Fast, Spectroscopy-Based Prediction of In Vitro Dissolution Profile of Extended Release Tablets Using Artificial Neural Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Experimental Design
2.2.2. Tablet Manufacturing on a Single Punch Tablet Press
2.2.3. Raman Spectroscopy
2.2.4. Fourier Transformation Near-Infrared Spectroscopy
2.2.5. In Vitro Dissolution Testing
2.3. Data Analysis
2.3.1. Experimental Design
2.3.2. Multivariate Data Analysis
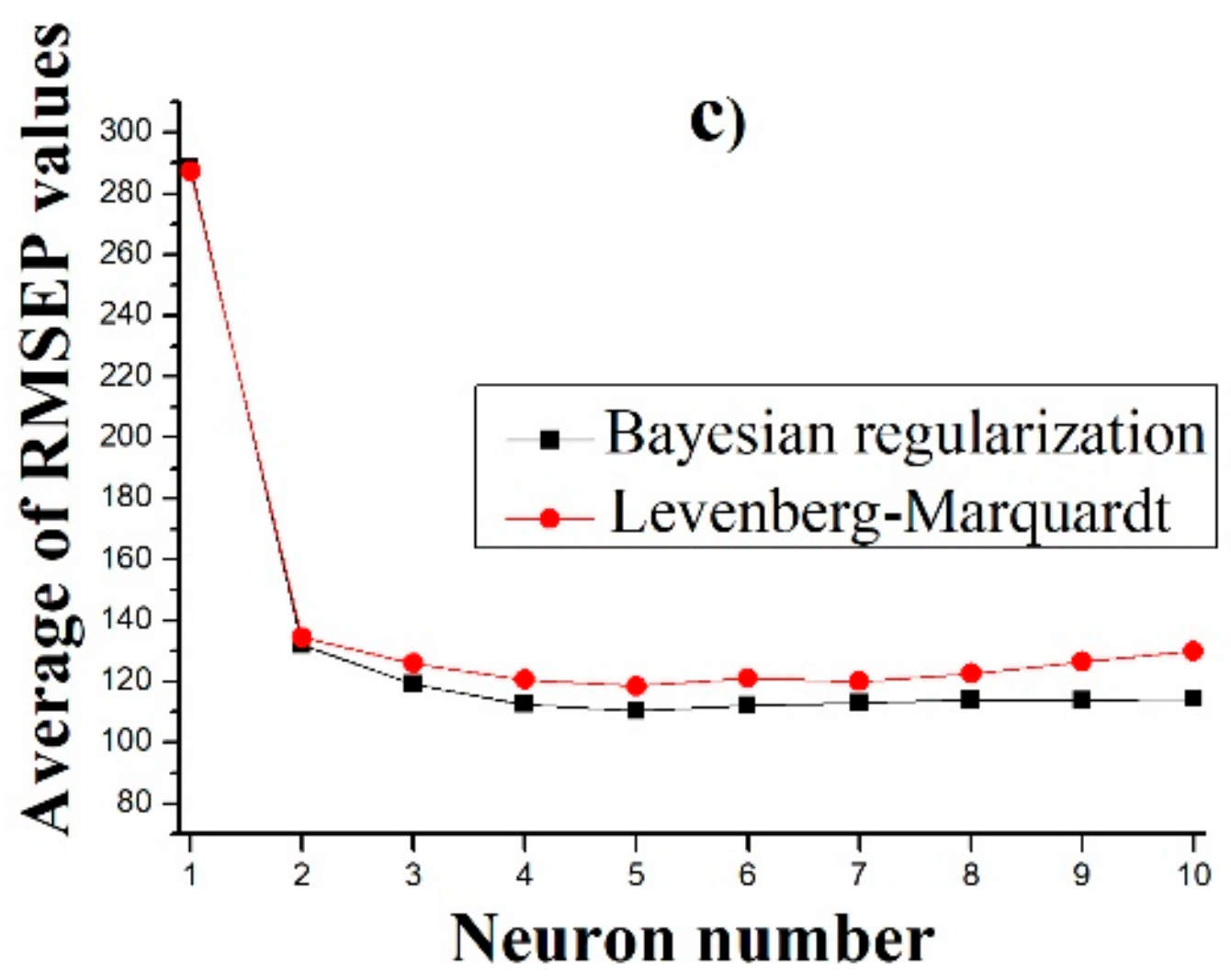
2.3.3. Artificial Neural Network Models
3. Results and Discussion
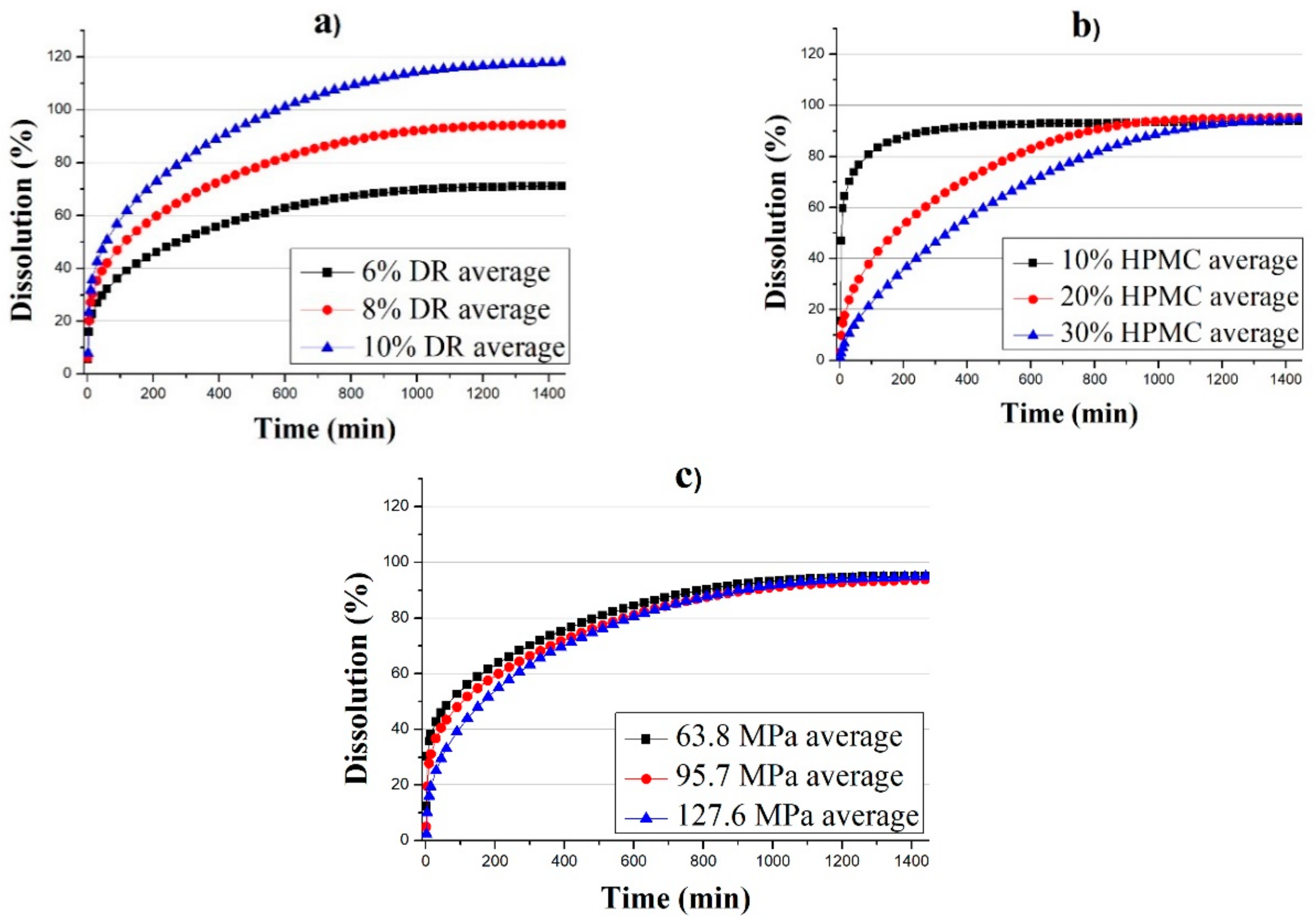
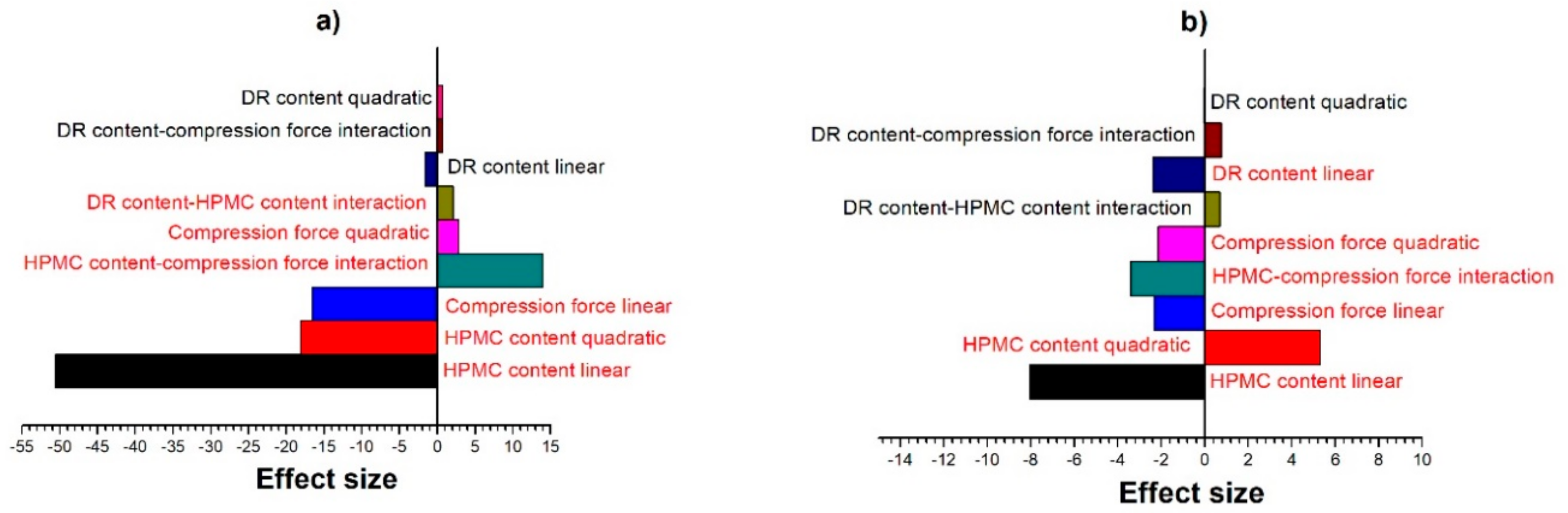
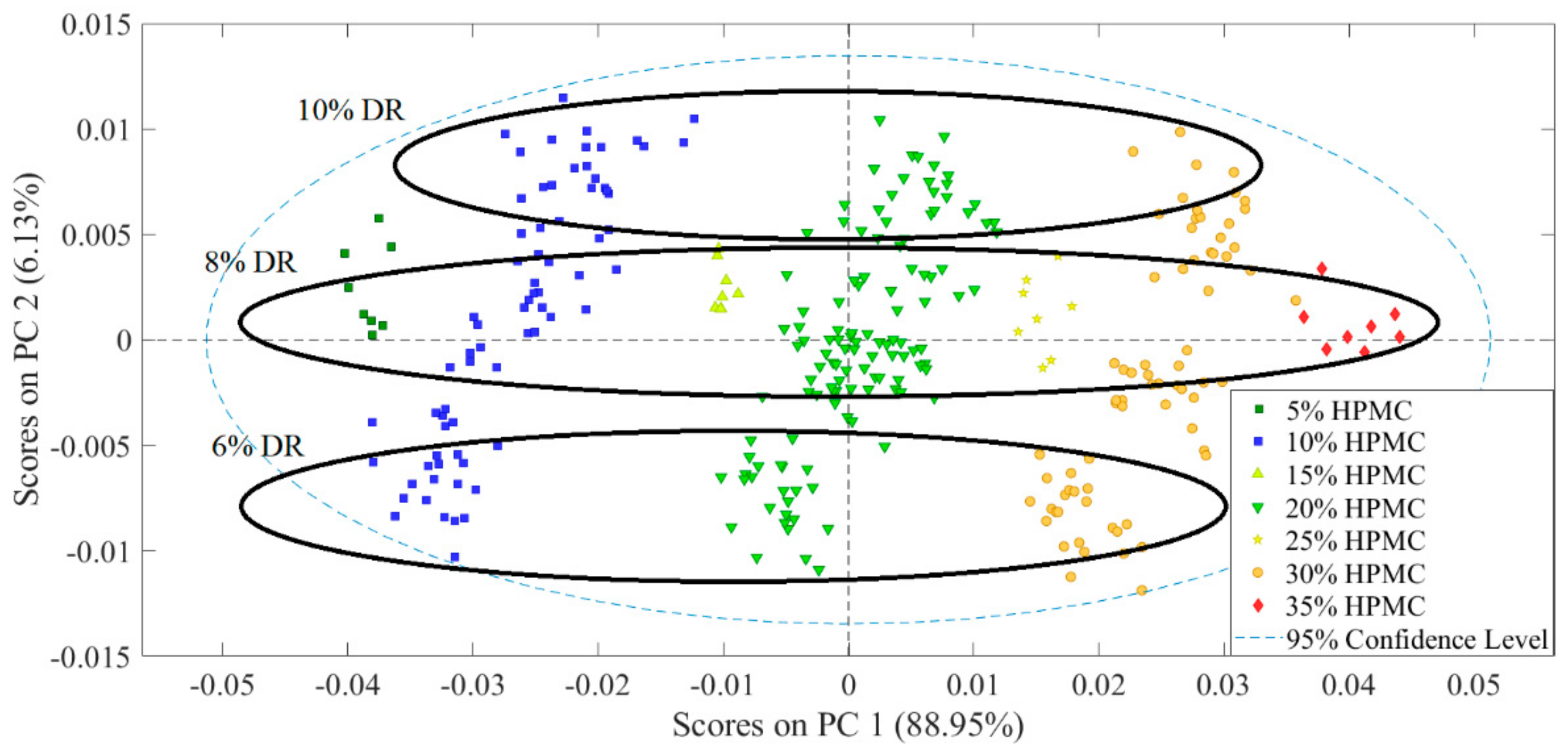
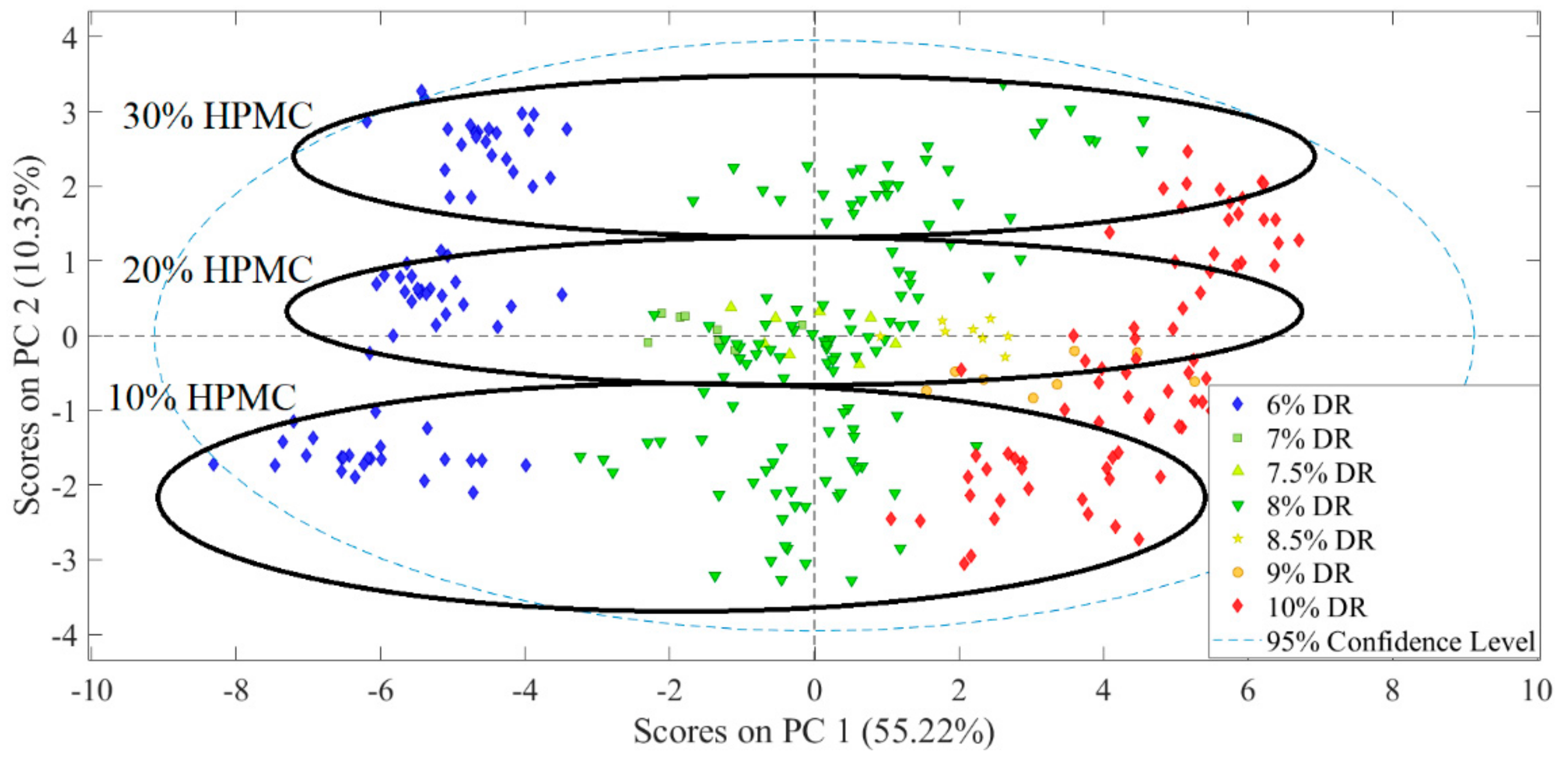
3.1. Evaluation of Experimental Design Results
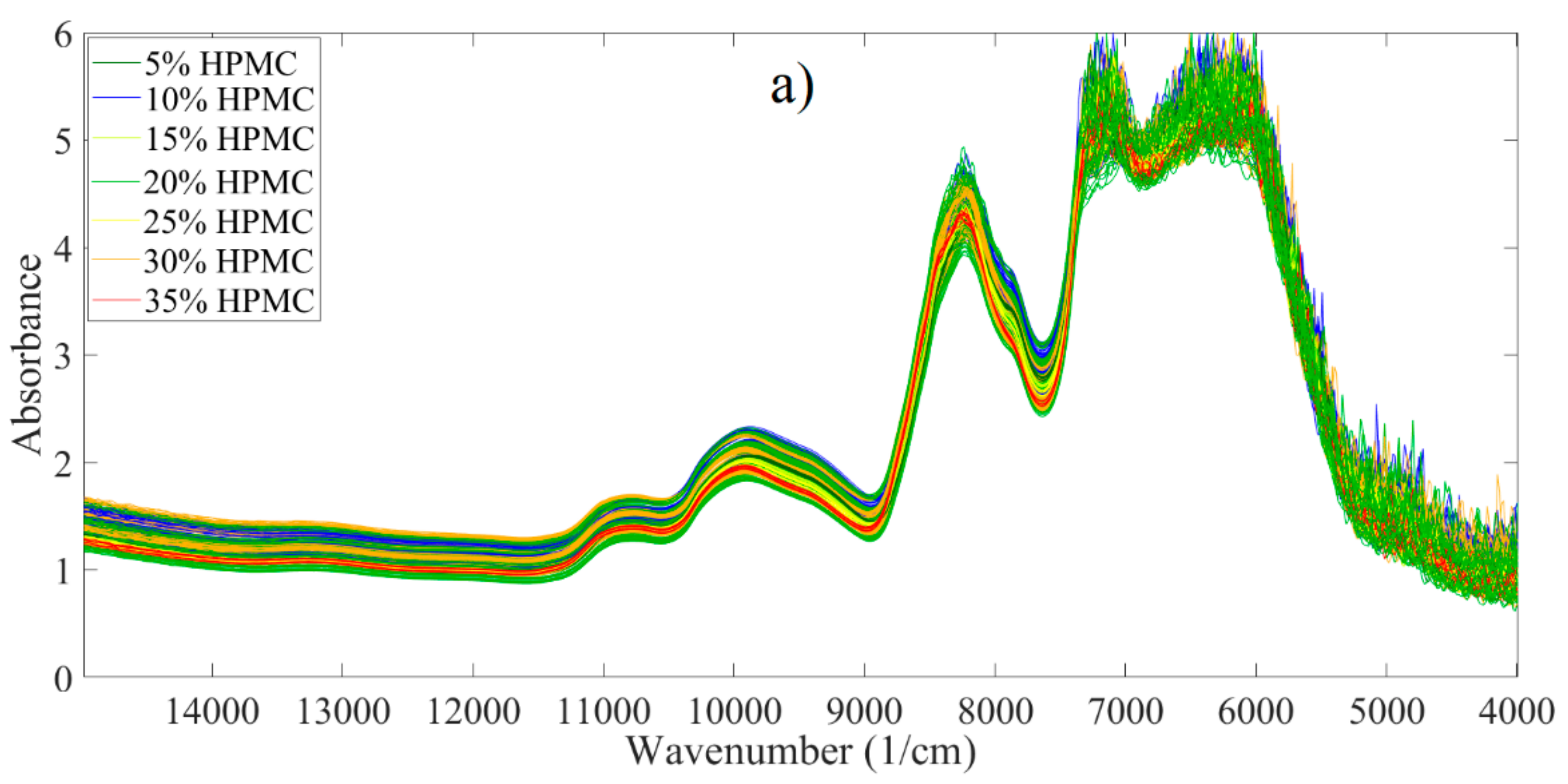
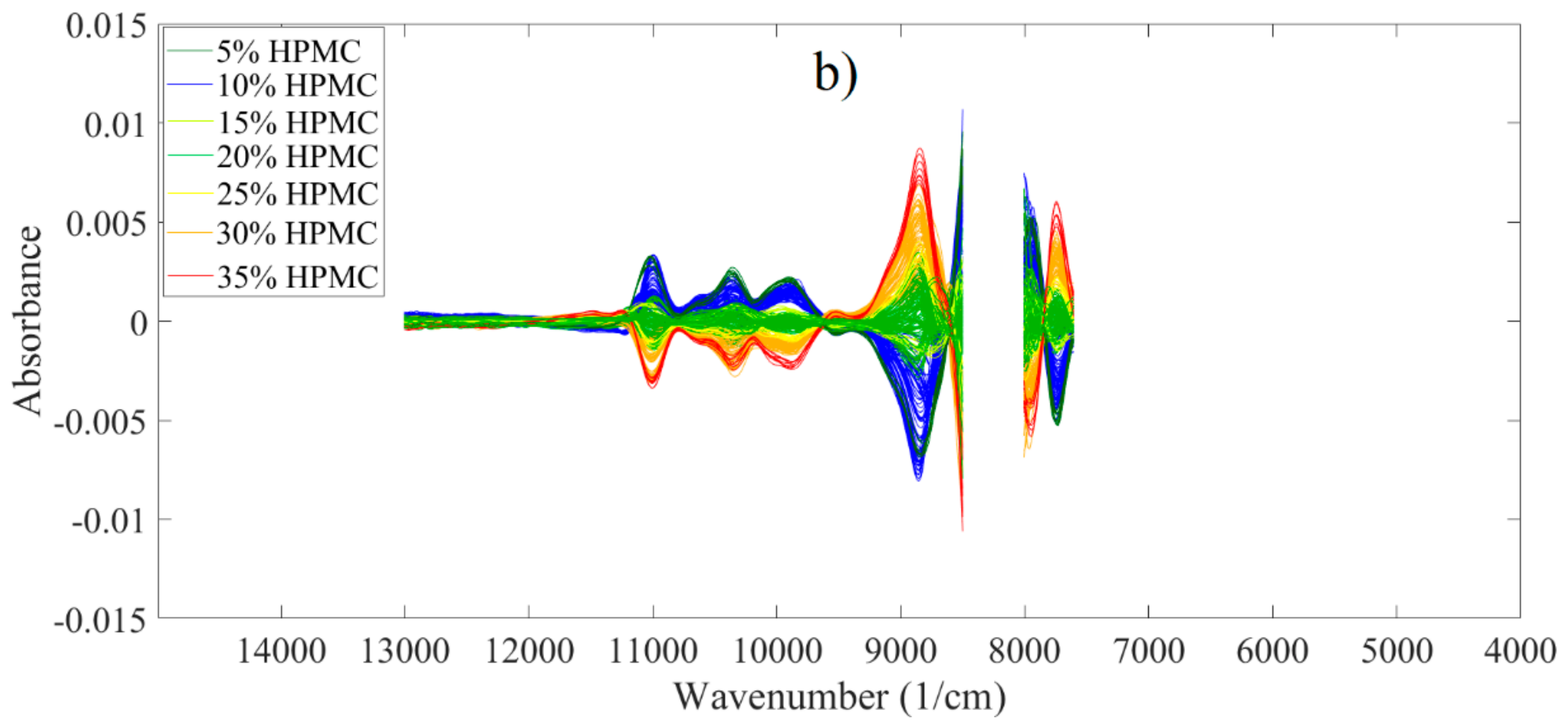
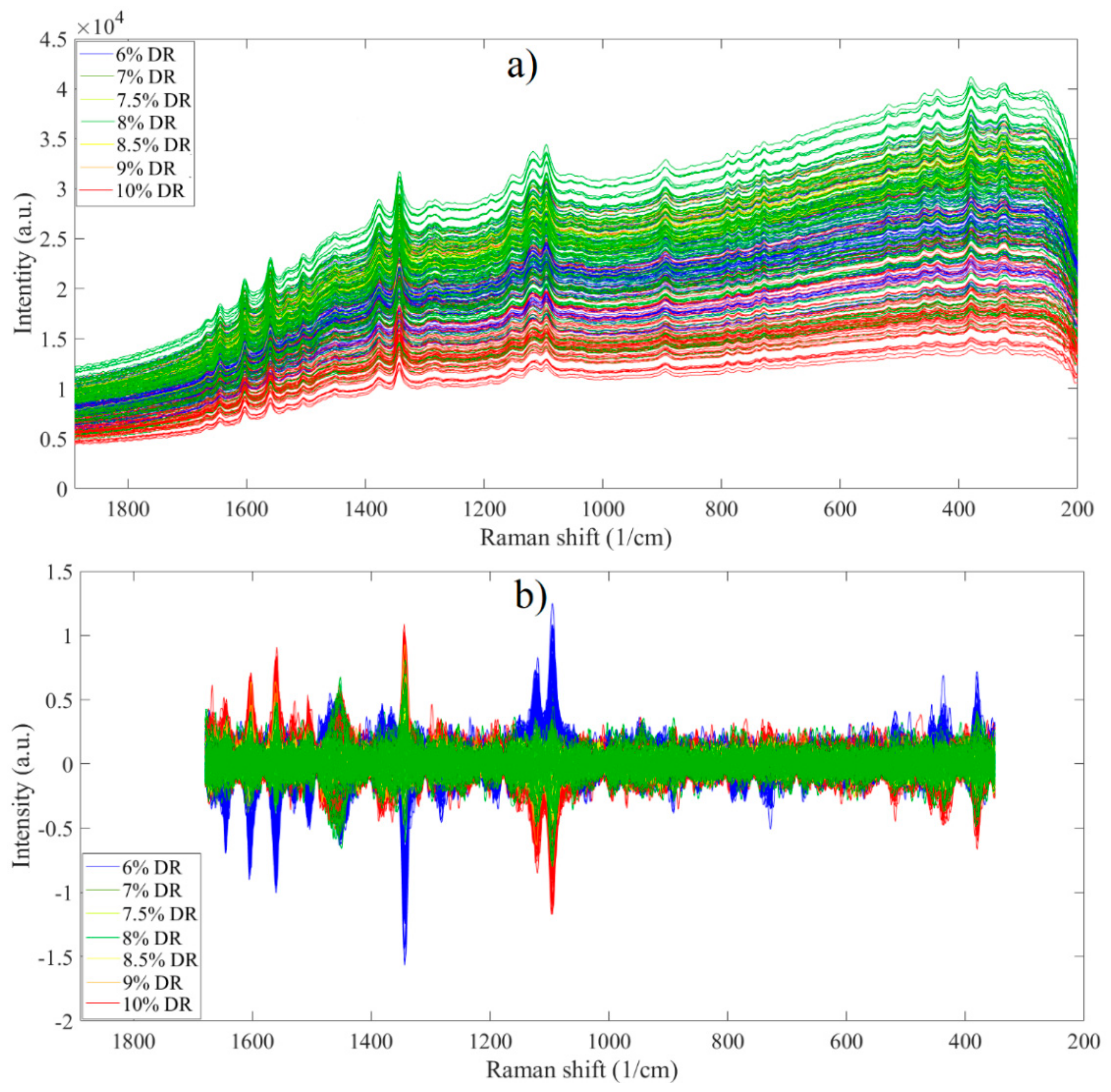
3.2. Analysis of Spectroscopic Data
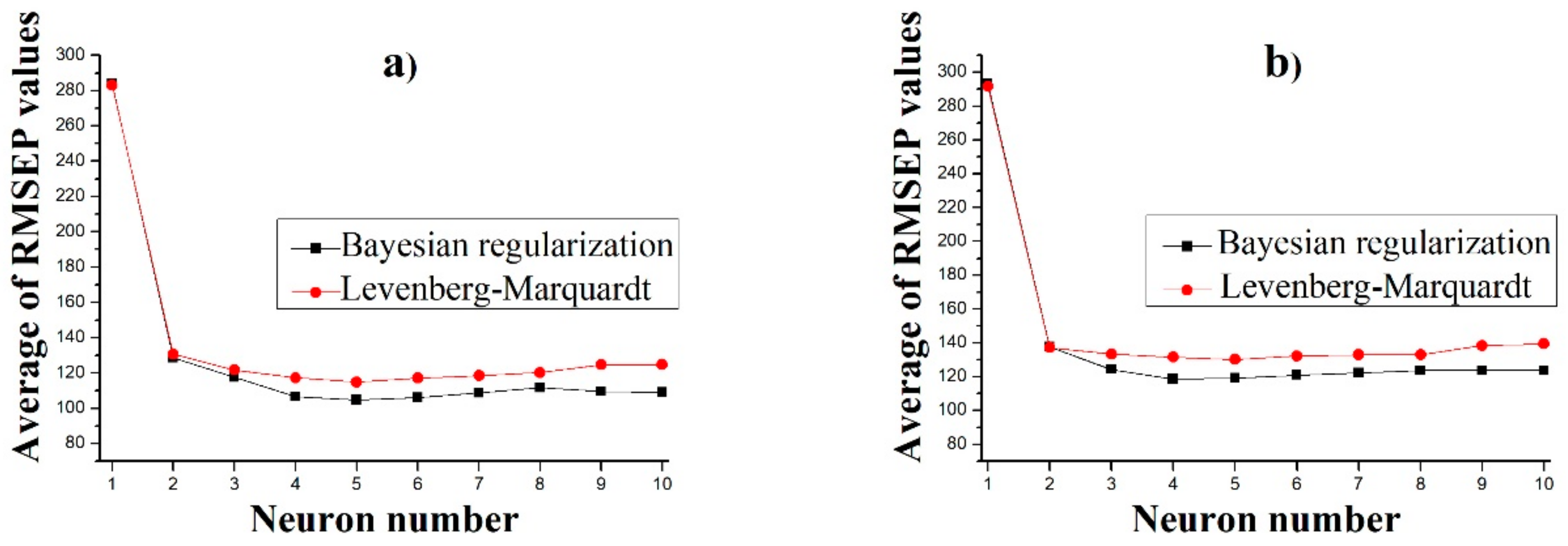
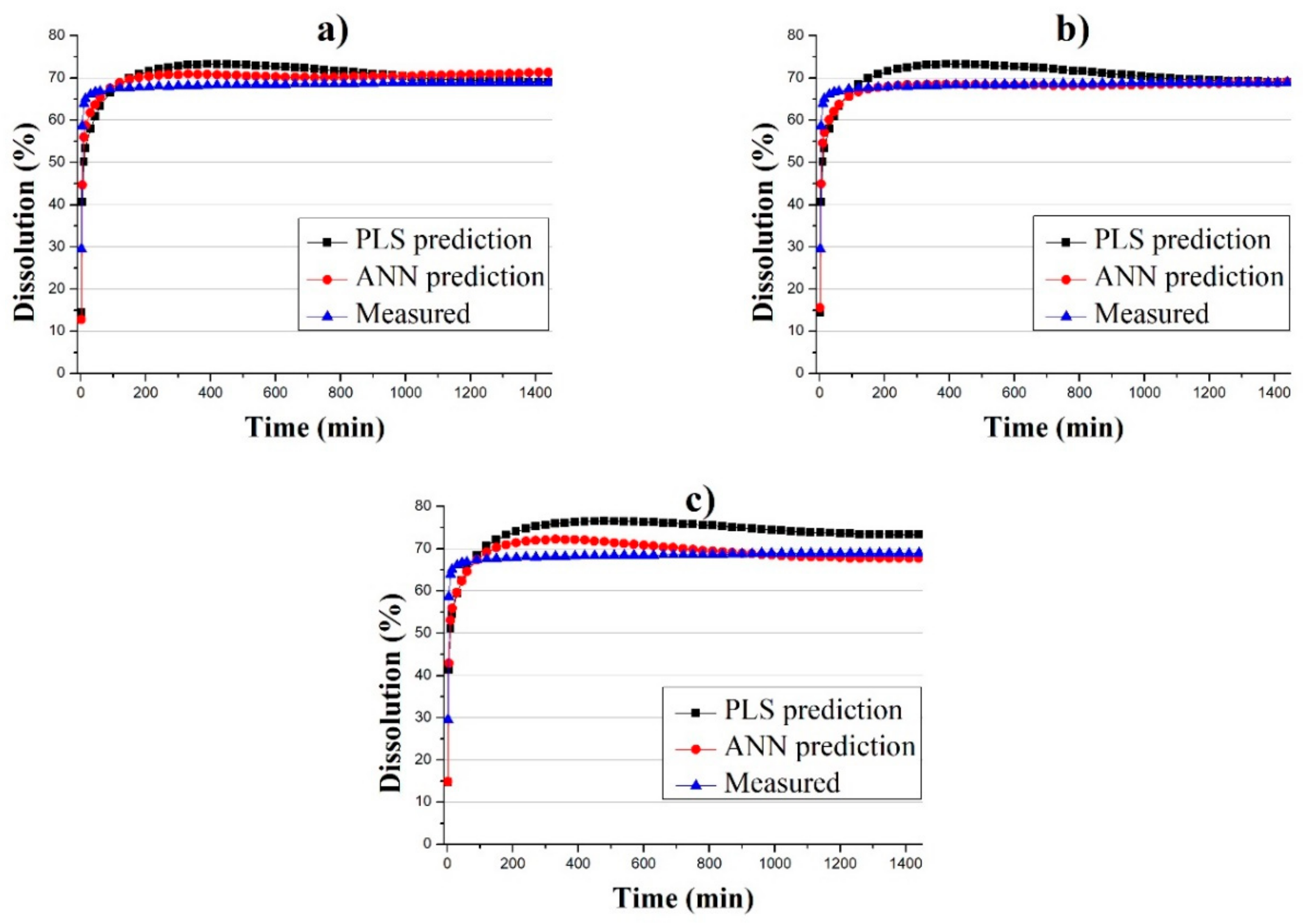
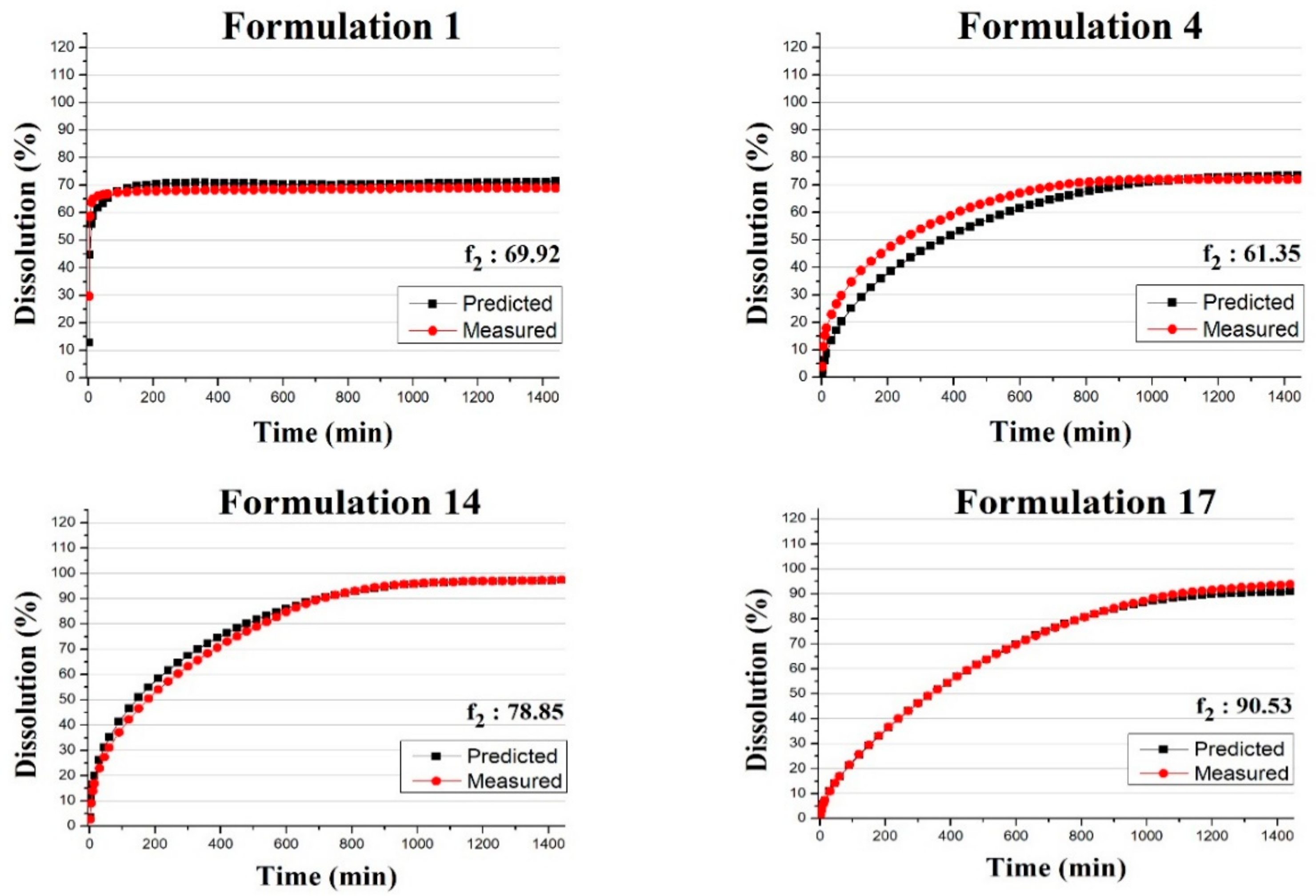
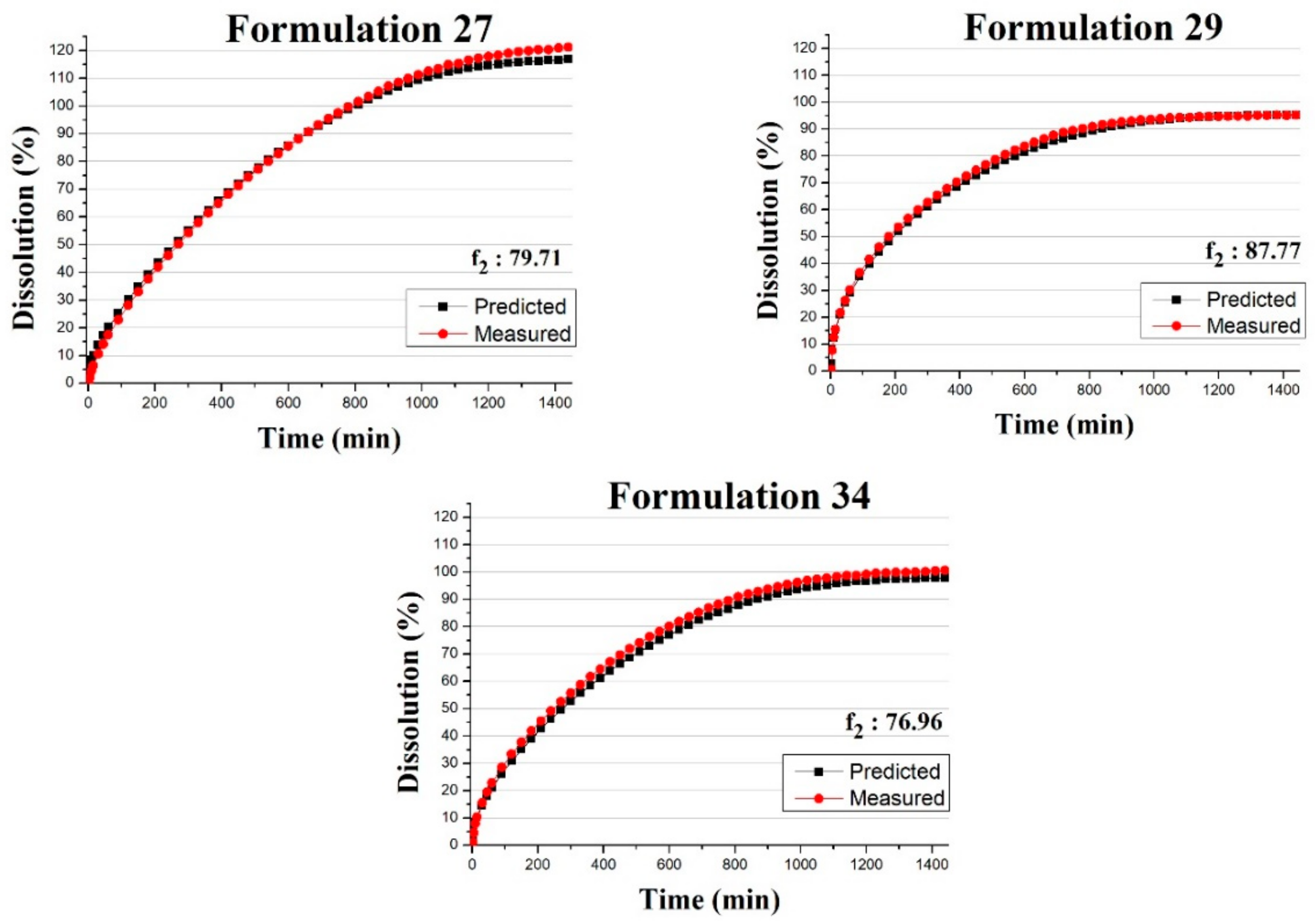
3.3. Predicting the Dissolution Profile by ANN
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hédoux, A. Recent developments in the Raman and infrared investigations of amorphous pharmaceuticals and protein formulations: A review. Adv. Drug Deliv. Rev. 2016, 100, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Porep, J.U.; Kammerer, D.R.; Carle, R. On-line application of near infrared (NIR) spectroscopy in food production. Trends Food Sci. Technol. 2015, 46, 211–230. [Google Scholar] [CrossRef]
- Esmonde-White, K.A.; Cuellar, M.; Uerpmann, C.; Lenain, B.; Lewis, I.R. Raman spectroscopy as a process analytical technology for pharmaceutical manufacturing and bioprocessing. Anal. Bioanal. Chem. 2017, 409, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Lohumi, S.; Lee, S.; Lee, H.; Cho, B.-K. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015, 46, 85–98. [Google Scholar] [CrossRef]
- Paudel, A.; Raijada, D.; Rantanen, J. Raman spectroscopy in pharmaceutical product design. Adv. Drug Deliv. Rev. 2015, 89, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Manley, M.; Baeten, V. Spectroscopic Technique: Near Infrared (NIR) Spectroscopy. In Modern Techniques for Food Authentication; Elsevier: Amsterdam, The Netherlands, 2018; pp. 51–102. [Google Scholar]
- Bumbrah, G.S.; Sharma, R.M. Raman spectroscopy–Basic principle, instrumentation and selected applications for the characterization of drugs of abuse. Egypt. J. Forensic Sci. 2016, 6, 209–215. [Google Scholar] [CrossRef]
- Das, R.S.; Agrawal, Y. Raman spectroscopy: Recent advancements, techniques and applications. Vib. Spectrosc. 2011, 57, 163–176. [Google Scholar] [CrossRef]
- Nagy, B.; Farkas, A.; Borbás, E.; Vass, P.; Nagy, Z.K.; Marosi, G. Raman Spectroscopy for Process Analytical Technologies of Pharmaceutical Secondary Manufacturing. AAPS PharmSciTech 2019, 20, 1. [Google Scholar] [CrossRef]
- Arruabarrena, J.; Coello, J.; Maspoch, S. Raman spectroscopy as a complementary tool to assess the content uniformity of dosage units in break-scored warfarin tablets. Int. J. Pharm. 2014, 465, 299–305. [Google Scholar] [CrossRef]
- Nagy, B.; Farkas, A.; Gyürkés, M.; Komaromy-Hiller, S.; Démuth, B.; Szabó, B.; Nusser, D.; Borbás, E.; Marosi, G.; Nagy, Z.K. In-line Raman spectroscopic monitoring and feedback control of a continuous twin-screw pharmaceutical powder blending and tableting process. Int. J. Pharm. 2017, 530, 21–29. [Google Scholar] [CrossRef]
- Riolo, D.; Piazza, A.; Cottini, C.; Serafini, M.; Lutero, E.; Cuoghi, E.; Gasparini, L.; Botturi, D.; Marino, I.G.; Aliatis, I. Raman spectroscopy as a PAT for pharmaceutical blending: Advantages and disadvantages. J. Pharm. Biomed. Anal. 2018, 149, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Harting, J.; Kleinebudde, P. Optimisation of an in-line Raman spectroscopic method for continuous API quantification during twin-screw wet granulation and its application for process characterisation. Eur. J. Pharm. Biopharm. 2019, 137, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B.; Farkas, A.; Magyar, K.; Démuth, B.; Nagy, Z.K.; Marosi, G. Spectroscopic characterization of tablet properties in a continuous powder blending and tableting process. Eur. J. Pharm. Sci. 2018, 123, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, L.; Yin, W.; Xu, D.; Zang, H. Near-infrared spectroscopy monitoring and control of the fluidized bed granulation and coating processes—A review. Int. J. Pharm. 2017, 530, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Pauli, V.; Elbaz, F.; Kleinebudde, P.; Krumme, M. Methodology for a variable rate control strategy development in continuous manufacturing applied to twin-screw wet-granulation and continuous fluid-bed drying. J. Pharm. Innov. 2018, 13, 247–260. [Google Scholar] [CrossRef]
- Dégardin, K.; Guillemain, A.; Guerreiro, N.V.; Roggo, Y. Near infrared spectroscopy for counterfeit detection using a large database of pharmaceutical tablets. J. Pharm. Biomed. Anal. 2016, 128, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Terra, L.A.; Poppi, R.J. Monitoring the polymorphic transformation on the surface of carbamazepine tablets generated by heating using near-infrared chemical imaging and chemometric methodologies. Chemom. Intell. Lab. Syst. 2014, 130, 91–97. [Google Scholar] [CrossRef]
- Pataki, H.; Csontos, I.; Nagy, Z.K.; Vajna, B.; Molnar, M.; Katona, L.; Marosi, G. Implementation of Raman signal feedback to perform controlled crystallization of carvedilol. Org. Process Res. Dev. 2012, 17, 493–499. [Google Scholar] [CrossRef]
- Pataki, H.; Markovits, I.; Vajna, B.z.; Nagy, Z.K.; Marosi, G.R. In-line monitoring of carvedilol crystallization using Raman spectroscopy. Cryst. Growth Des. 2012, 12, 5621–5628. [Google Scholar] [CrossRef]
- Sacré, P.-Y.; De Bleye, C.; Chavez, P.-F.; Netchacovitch, L.; Hubert, P.; Ziemons, E. Data processing of vibrational chemical imaging for pharmaceutical applications. J. Pharm. Biomed. Anal. 2014, 101, 123–140. [Google Scholar] [CrossRef]
- Vajna, B.; Patyi, G.; Nagy, Z.; Bódis, A.; Farkas, A.; Marosi, G. Comparison of chemometric methods in the analysis of pharmaceuticals with hyperspectral Raman imaging. J. Raman Spectrosc. 2011, 42, 1977–1986. [Google Scholar] [CrossRef]
- Farkas, A.; Vajna, B.; Sóti, P.L.; Nagy, Z.K.; Pataki, H.; Van der Gucht, F.; Marosi, G. Comparison of multivariate linear regression methods in micro-Raman spectrometric quantitative characterization. J. Raman Spectrosc. 2015, 46, 566–576. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Wold, H. Partial least squares. Encycl. Stat. Sci. 2004, 9, 1–10. [Google Scholar]
- Noyes, A.A.; Whitney, W.R. The rate of solution of solid substances in their own solutions. J. Am. Chem. Soc. 1897, 19, 930–934. [Google Scholar] [CrossRef]
- Dokoumetzidis, A.; Macheras, P. A century of dissolution research: From Noyes and Whitney to the biopharmaceutics classification system. Int. J. Pharm. 2006, 321, 1–11. [Google Scholar] [CrossRef]
- Ambrus, R.; Alshweiat, A.; Csóka, I.; Ovari, G.; Esmail, A.; Radacsi, N. 3D-printed electrospinning setup for the preparation of loratadine nanofibers with enhanced physicochemical properties. Int. J. Pharm. 2019, 567, 118455. [Google Scholar] [CrossRef]
- Zupančič, S.; Sinha-Ray, S.; Sinha-Ray, S.; Kristl, J.; Yarin, A.L. Controlled release of ciprofloxacin from core–Shell nanofibers with monolithic or blended core. Mol. Pharm. 2016, 13, 1393–1404. [Google Scholar] [CrossRef]
- Krupa, A.; Tabor, Z.; Tarasiuk, J.; Strach, B.; Pociecha, K.; Wyska, E.; Wroński, S.; Łyszczarz, E.; Jachowicz, R. The impact of polymers on 3D microstructure and controlled release of sildenafil citrate from hydrophilic matrices. Eur. J. Pharm. Sci. 2018, 119, 234–243. [Google Scholar] [CrossRef]
- Lawrence, X.Y. Pharmaceutical quality by design: Product and process development, understanding, and control. Pharm. Res. 2008, 25, 781–791. [Google Scholar]
- Pawar, P.; Wang, Y.; Keyvan, G.; Callegari, G.; Cuitino, A.; Muzzio, F. Enabling real time release testing by NIR prediction of dissolution of tablets made by continuous direct compression (CDC). Int. J. Pharm. 2016, 512, 96–107. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Health and Human Services; Food and Drug Administration; Center for Drug Evaluation and Research (CDER). Quality Considerations for Continuous Manufacturing Guidance for Industry (Draft Guidence); FDA: White Oak, MD, USA, 2019.
- Donoso, M.; Ghaly, E.S. Prediction of drug dissolution from tablets using near-infrared diffuse reflectance spectroscopy as a nondestructive method. Pharm. Dev. Technol. 2005, 9, 247–263. [Google Scholar] [CrossRef]
- Freitas, M.P.; Sabadin, A.; Silva, L.M.; Giannotti, F.M.; do Couto, D.A.; Tonhi, E.; Medeiros, R.S.; Coco, G.L.; Russo, V.F.; Martins, J.A. Prediction of drug dissolution profiles from tablets using NIR diffuse reflectance spectroscopy: A rapid and nondestructive method. J. Pharm. Biomed. Anal. 2005, 39, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, E.; Pawar, P.; Keyvan, G.; Wang, Y.; Velez, N.; Callegari, G.; Cuitino, A.; Michniak-Kohn, B.; Muzzio, F.J.; Romañach, R.J. Prediction of dissolution profiles by non-destructive near infrared spectroscopy in tablets subjected to different levels of strain. J. Pharm. Biomed. Anal. 2016, 117, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Sutariya, V.; Groshev, A.; Sadana, P.; Bhatia, D.; Pathak, Y. Artificial neural network in drug delivery and pharmaceutical research. Open Bioinform. J. 2013, 7, 49–62. [Google Scholar] [CrossRef]
- Yegnanarayana, B. Artificial Neural Networks; PHI Learning Pvt. Ltd.: Delhi, India, 2009. [Google Scholar]
- Potdar, A.; Longstaff, A.P.; Fletcher, S.; Mian, N.S. Application of Multi Sensor Data Fusion Based on Principal Component Analysis and Artificial Neural Network for Machine Tool Thermal Monitoring; EUSPEN: Bedford, UK, 2015. [Google Scholar]
- Edwards, C. Growing pains for deep learning. Commun. ACM 2015, 58, 14–16. [Google Scholar] [CrossRef]
- Ibrić, S.; Djuriš, J.; Parojčić, J.; Djurić, Z. Artificial neural networks in evaluation and optimization of modified release solid dosage forms. Pharmaceutics 2012, 4, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Ilić, M.; Đuriš, J.; Kovačević, I.; Ibrić, S.; Parojčić, J. In vitro–In silico–In vivo drug absorption model development based on mechanistic gastrointestinal simulation and artificial neural networks: Nifedipine osmotic release tablets case study. Eur. J. Pharm. Sci. 2014, 62, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Montañez-Godínez, N.; Martínez-Olguín, A.C.; Deeb, O.; Garduño-Juárez, R.; Ramírez-Galicia, G. QSAR/QSPR as an Application of Artificial Neural Networks. In Artificial Neural Networks; Springer: Berlin, Germany, 2015; pp. 319–333. [Google Scholar]
- Moss, G.P.; Sun, Y.; Wilkinson, S.C.; Davey, N.; Adams, R.; Martin, G.P.; Prapopopolou, M.; Brown, M.B. The application and limitations of mathematical modelling in the prediction of permeability across mammalian skin and polydimethylsiloxane membranes. J. Pharm. Pharmacol. 2011, 63, 1411–1427. [Google Scholar] [CrossRef]
- Lou, H.; Chung, J.I.; Kiang, Y.-H.; Xiao, L.-Y.; Hageman, M.J. The application of machine learning algorithms in understanding the effect of core/shell technique on improving powder compactability. Int. J. Pharm. 2019, 555, 368–379. [Google Scholar] [CrossRef]
- Millen, N.; Kovačević, A.; Djuriš, J.; Ibrić, S. Machine Learning Modeling of Wet Granulation Scale-up Using Particle Size Distribution Characterization Parameters. J. Pharm. Innov. 2019, 1–12. [Google Scholar] [CrossRef]
- Han, R.; Yang, Y.; Li, X.; Ouyang, D. Predicting oral disintegrating tablet formulations by neural network techniques. Asian J. Pharm. Sci. 2018, 13, 336–342. [Google Scholar] [CrossRef]
- Ebube, N.K.; McCall, T.; Chen, Y.; Meyer, M.C. Relating formulation variables to in vitro dissolution using an artificial neural network. Pharm. Dev. Technol. 1997, 2, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Peh, K.K.; Lim, C.P.; San Quek, S.; Khoh, K.H. Use of artificial neural networks to predict drug dissolution profiles and evaluation of network performance using similarity factor. Pharm. Res. 2000, 17, 1384–1389. [Google Scholar] [CrossRef] [PubMed]
- Helland, I. Partial Least Squares Regression. In Wiley StatsRef: Statistics Reference Online; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Moore, J.W. Mathematical comparison of dissolution profiles. Pharm. Technol. 1996, 20, 64–75. [Google Scholar]
- O’hara, T.; Dunne, A.; Butler, J.; Devane, J.; Group, I.C.W. A review of methods used to compare dissolution profile data. Pharm. Sci. Technol. Today 1998, 1, 214–223. [Google Scholar] [CrossRef]
- Démuth, B.; Nagy, Z.K.; Balogh, A.; Vigh, T.; Marosi, G.; Verreck, G.; Van Assche, I.; Brewster, M. Downstream processing of polymer-based amorphous solid dispersions to generate tablet formulations. Int. J. Pharm. 2015, 486, 268–286. [Google Scholar] [CrossRef]
| Formulation Number | DR Content (w/w %) | HPMC Content (w/w %) | Compression Force (MPa) |
|---|---|---|---|
| 1 | 6 | 10 | 63.8 |
| 2 | 8 | 10 | 63.8 |
| 3 | 10 | 10 | 63.8 |
| 4 | 6 | 20 | 63.8 |
| 5 | 8 | 20 | 63.8 |
| 6 | 10 | 20 | 63.8 |
| 7 | 6 | 30 | 63.8 |
| 8 | 8 | 30 | 63.8 |
| 9 | 10 | 30 | 63.8 |
| 10 | 6 | 10 | 95.7 |
| 11 | 8 | 10 | 95.7 |
| 12 | 10 | 10 | 95.7 |
| 13 | 6 | 20 | 95.7 |
| 14 | 8 | 20 | 95.7 |
| 15 | 10 | 20 | 95.7 |
| 16 | 6 | 30 | 95.7 |
| 17 | 8 | 30 | 95.7 |
| 18 | 10 | 30 | 95.7 |
| 19 | 6 | 10 | 127.6 |
| 20 | 8 | 10 | 127.6 |
| 21 | 10 | 10 | 127.6 |
| 22 | 6 | 20 | 127.6 |
| 23 | 8 | 20 | 127.6 |
| 24 | 10 | 20 | 127.6 |
| 25 | 6 | 30 | 127.6 |
| 26 | 8 | 30 | 127.6 |
| 27 | 10 | 30 | 127.6 |
| 28 | 7 | 20 | 63.8 |
| 29 | 7.5 | 20 | 63.8 |
| 30 | 8.5 | 20 | 63.8 |
| 31 | 9 | 20 | 63.8 |
| 32 | 8 | 5 | 63.8 |
| 33 | 8 | 15 | 63.8 |
| 34 | 8 | 25 | 63.8 |
| 35 | 8 | 35 | 63.8 |
| 36 | 8 | 20 | 31.9 |
| 37 | 8 | 20 | 159.5 |
| Type of Data | Raman Transmission (GA) | Raman Reflection (GA) | NIR Transmission (GA) | NIR Reflection (GA) |
|---|---|---|---|---|
| Pretreatment method a | bl, SNV, MC | SNV, MC | der, MSC, MC | der, MSC, MC |
| Spectral region (cm−1) | 350–1680 | 350–1680 | 7600–8000, 8500–13,000 | 4200–7400 |
| Number of LVs | 2 (3) | 4 (6) | 3 (3) | 6 (6) |
| R2c | 0.911 (0.943) | 0.893 (0.962) | 0.905 (0.934) | 0.750 (0.777) |
| R2cv | 0.894 (0.934) | 0.875 (0.928) | 0.876 (0.912) | 0.586 (0.700) |
| R2p | 0.913 (0.905) | 0.868 (0.778) | 0.856 (0.918) | 0.579 (0.444) |
| RMSEC (% w/w) | 0.428 (0.343) | 0.471 (0.281) | 0.443 (0.370) | 0.718 (0.680) |
| RMSECV (% w/w) | 0.468 (0.370) | 0.509 (0.386) | 0.506 (0.426) | 0.928 (0.789) |
| RMSEP (% w/w) | 0.386 (0.400) | 0.467 (0.602) | 0.500 (0.414) | 0.837 (0.977) |
| Type of Data | Raman Transmission (GA) | Raman Reflection (GA) | NIR Transmission (GA) | NIR Reflection (GA) |
|---|---|---|---|---|
| Pretreatment method a | bl, SNV, MC | SNV, MC | der, MSC, MC | der, MSC, MC |
| Spectral region (cm−1) | 350–1680 | 350–1680 | 7600–8000, 8500–13,000 | 4200–7400 |
| Number of LVs | 2 (5) | 4 (4) | 4 (4) | 4 (4) |
| R2c | 0.953 (0.986) | 0.958 (0.966) | 0.986 (0.988) | 0.924 (0.951) |
| R2cv | 0.947 (0.982) | 0.950 (0.959) | 0.983 (0.986) | 0.907 (0.947) |
| R2p | 0.956 (0.975) | 0.956 (0.942) | 0.982 (0.982) | 0.875 (0.909) |
| RMSEC (% w/w) | 1.753 (0.950) | 1.654 (1.500) | 0.949 (0.884) | 2.231 (1.610) |
| RMSECV (% w/w) | 1.862 (1.082) | 1.811 (1.643) | 1.049 (0.962) | 2.470 (1.861) |
| RMSEP (% w/w) | 1.381 (1.031) | 1.443 (1.630) | 0.861 (0.914) | 2.307 (2.068) |
| Modeling Method | Raman | NIR | NIR-Raman |
|---|---|---|---|
| ANN | 74.27 | 71.84 | 73.07 |
| PLS | 65.63 | 65.01 | 65.79 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galata, D.L.; Farkas, A.; Könyves, Z.; Mészáros, L.A.; Szabó, E.; Csontos, I.; Pálos, A.; Marosi, G.; Nagy, Z.K.; Nagy, B. Fast, Spectroscopy-Based Prediction of In Vitro Dissolution Profile of Extended Release Tablets Using Artificial Neural Networks. Pharmaceutics 2019, 11, 400. https://doi.org/10.3390/pharmaceutics11080400
Galata DL, Farkas A, Könyves Z, Mészáros LA, Szabó E, Csontos I, Pálos A, Marosi G, Nagy ZK, Nagy B. Fast, Spectroscopy-Based Prediction of In Vitro Dissolution Profile of Extended Release Tablets Using Artificial Neural Networks. Pharmaceutics. 2019; 11(8):400. https://doi.org/10.3390/pharmaceutics11080400
Chicago/Turabian StyleGalata, Dorián László, Attila Farkas, Zsófia Könyves, Lilla Alexandra Mészáros, Edina Szabó, István Csontos, Andrea Pálos, György Marosi, Zsombor Kristóf Nagy, and Brigitta Nagy. 2019. "Fast, Spectroscopy-Based Prediction of In Vitro Dissolution Profile of Extended Release Tablets Using Artificial Neural Networks" Pharmaceutics 11, no. 8: 400. https://doi.org/10.3390/pharmaceutics11080400
APA StyleGalata, D. L., Farkas, A., Könyves, Z., Mészáros, L. A., Szabó, E., Csontos, I., Pálos, A., Marosi, G., Nagy, Z. K., & Nagy, B. (2019). Fast, Spectroscopy-Based Prediction of In Vitro Dissolution Profile of Extended Release Tablets Using Artificial Neural Networks. Pharmaceutics, 11(8), 400. https://doi.org/10.3390/pharmaceutics11080400






