Current Advances in the Development of Diagnostic Tests Based on Aptamers in Parasitology: A Systematic Review
Abstract
1. Introduction
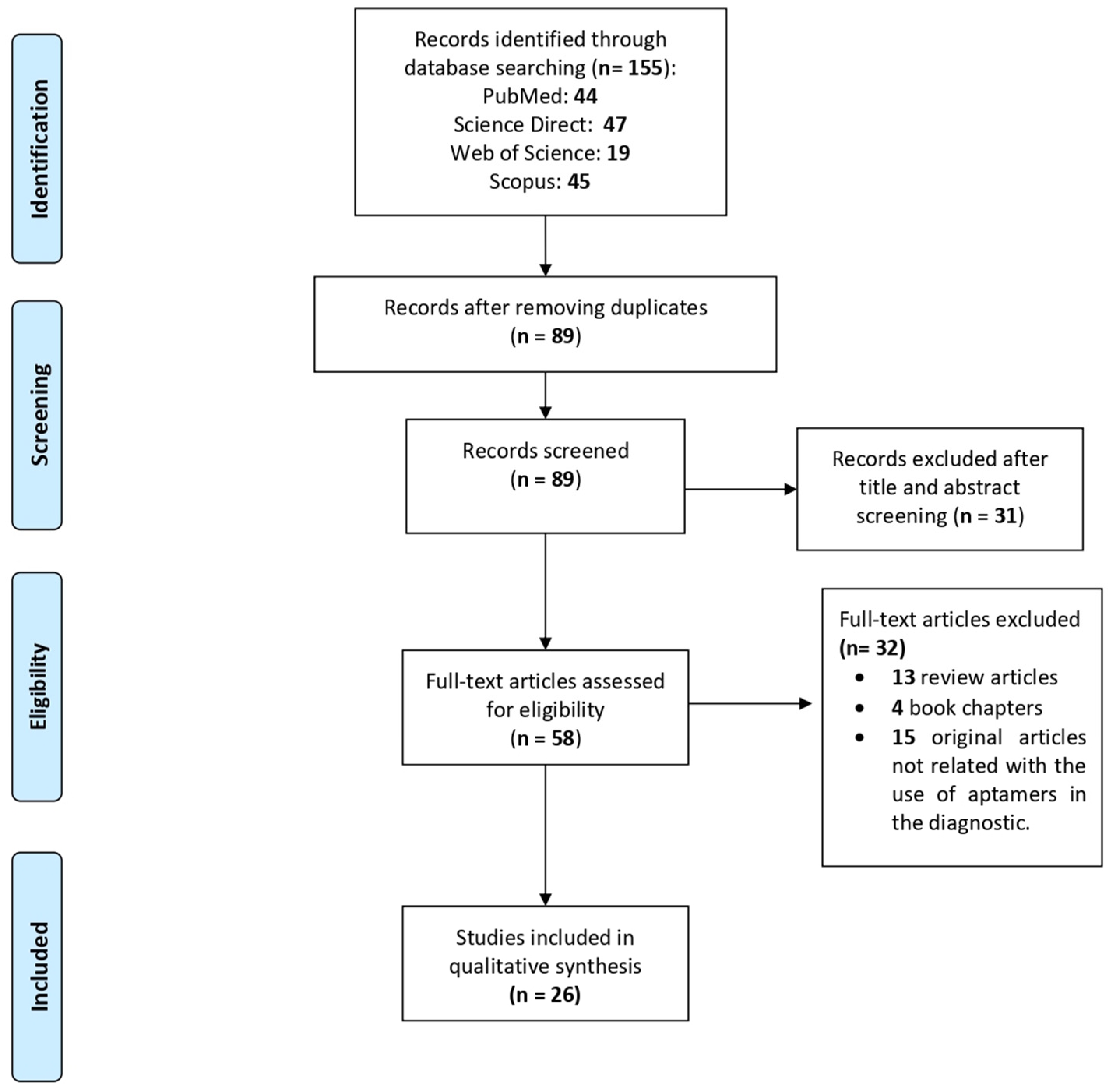
2. Materials and Methods
3. Results
3.1. Article Selection
3.2. Plasmodium spp.
3.2.1. Aptasensors Using the pL1 Aptamer to Detect the Parasite Lactate Dehydrogenase
3.2.2. Aptasensors Using the 2008a Aptamer to Detect the Parasite Lactate Dehydrogenase
3.2.3. Aptasensors Using the P38 Aptamer to Detect the Parasite Lactate Dehydrogenase
3.2.4. Aptasensors Using the NG3 Aptamer to Detect the Parasite Glutamate Dehydrogenase
3.3. Leishmania spp.
3.3.1. Aptamers against Parasite Histone Proteins
3.3.2. Aptasensor Using Aptamers against the Parasite Hydrophilic Surface Protein
3.4. Trypanosoma cruzi
3.5. Cryptosporidium spp.
3.6. Toxoplasma gondii
3.7. Trichomonas vaginalis
4. Discussion
- For years, all antibody-based diagnostic methods have focused on the identification of immunogenic proteins. In contrast, a good target for an aptamer does not need to stimulate an immune response. Some of the main criteria for a good target for an aptamer include aqueous solubility, stability, purity, and cost. Therefore, clever approaches are necessary for the identification of new targets with potential for aptamer-based diagnosis.
- The SELEX protocol for the design of aptamers was protected by a patent until 2016 and the window of research opportunity has not been open for very long.
- Although aptamers have numerous advantages over antibodies, the development of commercial products using aptamers takes decades from the identification of a specific aptamer in the laboratory to the development of an aptasensor for parasite identification in patients.
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 6-MCH | 6-mercapto-1-hexanol |
| AP65 | adhesion protein 65 |
| APTEC | aptamer-tethered enzyme capture |
| ARMD | age-related macular degeneration |
| AuNPs | gold nanoparticles |
| AUR | amplex ultra red |
| BSA | bovine serum albumin |
| CL | cutaneous leishmaniasis |
| CTAB | cationic surfactant hexadecyltrimethylammonium bromide |
| dCTP | deoxycytidine triphosphate |
| DEAE | diethylaminoethyl |
| DSM | DEAE-cellulose surface modified |
| dUTP | deoxyuridine triphosphate |
| ELAA | enzyme linked aptamer assay |
| ELASA | enzyme linked apta-sorbent assay |
| ELISA | enzyme linked immunosorbent assay |
| EMSA | electrophoretic mobility shift assay |
| FAM | fluorescein amidites |
| FDA | food and drug administration |
| FET | field effect transistor |
| FLASH | fluorescent assay sensor handheld |
| FRET | fluorescence resonance energy transfer |
| GNPS-SPCE | gold nanoparticles-modified screen-printed carbon electrode |
| GO | graphene oxide |
| HBsAg | hepatitis B surface antigen |
| IDμE | inter-digitated gold microelectrode |
| ITC | isothermal titration calorimetry |
| LDH | lactate dehydrogenase |
| LOD | limits of detection |
| ML | mucous leishmaniasis |
| MMP | magnetic microparticles |
| NTB | nitrotetrazolium blue chloride |
| PAH | poly Allylamine Hydrochloride |
| PCR | polymerase chain reaction |
| PDDA | poly (diallyldimethylammonium chloride) |
| PEG | polyethylene glycol |
| PfGDH | P. falciparum glutamate dehydrogenase |
| PfLDH | P. falciparum lactate dehydrogenase |
| PRISMA | preferred reporting items for systematic reviews and meta-analyses |
| PVDF | polyvinylidene difluoride |
| PvLDH | P. vivax lactate dehydrogenase |
| QD | quantum dots |
| RDT | rapid diagnostic test |
| rHSP | recombinant hydrophilic surface protein |
| rLiH3 | recombinant L. infantum H3 |
| rROP18 | recombinant rhoptry protein 18 |
| SELEX | systematic evolution of ligands by exponential enrichment |
| SPR | surface plasmon resonance |
| ss | single stranded |
| TEM | transmission electron microscopy |
| TESA | trypomastigote excreted-secreted antigens |
| TnI | troponin I |
| VL | visceral leishmaniasis |
References
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S.; Mori, Y.; Nakamura, Y. Evolution of an Inhibitory RNA Aptamer against T7 RNA Polymerase. FEBS Open Bio 2012, 2, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Raducanu, V.-S.; Rashid, F.; Zaher, M.S.; Li, Y.; Merzaban, J.S.; Hamdan, S.M. A Direct Fluorescent Signal Transducer Embedded in a DNA Aptamer Paves the Way for Versatile Metal-Ion Detection. Sens. Actuators B Chem. 2020, 304, 127376. [Google Scholar] [CrossRef]
- Shiratori, I.; Akitomi, J.; Boltz, D.A.; Horii, K.; Furuichi, M.; Waga, I. Selection of DNA Aptamers That Bind to Influenza A Viruses with High Affinity and Broad Subtype Specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. [Google Scholar] [CrossRef]
- Liu, F.; Ding, A.; Zheng, J.; Chen, J.; Wang, B. A Label-Free Aptasensor for Ochratoxin a Detection Based on the Structure Switch of Aptamer. Sensors 2018, 18, 1769. [Google Scholar] [CrossRef]
- Aptekar, S.; Arora, M.; Lawrence, C.L.; Lea, R.W.; Ashton, K.; Dawson, T.; Alder, J.E.; Shaw, L. Selective Targeting to Glioma with Nucleic Acid Aptamers. PLoS ONE 2015, 10, e0134957. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef]
- Xiang, D.; Zheng, C.; Zhou, S.-F.; Qiao, S.; Tran, P.H.-L.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a Targeted Anti-VEGF Aptamer for Ocular Vascular Disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, N.; Zeng, Z.; Feng, Y.; Tung, C.-H.; Chang, C.-C.; Zu, Y. Using an RNA Aptamer Probe for Flow Cytometry Detection of CD30-Expressing Lymphoma Cells. Lab. Investig. 2009, 89, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Zourob, M. Aptamer-Based Label-Free Electrochemical Biosensor Array for the Detection of Total and Glycated Hemoglobin in Human Whole Blood. Sci. Rep. 2017, 7, 1016. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef]
- Srivastava, M.; Nirala, N.R.; Srivastava, S.K.; Prakash, R. A Comparative Study of Aptasensor Vs Immunosensor for Label-Free PSA Cancer Detection on GQDs-AuNRs Modified Screen-Printed Electrodes. Sci. Rep. 2018, 8, 1923. [Google Scholar] [CrossRef]
- Li, P.; Zhou, L.; Wei, J.; Yu, Y.; Yang, M.; Wei, S.; Qin, Q. Development and Characterization of Aptamer-Based Enzyme-Linked Apta-Sorbent Assay for the Detection of Singapore Grouper Iridovirus Infection. J. Appl. Microbiol. 2016, 121, 634–643. [Google Scholar] [CrossRef]
- Negahdary, M.; Behjati-Ardakani, M.; Sattarahmady, N.; Heli, H. An Aptamer-Based Biosensor for Troponin I Detection in Diagnosis of Myocardial Infarction. J. Biomed. Phys. Eng. 2018, 8, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhao, J.; Huang, Y.; Zhao, S.; Liu, Y.-M. Aptamer-Based Microchip Electrophoresis Assays for Amplification Detection of Carcinoembryonic Antigen. Clin. Chim. Acta 2015, 450, 304–309. [Google Scholar] [CrossRef]
- Kim, D.T.H.; Bao, D.T.; Park, H.; Ngoc, N.M.; Yeo, S.-J. Development of a Novel Peptide Aptamer-Based Immunoassay to Detect Zika Virus in Serum and Urine. Theranostics 2018, 8, 3629–3642. [Google Scholar] [CrossRef]
- Xi, Z.; Gong, Q.; Wang, C.; Zheng, B. Highly Sensitive Chemiluminescent Aptasensor for Detecting HBV Infection Based on Rapid Magnetic Separation and Double-Functionalized Gold Nanoparticles. Sci. Rep. 2018, 8, 9444. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- World Health Organization. Available online: https://www.who.int/malaria/publications/world-malaria-report-2015/report/en/ (accessed on 15 August 2020).
- Cunningham, J.; Jones, S.; Gatton, M.L.; Barnwell, J.W.; Cheng, Q.; Chiodini, P.L.; Glenn, J.; Incardona, S.; Kosack, C.; Luchavez, J.; et al. A review of the WHO malaria rapid diagnostic test product testing programme (2008–2018): Performance, procurement and policy. Malar. J. 2019, 18, 387. [Google Scholar] [CrossRef]
- Lee, S.; Song, K.-M.; Jeon, W.; Jo, H.; Shim, Y.-B.; Ban, C. A Highly Sensitive Aptasensor towards Plasmodium Lactate Dehydrogenase for the Diagnosis of Malaria. Biosens. Bioelectron. 2012, 35, 291–296. [Google Scholar] [CrossRef]
- Jeon, W.; Lee, S.; Dh, M.; Ban, C. A Colorimetric Aptasensor for the Diagnosis of Malaria Based on Cationic Polymers and Gold Nanoparticles. Anal. Biochem. 2013, 439, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Manjunatha, D.H.; Jeon, W.; Ban, C. Cationic Surfactant-Based Colorimetric Detection of Plasmodium Lactate Dehydrogenase, a Biomarker for Malaria, Using the Specific DNA Aptamer. PLoS ONE 2014, 9, e100847. [Google Scholar] [CrossRef] [PubMed]
- Geldert, A.; Lim, C.T. Paper-Based MoS2 Nanosheet-Mediated FRET Aptasensor for Rapid Malaria Diagnosis. Sci. Rep. 2017, 7, 17510. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.-W.; Kwok, J.; Law, A.W.L.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer. Proc. Natl. Acad. Sci. USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Dirkzwager, R.M.; Kinghorn, A.B.; Richards, J.S.; Tanner, J.A. APTEC: Aptamer-Tethered Enzyme Capture as a Novel Rapid Diagnostic Test for Malaria. Chem. Commun. 2015, 51, 4697–4700. [Google Scholar] [CrossRef]
- Cheung, Y.-W.; Dirkzwager, R.M.; Wong, W.-C.; Cardoso, J.; D’Arc Neves Costa, J.; Tanner, J.A. Aptamer-Mediated Plasmodium-Specific Diagnosis of Malaria. Biochimie 2018, 145, 131–136. [Google Scholar] [CrossRef]
- Fraser, L.A.; Kinghorn, A.B.; Dirkzwager, R.M.; Liang, S.; Cheung, Y.-W.; Lim, B.; Shiu, S.C.-C.; Tang, M.S.L.; Andrew, D.; Manitta, J.; et al. A Portable Microfluidic Aptamer-Tethered Enzyme Capture (APTEC) Biosensor for Malaria Diagnosis. Biosens. Bioelectron. 2018, 100, 591–596. [Google Scholar] [CrossRef]
- Godonoga, M.; Lin, T.-Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.L.; Cheung, Y.-W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.; Kuzuya, A.; et al. A DNA Aptamer Recognising a Malaria Protein Biomarker Can Function as Part of a DNA Origami Assembly. Sci. Rep. 2016, 6, 21266. [Google Scholar] [CrossRef]
- Figueroa-Miranda, G.; Feng, L.; Shiu, S.C.-C.; Dirkzwager, R.M.; Cheung, Y.-W.; Tanner, J.A.; Schöning, M.J.; Offenhäusser, A.; Mayer, D. Aptamer-Based Electrochemical Biosensor for Highly Sensitive and Selective Malaria Detection with Adjustable Dynamic Response Range and Reusability. Sens. Actuators B Chem. 2018, 255, 235–243. [Google Scholar] [CrossRef]
- Figueroa-Miranda, G.; Wu, C.; Zhang, Y.; Nörbel, L.; Lo, Y.; Tanner, J.A.; Elling, L.; Offenhäusser, A.; Mayer, D. Polyethylene Glycol-Mediated Blocking and Monolayer Morphology of an Electrochemical Aptasensor for Malaria Biomarker Detection in Human Serum. Bioelectrochemistry 2020, 136, 107589. [Google Scholar] [CrossRef]
- Kim, C.; Searson, P.C. Detection of Plasmodium Lactate Dehydrogenase Antigen in Buffer Using Aptamer-Modified Magnetic Microparticles for Capture, Oligonucleotide-Modified Quantum Dots for Detection, and Oligonucleotide-Modified Gold Nanoparticles for Signal Amplification. Bioconjug. Chem. 2017, 28, 2230–2234. [Google Scholar] [CrossRef]
- Jain, P.; Das, S.; Chakma, B.; Goswami, P. Aptamer-Graphene Oxide for Highly Sensitive Dual Electrochemical Detection of Plasmodium Lactate Dehydrogenase. Anal. Biochem. 2016, 514, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Arya, S.K.; Estrela, P.; Goswami, P. Capacitive Malaria Aptasensor Using Plasmodium falciparum Glutamate Dehydrogenase as Target Antigen in Undiluted Human Serum. Biosens. Bioelectron. 2018, 117, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Thungon, P.D.; Estrela, P.; Goswami, P. Development of an Aptamer-Based Field Effect Transistor Biosensor for Quantitative Detection of Plasmodium falciparum Glutamate Dehydrogenase in Serum Samples. Biosens. Bioelectron. 2019, 123, 30–35. [Google Scholar] [CrossRef]
- Singh, N.K.; Jain, P.; Das, S.; Goswami, P. Dye Coupled Aptamer-Captured Enzyme Catalyzed Reaction for Detection of Pan Malaria and P. falciparum Species in Laboratory Settings and Instrument-Free Paper-Based Platform. Anal. Chem. 2019, 91, 4213–4422. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: www.who.int/es/news-room/fact-sheets/detail/leishmaniasis (accessed on 18 August 2020).
- Ramos, E.; Piñeiro, D.; Soto, M.; Abanades, D.R.; Martín, M.E.; Salinas, M.; González, V.M. A DNA Aptamer Population Specifically Detects Leishmania Infantum H2A Antigen. Lab. Investig. 2007, 87, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.; Moreno, M.; Martín, M.E.; Soto, M.; Gonzalez, V.M. In Vitro Selection of Leishmania Infantum H3-Binding SsDNA Aptamers. Oligonucleotides 2010, 20, 207–213. [Google Scholar] [CrossRef]
- Frezza, V.; Pinto-Díez, C.; Fernández, G.; Soto, M.; Martín, M.E.; García-Sacristán, A.; González, V.M. DNA Aptamers Targeting Leishmania Infantum H3 Protein as Potential Diagnostic Tools. Anal. Chim. Acta 2020, 1107, 155–163. [Google Scholar] [CrossRef]
- Bruno, J.G.; Richarte, A.M.; Phillips, T.; Savage, A.A.; Sivils, J.C.; Greis, A.; Mayo, M.W. Development of a Fluorescent Enzyme-Linked DNA Aptamer-Magnetic Bead Sandwich Assay and Portable Fluorometer for Sensitive and Rapid Leishmania Detection in Sandflies. J. Fluoresc. 2014, 24, 267–277. [Google Scholar] [CrossRef]
- Schijman, A.G.; Bisio, M.; Orellana, L.; Sued, M.; Duffy, T.; Jaramillo, A.M.M.; Cura, C.; Auter, F.; Veron, V.; Qvarnstrom, Y.; et al. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl. Trop. Dis. 2011, 5, e931. [Google Scholar] [CrossRef] [PubMed]
- Nagarkatti, R.; Bist, V.; Sun, S.; Fortes de Araujo, F.; Nakhasi, H.L.; Debrabant, A. Development of an Aptamer-Based Concentration Method for the Detection of Trypanosoma Cruzi in Blood. PLoS ONE 2012, 7, e43533. [Google Scholar] [CrossRef] [PubMed]
- Nagarkatti, R.; de Araujo, F.F.; Gupta, C.; Debrabant, A. Aptamer Based, Non-PCR, Non-Serological Detection of Chagas Disease Biomarkers in Trypanosoma Cruzi Infected Mice. PLoS Negl. Trop. Dis. 2014, 8, e2650. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Chaudhary, P. Laboratory diagnosis of cryptosporidiosis. Trop. Parasitol. 2018, 8, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Labib, M.; Muharemagic, D.; Sattar, S.; Dixon, B.R.; Berezovski, M.V. Detection of Cryptosporidium Parvum Oocysts on Fresh Produce Using DNA Aptamers. PLoS ONE 2015, 10, e013745. [Google Scholar] [CrossRef]
- Iqbal, A.; Liu, J.; Dixon, B.; Zargar, B.; Sattar, S.A. Development and Application of DNA-Aptamer-Coupled Magnetic Beads and Aptasensors for the Detection of Cryptosporidium Parvum Oocysts in Drinking and Recreational Water Resources. Can. J. Microbiol. 2019, 65, 851–857. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.D.; Huang, S.Y.; Zhu, X.Q. Diagnosis of toxoplasmosis and typing of Toxoplasma gondii. Parasites Vectors 2015, 8, 292. [Google Scholar] [CrossRef]
- Vargas-Montes, M.; Cardona, N.; Moncada, D.M.; Molina, D.A.; Zhang, Y.; Gómez-Marín, J.E. Enzyme-Linked Aptamer Assay (ELAA) for Detection of Toxoplasma ROP18 Protein in Human Serum. Front. Cell. Infect. Microbiol. 2019, 9, 386. [Google Scholar] [CrossRef]
- Edwards, T.; Burke, P.; Smalley, H.; Hobbs, G. Trichomonas vaginalis: Clinical relevance, pathogenicity and diagnosis. Crit. Rev. Microbiol. 2016, 42, 406–417. [Google Scholar] [CrossRef]
- Espiritu, C.A.L.; Justo, C.A.C.; Rubio, M.J.; Svobodova, M.; Bashammakh, A.S.; Alyoubi, A.O.; Rivera, W.L.; Rollon, A.P.; O’Sullivan, C.K. Aptamer Selection against a Trichomonas vaginalis Adhesion Protein for Diagnostic Applications. ACS Infect. Dis. 2018, 4, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Villa, J.D.; Dufour, A.; Weber, C.; Ramirez-Moreno, E.; Zamorano-Carrillo, A.; Guillen, N.; Lopez-Camarillo, C.; Marchat, L.A. Targeting the polyadenylation factor EhCFIm25 with RNA aptamers controls survival in Entamoeba histolytica. Sci. Rep. 2018, 8, 5720. [Google Scholar] [CrossRef]
- Tanner, J.A.; Cheung, J.W.; Kotaka, M. Nucleic Acid Aptamers against Plasmodium Lactate Dehydrogenase and Histidine-Rich Protein II and Uses Thereof for Malaria Diagnosis. EP2812453A1, 08 February 2013. [Google Scholar]
- Tanner, J.A.; Cheung, J.W. Sandwich and Species-Specific Nucleic Acid Aptamers against Plasmodium Lactate Dehydrogenase for Malaria Diagnosis. WO2019113827A1, 20 June 2019. [Google Scholar]
- Ban, C.; Jeong, W.; Lee, S. DNA Aptamer Specifically Binding to pLDH. JP2012254074A, 27 December 2012. [Google Scholar]
- González, V.M.; Palma, M.E.; Piñeiro Del Río, D.; Ramos Muñoz, E.; Salinas Aracil, M. Specific Aptamers for Histones of Leishmania. ES2356439B1, 22 February 2012. [Google Scholar]

| SELEX | Aptamer | Diagnosis Method | Reference | ||||
|---|---|---|---|---|---|---|---|
| SELEX Type | Nucleic Acid | Target | Name | Modification | Kd(nM)/Method | ||
| Plasmodium | |||||||
| Standard SELEX | DNA | PvLDH | pL1 | ns | 16.8 ± 0.6/Fluorescence assay | pLDH aptasensor | [23] |
| Colorimetric aptasensors | [24,25] | ||||||
| PfLDH | 38.27 ± 1.3/Fluorescence assay | ||||||
| FRET-based paper aptasensor | [26] | ||||||
| Standard SELEX | DNA | PfLDH | 2008s | ns | 56 ± 18/EMSA 42/ITC 59/SPR | Colorimetric assay | [27] |
| APTEC assay | [28] | ||||||
| APTEC assay | [29] | ||||||
| 3D printed microfluidic biosensor | [30] | ||||||
| Aptamer ligated to a polyT linker sequence | 1090 ± 183 to 647 ± 128/EMSA | DNA origami | [31] | ||||
| Aptamer with a 5′-Thiol–(CH2)6 group | 56 ± 18/EMSA 42/ITC 59/SPR | 6-MCH-Electrochemical impedance aptasensor | [32] | ||||
| PEG-Electrochemical aptasensor | [33] | ||||||
| ns | Aptabiosensor | [34] | |||||
| Standard SELEX | DNA | PfLDH | P38 | ns | 0.35 μM/EMSA | Aptamer-graphene oxide aptasensor | [35] |
| Standard SELEX | DNA | PfGDH | NG3 | Aptamers with a 5′-Thiol–(CH2)6 | 79.16 ± 1.58/SPR | Capacitive aptasensor | [36] |
| AptaFET biosensor | [37] | ||||||
| Standard SELEX | DNA | PfGDH | 79.16 ± 1.58/SPR | Dye Coupled Aptamer-Captured Enzyme Catalyzed Reaction | [38] | ||
| PLDH | P38 | ns | 0.35 μM/EMSA | ||||
| Leishmania | |||||||
| Standard SELEX | DNA | H2A histone | SELH2A | ns | 2.065 ± 0.652/ELONA | ELONA, Slot blot, Western blot | [40] |
| Standard SELEX | DNA | H3 histone | SELH3 | ns | 0.94 ± 0.19/ELONA | [41] | |
| Standard SELEX | DNA | H3 histone | AptLiH3#4 | ns | 0.52 ± 0.05/ELONA | [42] | |
| AptLiH3#10 | 0.37 ± 0.05/ELONA | ||||||
| Cell-SELEX | DNA | Live promastigotes | Capture LmWC-25R | ns | ns | FLASH | [43] |
| Standard SELEX | HSP | Reporter LmHSP-7b/11R. | |||||
| Trypanosoma | |||||||
| Cell-SELEX | RNA | Live trypomastigotes | Apt68 | Aptamers with fluorinated deoxynucleotides (2′ F-dUTP and 2′ F-dCTP) | 7.62 ± 1.63/Binding assay | A concentration method using streptavidin paramagnetic beads | [45] |
| Standard SELEX | RNA | TESA | Apt-L44 | ns | ELAA | [46] | |
| Cryptosporidium | |||||||
| Cell-SELEX | DNA | Oocyst | R4-6 | ns | 177.5 ± 6.1 μA/Flow cytometric | Aptamer-based electrochemical biosensor | [48] |
| DNA- aptasensor coupled with magnetic beads | [49] | ||||||
| Toxoplasma | |||||||
| Standard SELEX | DNA | ROP18 | AP001 | ns | 62.7 ± 17.27/ELAA | Direct or sandwich ELAA | [51] |
| AP002 | 97.7 ± 22.20/ELAA | ||||||
| Trichomonas | |||||||
| p-SELEX | DNA | AP65 | AP65_A1 | ns | 56/SPR 1.07/ELAA | ELAA | [53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ospina-Villa, J.D.; Cisneros-Sarabia, A.; Sánchez-Jiménez, M.M.; Marchat, L.A. Current Advances in the Development of Diagnostic Tests Based on Aptamers in Parasitology: A Systematic Review. Pharmaceutics 2020, 12, 1046. https://doi.org/10.3390/pharmaceutics12111046
Ospina-Villa JD, Cisneros-Sarabia A, Sánchez-Jiménez MM, Marchat LA. Current Advances in the Development of Diagnostic Tests Based on Aptamers in Parasitology: A Systematic Review. Pharmaceutics. 2020; 12(11):1046. https://doi.org/10.3390/pharmaceutics12111046
Chicago/Turabian StyleOspina-Villa, Juan David, Alondra Cisneros-Sarabia, Miryan Margot Sánchez-Jiménez, and Laurence A. Marchat. 2020. "Current Advances in the Development of Diagnostic Tests Based on Aptamers in Parasitology: A Systematic Review" Pharmaceutics 12, no. 11: 1046. https://doi.org/10.3390/pharmaceutics12111046
APA StyleOspina-Villa, J. D., Cisneros-Sarabia, A., Sánchez-Jiménez, M. M., & Marchat, L. A. (2020). Current Advances in the Development of Diagnostic Tests Based on Aptamers in Parasitology: A Systematic Review. Pharmaceutics, 12(11), 1046. https://doi.org/10.3390/pharmaceutics12111046





