google Scholar] [CrossRef]
- Klein, M.E.; Parvez, M.M.; Shin, J.-G. Clinical Implementation of Pharmacogenomics for Personalized Precision Medicine: Barriers and Solutions. J. Pharm. Sci. 2017, 106, 2368–2379. [Google Scholar] [CrossRef] [Green Version]
- Becquemont, L.; Alfirevic, A.; Amstutz, U.; Brauch, H.; Jacqz-Aigrain, E.; Laurent-Puig, P.; Molina, M.A.; Niemi, M.; Schwab, M.; Somogyi, A.A.; et al. Practical Recommendations for Pharmacogenomics-Based Prescription: 2010 ESF-UB Conference on Pharmacogenetics and Pharmacogenomics. Pharmacogenomics 2011, 12, 113–124. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Moyer, A.M.; Caraballo, P.J. The Challenges of Implementing Pharmacogenomic Testing in the Clinic. Expert Rev. Pharm. Outcomes Res. 2017, 17, 567–577. [Google Scholar] [CrossRef]
| Resource | Description | Reference |
|---|---|---|
| Coursera | Online personalized medicine course that provides short educational courses in genetics and mechanisms determining the variability of response to drugs; development of ethical issues and objections related to implementation and introduction of wide-scale genome-sequencing into clinical practice. | https://www.coursera.org/learn/personalizedmed |
| CPIC | An international consortium that specializes in publishing genotype-based drug guidelines to help clinicians understand the usability of the available genetic test results in optimizing drug therapy. | https://www.cpicpgx.org [50,51] |
| eMERGE | Funded by the NIH. This network brings together researchers with a wide range of expertise in genomics, statistics, ethics, informatics, and clinical medicine from leading medical research institutions across the country to research in genomics including the discovery, clinical implementation, and public resources. | https://www.emerge-network.org |
| GTR | Free of charge resource that provides generalized datastore of the exhaustive information about genetic tests which is provided and supported by vendors; main auditory is clinicians and researchers. | https://www.ncbi.nlm.nih.gov/gtr [52] |
| IGNITE | Was developed to enhance the use of genomic medicine by supporting the incorporation of genomic information into clinical practice and exploring methods for effective implementation, diffusion, and sustainability in various clinical settings. | https://www.gmkb.org |
| My Drug Genome | A portal to study how genetics affects drug response and how results of genetic testing can be implemented into healthcare. | https://www.mydruggenome.org |
| PharmGKB | Online knowledge base responsible for the aggregation, curation, integration, and distribution of data on the influence of genetic variation on the drug response in humans. | https://www.pharmgkb.org [53] |
| Companies | Main Activity | Reference |
|---|---|---|
| Ayass BioScience | A disease monitoring system for implementation in modern molecular medicine and daily clinical practice. Emphasizes genetic, epigenetic, proteomic, and metabolomic profiling, for data collection and interpretation using bioinformatics and biostatistics. | https://ayassbioscience.com |
| Biocerna | This company elaborated a PGx360™ test which is a panel of 22 genes and 62 associated variants to provide an opportunity for clinicians in the selecting of a proper drug. “Biocerna” also provides specific testing of translocations used to monitor a patient’s response to chemotherapy strategy. | http://www.biocerna.com |
| Coriell Life Science (Gene Dose) | Provides data analysis of and reports on clinical laboratory pharmacogenomic assays. Coriell Life Sciences PGx elucidates results of PGx assays in association with drug-related risks to improve patient health to provide a complete, safe, and personalized drug therapy strategy. | https://www.coriell.com |
| Diatech Pharmacogenetics | Develops pharmacogenetic tests for precision cancer medicine and produces two groups of cancer therapy products: (1) pyrosequencing technology in pharmacogenetics of anti-EGFR therapy, and (2) pyrosequencing technology in pharmacogenetics of chemo- and radiotherapy. | https://www.diatechpharmacogenetics.com |
| Dynamic DNA Laboratories | This company provides a wide variety of gene testing services, including pharmacogenomic testing, drug discovery, DNA expression, and also some customized testing and DNA testing services. The main PGx product is the predictive Comprehensive PGx Test for over 150 different drugs. | https://dynamicdnalabs.com |
| Eurofins Genomics | Leader in food, environmental, pharmaceutical, and cosmetic testing. Specializes in pharmacogenetics and PGx research, offers a comprehensive package of services for the drug development process. | https://www.eurofins.com |
| Exceltox Laboratories | A CAP and CLIA accredited laboratory that offers advanced clinical, PGx, and toxicological analysis. | http://exceltox.com |
| GeneDx | Leader in genomics, including research on rare genetic diseases. PharmacoDx targets sequence variants in genes that contribute to drug metabolism. PharmacoDx is a comprehensive pharmacogenetic panel with over 100 genetic variants. | https://www.genedx.com |
| Genentech | A biotechnology company that pioneers research in and develops medications for patients with severe and life-threatening diseases. | https://www.gene.com |
| Genewiz | A leading international company that offers a wide range of services in genomic technologies, including NGS, classic Sanger sequencing, elaboration of synthetic genes, and bioinformatic data analysis support. | https://www.genewiz.com/en-GB |
| HudsonAlpha Institute for Biotechnology | Initiation and support of scientific research programs related to human health and well-being; supports the introduction of genomic medicine into clinical practice and promotes entrepreneurship in life sciences. Developing of work programs for specialists in genomics. Conducting extensive elaboration in the pharmacogenetic testing platform in collaboration with Kailos Genetics. | https://hudsonalpha.org |
| Integrated DNA Technologies | Develops and manufactures nucleic acid products. Areas of activity include scientific and commercial research, agriculture, medical diagnostics, pharmaceutical development, and synthetic biology. | https://www.idtdna.com |
| Myriad Genetics | Pioneering researches and innovations in molecular diagnostic testing aimed to improve personalized medicine. | https://myriad.com |
| Pathway Genomics | A company private that offers customized tests for the screening of diet, weight loss, and metabolic response to numerous commonly prescribed medications. The information can be securely delivered to patients and physicians through any mobile device in a comprehensive form using the in-house developed application. The company produces several PGx products, including “OmePsychiatricMeds” (genetic test for mental health medication efficacy) and “OmePainMeds” (genetic test for pain management medication efficacy). | https://www.pathway.com/about |
| Phenomics Health | This is a bioinformatic platform for precision medicine that transforms large health data sets of patients and even populations into certain products and services to support decision-making about pharmacological treatment. | https://www.phenomicshealth.com/ |
| Quantigen | Development of gene expression and gene variation tests, methods validation, and other services related to clinical assays and PGx researches. | http://www.quantigen.com |
| RxGenomix | Developed a new highly secured and compatible data-concentrator RxGenomix that provides genomic data exchange through distinct healthcare IT services including laboratory management systems, electronic clinical records, and pharmacy operation systems. | https://www.rxgenomix.com |
| Sema4 | This is an interdisciplinary partnership of scientists, clinicians, engineers, and genetic consultants. It is a unique consortium with a solid basis of more than 160 years of clinical experience, world-class academic research, and groundbreaking information technology. | https://sema4.com |
| Sorenson Genomics | This company is mainly focused on DNA testing in forensic and research projects. The main areas of interest are DNA genotyping, DNA sequencing and analysis of fragments in population genetics, and human genotyping. Offers: LEAD Local Entry Access DNA Database—a reliable, proven software that allows fast and secure storage, search, and analysis of millions of DNA profiles. | https://sorensongenomics.com |
| Transgenomic | Development of molecular technologies for personalized medicine specifically in cardiology and oncology. This company is an international leader in pharmacogenetic testing and offers a variety of products designed to detect specific mutations in a certain gene that can indicate a specific heart disease and risk of heart failure. | http://www.transgenomic.com |
| Translational Software | A leader in the use of genetic data purposed to support decision-making in precision medicine. Developed software that enables laboratories and clinicians to incorporate PGx data into treatment strategies to improve a personalized approach such as “PGxAPI”—a knowledge base purposed to include PGx data into healthcare and laboratory systems. PGxPortal offers an HL7 interface for receiving data and checking the quality of test results. This portal enables clinicians to deliver better patient care by providing clinically relevant pharmacogenomic information. | https://www.translationalsoftware.com |
| Xact Laboratories | A molecular diagnostics laboratory with a sophisticated research approach that provides a wide range of custom-centered distinctive tests for clinicians and healthcare providers. | https://xactlaboratories.com |
| 23andMe | A biotechnology company that provides customers with information on their disease susceptibility. Pharmacogenetic studies include analysis of CYP2C19, DPYD, SLCO1B1. | https://www.23andme.com |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malsagova, K.A.; Butkova, T.V.; Kopylov, A.T.; Izotov, A.A.; Potoldykova, N.V.; Enikeev, D.V.; Grigoryan, V.; Tarasov, A.; Stepanov, A.A.; Kaysheva, A.L. Pharmacogenetic Testing: A Tool for Personalized Drug Therapy Optimization. Pharmaceutics 2020, 12, 1240. https://doi.org/10.3390/pharmaceutics12121240
Malsagova KA, Butkova TV, Kopylov AT, Izotov AA, Potoldykova NV, Enikeev DV, Grigoryan V, Tarasov A, Stepanov AA, Kaysheva AL. Pharmacogenetic Testing: A Tool for Personalized Drug Therapy Optimization. Pharmaceutics. 2020; 12(12):1240. https://doi.org/10.3390/pharmaceutics12121240
Chicago/Turabian StyleMalsagova, Kristina A., Tatyana V. Butkova, Arthur T. Kopylov, Alexander A. Izotov, Natalia V. Potoldykova, Dmitry V. Enikeev, Vagarshak Grigoryan, Alexander Tarasov, Alexander A. Stepanov, and Anna L. Kaysheva. 2020. "Pharmacogenetic Testing: A Tool for Personalized Drug Therapy Optimization" Pharmaceutics 12, no. 12: 1240. https://doi.org/10.3390/pharmaceutics12121240
APA StyleMalsagova, K. A., Butkova, T. V., Kopylov, A. T., Izotov, A. A., Potoldykova, N. V., Enikeev, D. V., Grigoryan, V., Tarasov, A., Stepanov, A. A., & Kaysheva, A. L. (2020). Pharmacogenetic Testing: A Tool for Personalized Drug Therapy Optimization. Pharmaceutics, 12(12), 1240. https://doi.org/10.3390/pharmaceutics12121240






