Abstract
A family of bis(2-pyridyl)amino-modified poly(amidoamine) dendrimer Cu complexes was prepared, and their chemical nuclease activities and binding affinity (Kb) levels for DNA plasmid were investigated. The Kb values of the G2 to G6 apodendrimers for DNA plasmid were found to be 7.4, 23, 48, 70, and 280 µM−1, respectively, using ethidium bromide (EtBr) displacement experiments. The chemical nuclease activities of the corresponding complexes were determined by gel electrophoresis, and a clear positive dendritic effect was observed. Further analysis indicated a linear correlation between the Kb values of the G2 to G5 apodendrimers and the nuclease activity of the corresponding complexes. This observation indicated the importance of substrate binding affinity for macromolecular nuclease activity. In addition, an experiment using 3′-(p-hydroxyphenyl) fluorescein suggested that hydroxyl radicals formed under the tested conditions. Subsequently performed inhibition studies indicated that the hydroxyl radical was the active species responsible for the plasmid cleavage.
1. Introduction
Metallodendrimers have attracted considerable interest in the past few decades because they have the advantages of organic materials while also possessing the properties of metallic ions [1,2,3]. In contrast to small-molecule complexes, metallodendrimers provide unique complexation environments and contain many coordination sites as well as a protective environment for the metallic ions. As a result, metallodendrimers have characteristics not displayed by small-molecule complexes and have been used in various applications [4,5,6,7,8,9].
One representative application of metallodendrimers has been their use as mimics of natural metallomacromolecules, such as metalloenzymes. Metallonucleases are metalloenzymes that contribute to the regulation of the cell cycle by participating in the editing of nucleic acids [10]. In such nucleases, catalytic metallic ions have been shown to function by either of two working mechanisms: hydrolytic or oxidative cleavage. For oxidative nucleases, the coordination environment [11], molecular charge [12], and size of the ligands [13] have been explored, and determined to be vital factors in this catalytic process [14]. Various cofactors, such as H2O2, are usually necessary to break down nucleic acid chains: these cofactors generate reactive oxygen species during oxidative cleavage by the nuclease [15]. By contrast, hydrolytic nucleases operate using a catalytic mechanism involving assistance by a Lewis acid [16,17]. Although several types of metal ions have been shown to form metallonucleases [18,19], copper ions can act as either Lewis acids or oxidizing agents in nucleases. This unique character makes copper very attractive for the study of both natural and artificial nucleases [20].
In addition to proteins, numerous synthetic molecules, known as chemical nucleases, are capable of excising polynucleic acid backbones, and have been used for biomedical investigations and clinical applications [21,22]. These synthetic analogues of natural metalloenzymes demonstrate programmable nuclease activities and tolerate a wide range of conditions, features necessary for various investigations and applications. Various ligands, therefore, have been developed and characterized [23]. In contrast to aliphatic ligands, aromatic heterocycles can engage in π–π interactions with nucleic acids, and such interactions have been shown to improve the efficiency of the nuclease [24]. Of the various complexes of aromatic heterocycles with metal ions, aminopyridine metal complexes are particularly efficient catalysts due to their structural flexibility [25].
Macromolecular nucleases have been shown to provide advantages over their small-molecule analogues [26]. However, relatively few investigations of macromolecular nucleases have been carried out, due to the tedious nature of preparing monodisperse macromolecules and the difficulty in characterizing their active centers. Both issues have in particular hindered detailed investigations of macromolecular nucleases and their potential biomedical applications. Moreover, while the substrate binding affinities of small-molecule nucleases have been investigated, and reported to be a critical factor for the activity levels of these nucleases [27], the relationship between the activity levels of macromolecular nucleases and their binding affinities for nucleic acids has not been characterized [17] In contrast to other macromolecules, dendrimers are suitable candidates as nucleases because they are monodisperse and well characterized. These characteristic properties endow dendrimers with a well-controlled topology and make them very good models for investigation.
Herein, we report our research results on the use of metallodendrimers as chemical nucleases. G2–G6 poly(amidoamine) (PAMAM) dendrimers with peripheral bis(2-pyridyl)amino groups were synthesized and then coordinated with copper (Cu) ions to obtain nuclease complexes. These Cu complexes were tested for their nuclease activities, and the results were analyzed to reveal the relationship between the metallodendrimer nuclease activity and binding affinity to DNA.
2. Materials and Methods
2.1. DNA Binding Affinity
Ethidium bromide (EtBr) displacement assay studies were used to measure the binding affinities between dendrimer 2a to 2e and CT-DNA. The CT-DNA solution (0.46 mM) and ethidium bromide (EtBr, TCI, Tokyo, Japan) (50 µM) were dissolved in buffer (tris buffer 24 mM, pH = 7.24) and mixed in a florescence cuvette. A series of experiments were conducted by adding 0–14 µM of the compounds 2a–2e individually to the EtBr-bound CT-DNA in the florescence cuvette, and the florescence intensities were measured over time. The normalized florescence intensity was then plotted against the dendrimer concentration.
2.2. Nuclease Activity
The cleavage activity was investigated by agarose gel electrophoresis. Complexes 3a (1.67 µM, 3.33 µM, 6.67 µM, 8.33 µM, 10.00 µM, 16.67 µM, 20.00 µM) were mixed with plasmid DNA in the presence of 1,4-dithiothreitol (DTT) (0.66 mM) in Tris buffer (pH = 7.24, 24 mM), and the fluorescence was monitored over 45 min. Thereafter, the plasmid DNA was stained with EtBr and separated on a 0.8% agarose gel in standard Tris/borate /EDTA (TBE) buffer at 110 V over 35 min. The fluorescence intensity of form II under various conditions was obtained by using the ImageJ software.
2.3. Reactive Oxygen Species (ROS) Responsible for DNA Cleavage
Radical scavengers or metal chelators, such as sodium formate (53 mM), neocuproine (0.53 mM), EDTA (13 mM), and sodium azide (53 mM), were treated with 5e (33 nM) and DTT (0.66 mM). Complex 5e was mixed with DTT and various radical scavengers and incubated for 2 h. The solution was then separated on a 0.8% agarose gel in the standard Tris/borate/EDTA (TBE) buffer at 110 V over 35 min.
2.4. Determination of Hydroxyl Radicals
Tris buffer (24 mM, pH = 7.24), was degassed by bubbling nitrogen through the solution over two h to form an anaerobic environment. Compound 5e chelated 57 Equations copper ions to form a copper complex. The copper complex (2.60 µM) and DTT (161.00 µM) were mixed in the degassed Tris buffer and transferred to a fluorescence cuvette. H2O2 (0.27 mM, TCI) and 3′-(p-hydroxyphenyl) fluorescein (HPF) dye (Thermo Fisher, Waltham, MA, USA) were added to the fluorescence cuvette to obtain the reduced copper complex. The fluorescence intensity was used to determine the presence of hydrolytic radicals.
3. Results and Discussion
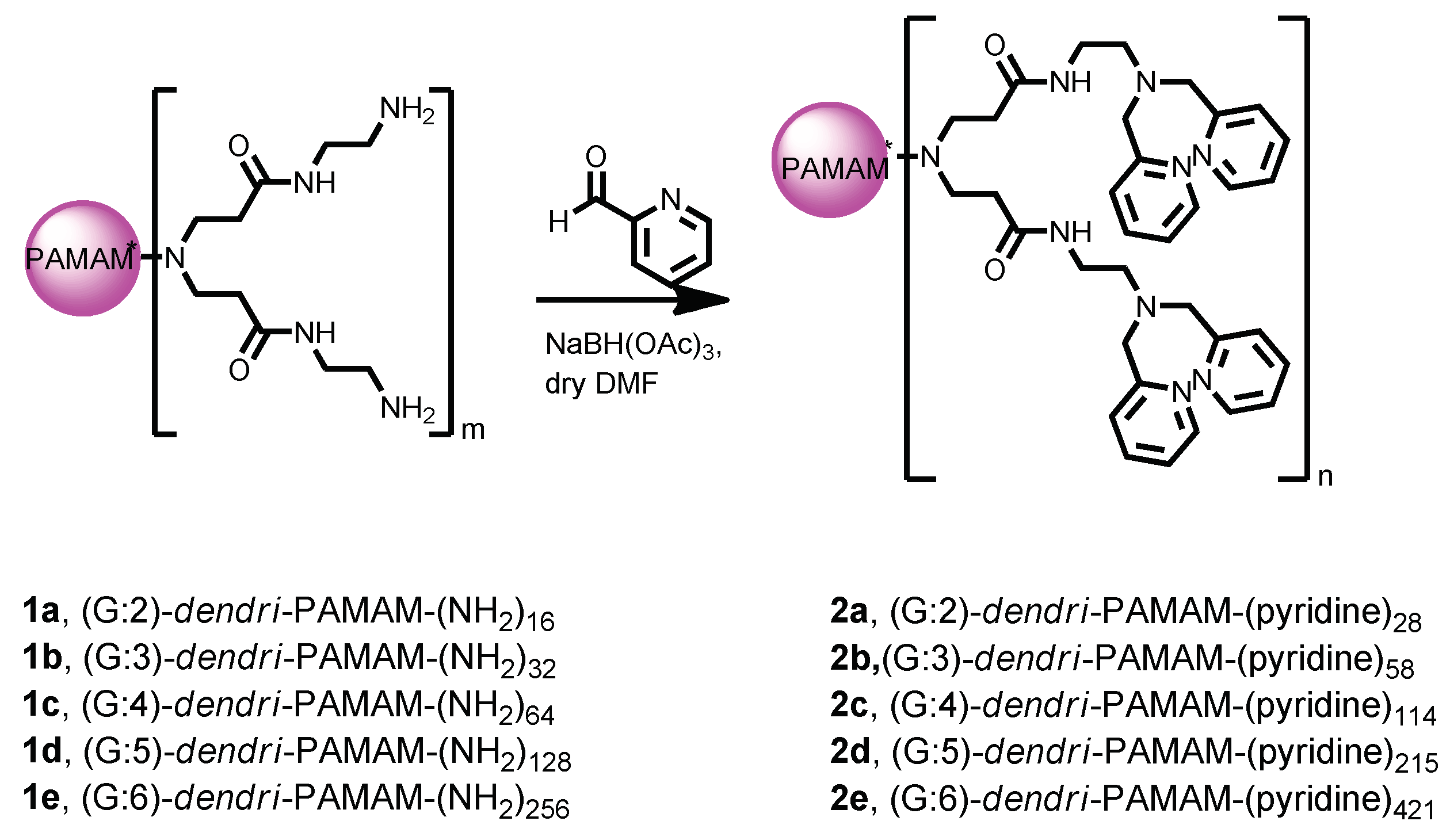
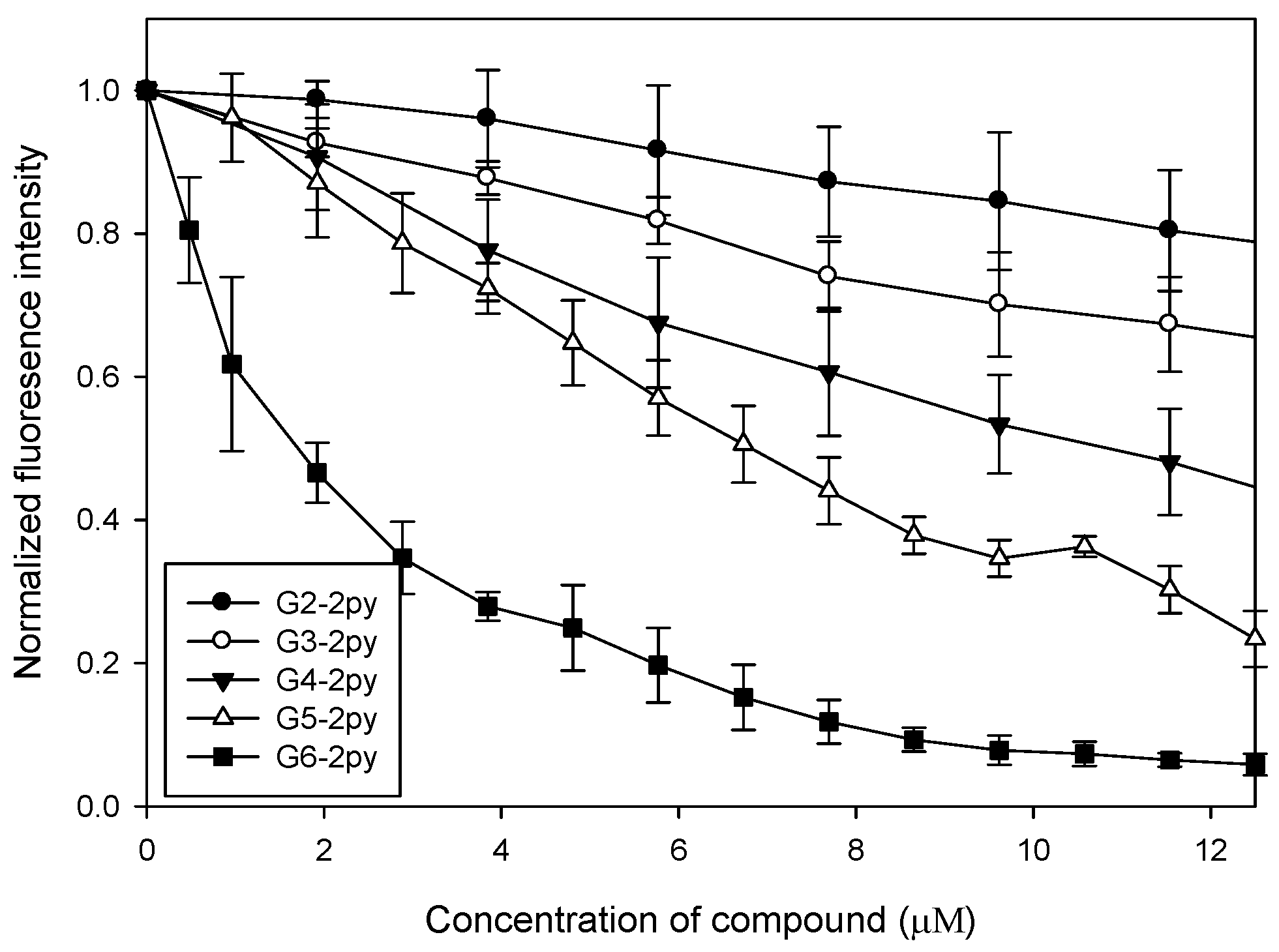
The bis(2-pyridyl)amino-modified (PAMAM) dendrimers (2) were prepared according to the literature [28] (Scheme 1). To study their binding affinity, an ethidium bromide (EtBr) displacement assay was used and the results indicated a clear negative correlation between the fluorescence intensity of EtBr and the concentration of each of the synthetic dendrimers 2. For any given quantity of the dendrimer, the high-generation dendrimers displayed lower fluorescence intensity levels than the low-generation ones. This observation suggested that the high-generation dendrimers bound more strongly to the nucleic acids (Figure 1). From these data we computed binding constants of 7.4, 23, 48, 70, and 280 µM−1 for 2a–2e, respectively [19], i.e., the larger the dendrimer, the stronger its binding of the nucleic acid.
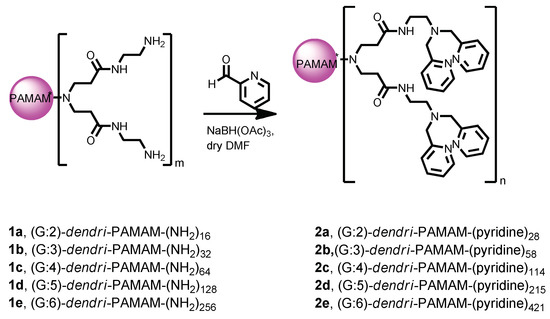
Scheme 1.
Synthesis of compounds 2.
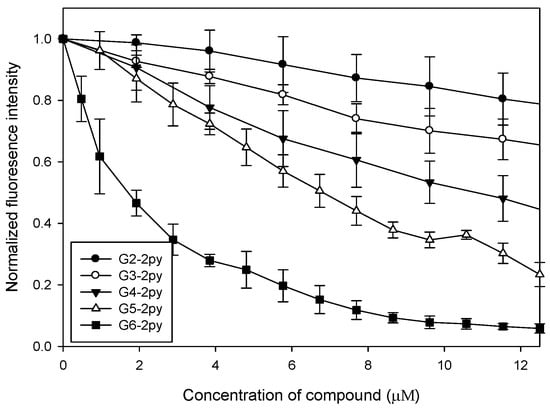
Figure 1.
Ethidium bromide (EtBr) displacement assay.
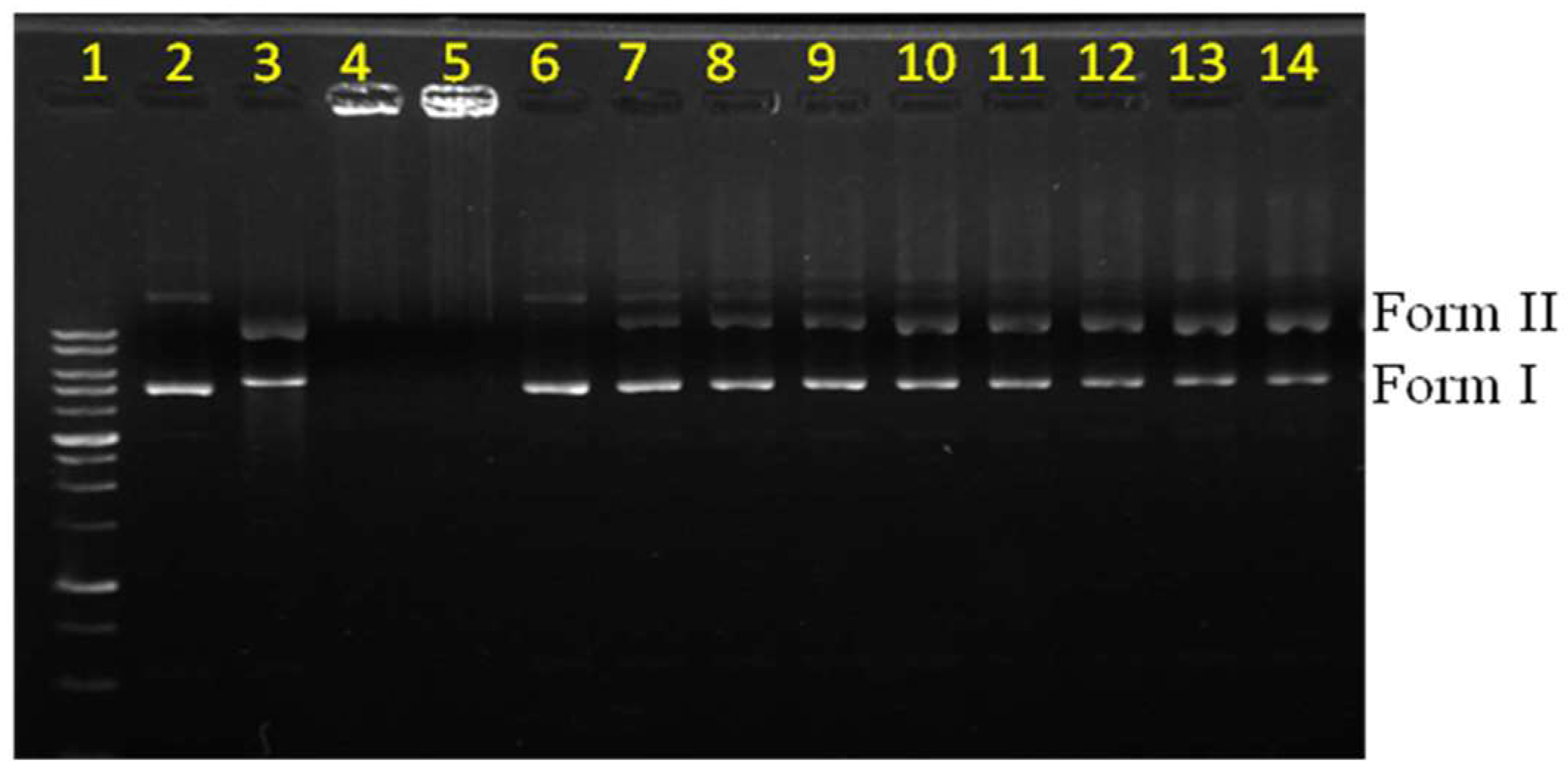
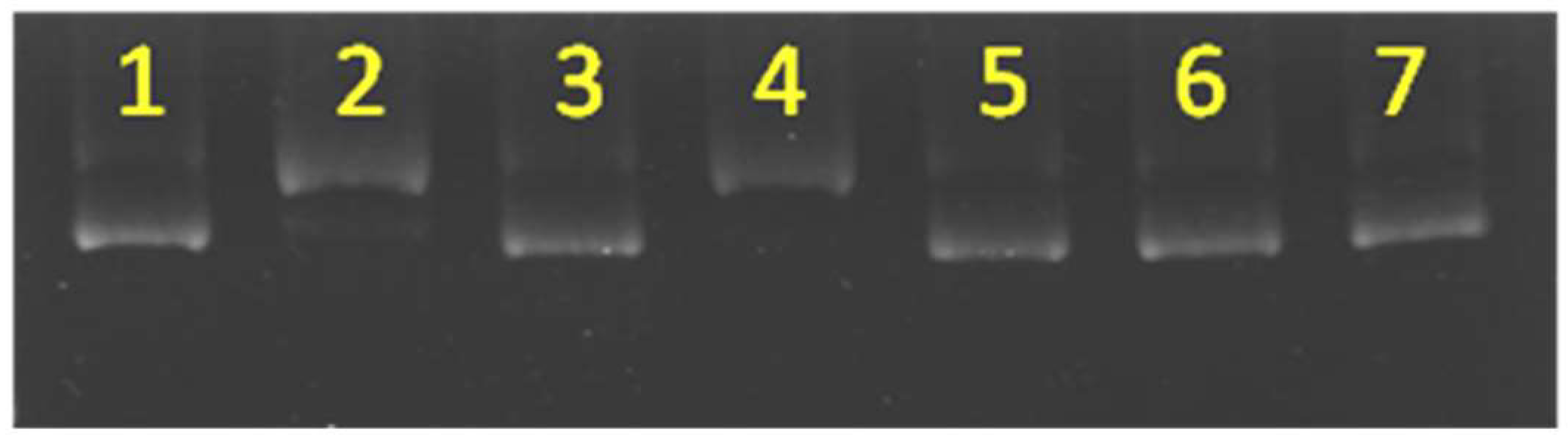
To investigate the nuclease activity, we first saturated dendrimer 2a with Cu and then subjected the resulting complex 3a to a DNA cleavage experiment. 3a displayed nuclease activity toward the supercoiled form (form I) of the plasmid to produce the circular form (form II) in the presence of 1,4-dithiothreitol (DTT) as a reducing agent (Figure 2). In this investigation, bleomycin was used as a reference compound (lane 3). Different amounts of the complex (3a) were used to study its nuclease activity, and the results are indicated in lanes 7–14. The quantity of the circular form (form II) of the DNA plasmid increased in the presence of larger quantities of the complex. The concentration-dependent nuclease activity was used to characterize the catalytic efficiency of this complex. Complex 3a alone showed no nuclease effect (lane 6). This observation implied that no role was played by the hydrolytic mechanism in this reaction. To understand the possible effect of ligand and copper alone, a comparable amount of bipyridine analogue (4, Figure S1) and CuSO4 were subjected to similar experiments and the results are shown in lanes 4 and 5; here the plasmid remained in the loading well. Presumably, the negative charges of the plasmid were neutralized by the excess quantity of copper ions, making the complex stationary on the gel. The monomeric dendrimer and the free copper ions have been reported to show no ROS generation activity under the given conditions [28]. Therefore, the monomer was assumed to have no nuclease activity in the present studies. The observed activity was therefore concluded to have arisen from the macromolecular Cu complex rather than from the free Cu ions in the medium.
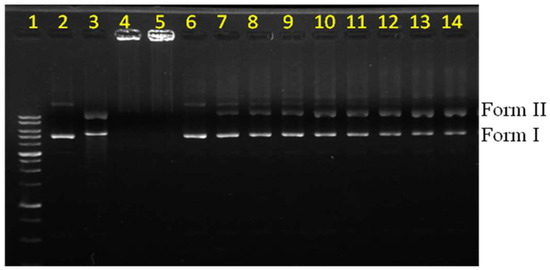
Figure 2.
Electrophoresis experiments were performed to characterize the conditions under which reactive oxygen species (ROS) radicals were generated and nuclease activity was observed. Lane 1: marker; Lane 2: DNA; Lane 3: bleomycin (10.00 µM); Lane 4: CuSO4 (1.6 mM) + 4 (16.7 µM) + DTT (0.66 mM); Lane 5: CuSO4 (1.6 mM) + DTT (0.66 mM); Lane 6: 3a (1.67 µM); Lane 7: DTT (0.66 mM); Lane 8: 3a (1.67 µM) + DTT (0.66 mM); Lane 9: 3a (3.33 µM) + DTT (0.66 mM); Lane 10: 3a (6.67 µM) + DTT (0.66 mM); Lane 11: 3a (8.33 µM) + DTT (0.66 mM); Lane 12: 3a (10.00 µM) + DTT (0.66 mM); Lane 13: 3a (16.67 µM) + DTT (0.66 mM); Lane 14: 3a (20.00 µM) + DTT (0.66 mM). DTT = 1,4-dithiothreitol.
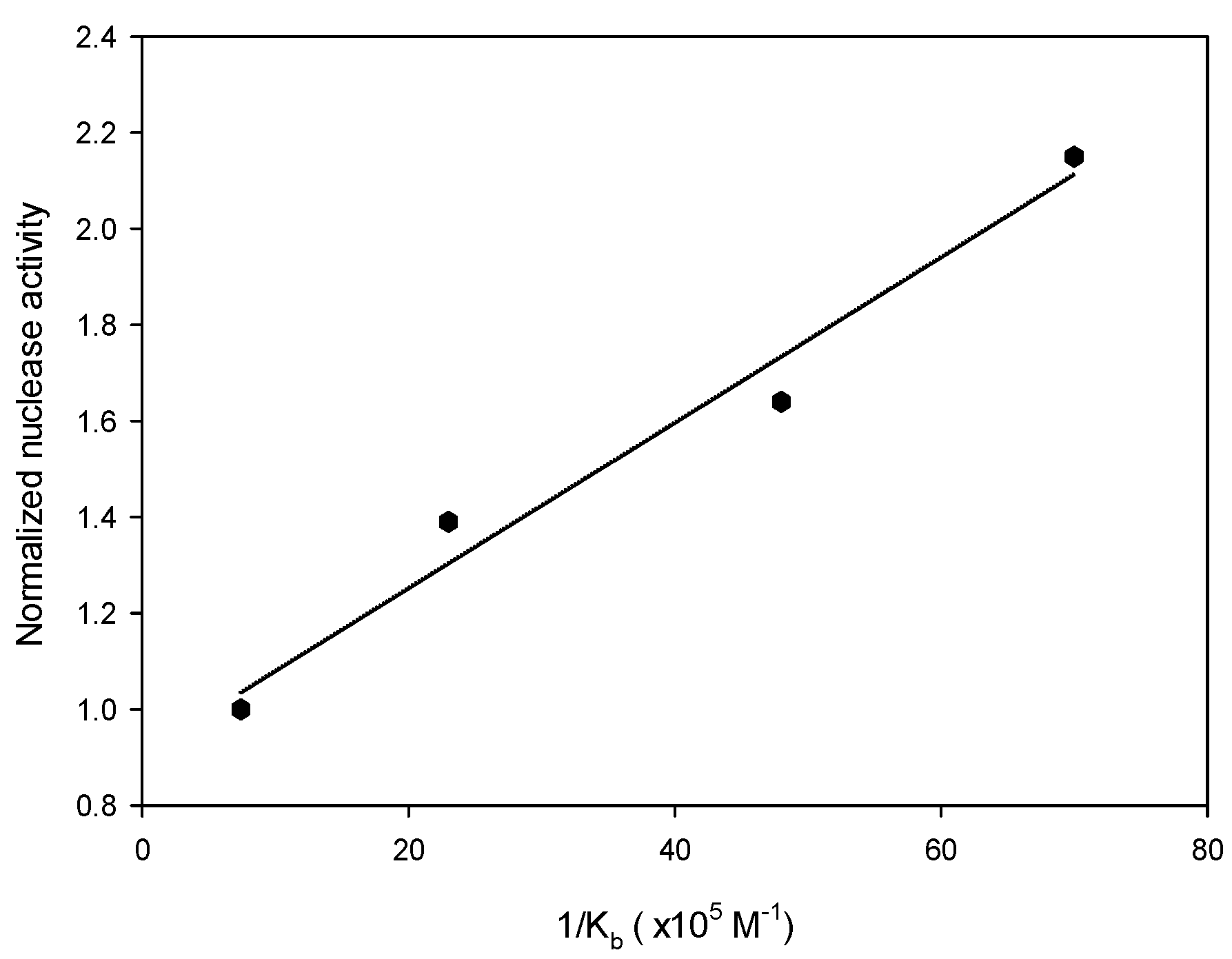
To explore the relationship between dendrimer binding affinity and nuclease activity, the number of Cu ions in each dendrimer must be the same to prevent any possible bias associated with a different number of Cu ions in each dendrimer. In previous work [29], each G2-dendrimer (2a) molecule was found to bind to no more than six copper ions. Accordingly, six equivalents of copper ions were coordinated to each dendrimer generation (2a–2e) to form hexaCu complexes 5a–5e, respectively, and these complexes were subjected to nuclease activity assays. The fluorescence intensities obtained from form II indicated that the nuclease activity was positively correlated with the dendrimer size (Figure S2). High-generation dendrimers exhibited stronger plasmid DNA cutting activities than did the low-generation analogues. Dendrimers 5d and 5e exhibited, respectively, 2.15-fold and 2.02-fold higher nuclease activities than did 5a. Meanwhile, 5b and 5c exhibited, respectively, only 1.39-fold and 1.64-fold higher nuclease activities than did 5a. Surprisingly, the normalized nuclease activities of metallodendrimers G2 through G5 were found to be highly positively correlated (R2 = 0.97) with the Kb values of the corresponding apodendrimers for the DNA plasmid (Figure 3). These results indicated that the binding affinity of the dendrimer for the DNA plasmid directly contributed to its nuclease activity. While 5e was expected, based on the trend of the results for 5a–5d, to exhibit higher activity than 5d, it was obser ved to display only 94% of the activity towards form II of 5d. (Table S1) This observation may have been due to the greater degradation of form II in the presence of complex 5e.
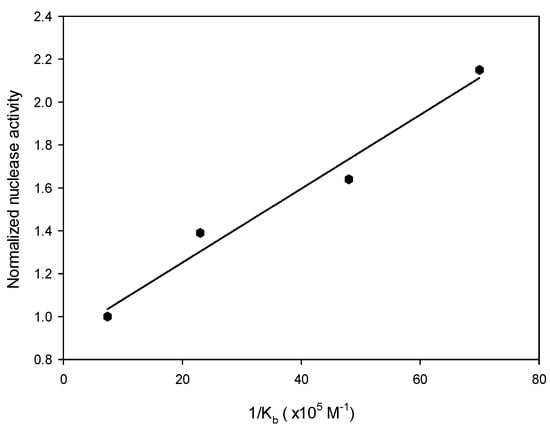
Figure 3.
Plot of the normalized nuclease activities of hexaCu complexes 5a–5d as a function of 1/Kb for apodendrimers 2a–2d toward the plasmid. The fluorescence intensity of form II in the presence of each compound 5 was obtained by using ImageJ software.
Identifying the active species responsible for the nuclease activity is also an important issue for developing biomedical applications of the nuclease. To determine the major active species of the synthesized dendrimers responsible for their cleavage of plasmid, we only considered oxygen-derived species, since hydrolytic cleavage is not possible here [26]. Of the various possible reactive oxygen species (ROS), the hydroxyl radical has been found to be responsible for the activities of various nucleases [27]. Therefore, the hydroxyl radical was hypothesized to be involved in the activity of the dendrimer nuclease. This hypothesis was tested by determining the presence of hydroxyl radicals using 3′-(p-hydroxyphenyl) fluorescein (HPF), which reacts predominantly with hydroxyl radicals or superoxide anion radicals via dearylation to form a fluorescent product. Superoxide, however, can induce re-oxidation reactions that disturb the identification of the hydroxyl radical. Tris buffer was, therefore, degassed to avoid the formation of metallodendrimer-generated superoxide anion radicals in the aerobic environment used. Instead, H2O2 was used as the oxygen source and mixed with 5e in an anoxic environment, and the formation of hydroxyl radicals was monitored using HPF. The fluorescence intensity was much higher in the presence of the metallodendrimer than in the absence of the metallodendrimer (Figure S3). This observation using complex 5e indicated that this dendrimer, and hence likely 5a–5d as well, can catalyze the formation of the hydroxyl radical.
To further confirm the presence of the hydroxyl radical under the given conditions, we carried out inhibition studies using various ROS scavengers or metal chelators, including EDTA, sodium azide, neocuproine, and sodium formate, which served as a copper chelator, singlet oxygen inhibitor/copper chelator, specific copper (I) chelator, and hydroxyl radical scavenger, respectively. The inhibition activities of these compounds were examined by introducing each inhibitor into the DNA cleavage experiments and measuring the resulting nuclease activities. As shown in Figure 4, in lanes 3, 5, and 6, the plasmid remained in form I when sodium formate, EDTA, and sodium azide, respectively, were included, clearly indicating that each of them inhibited DNA cutting (Figure 4). EDTA removed metal ions from the dendrimers. The nuclease activity results indicated that the copper ions were crucial to the nuclease activity. Also note that azide has been shown to chelate copper ions and abrogate copper re-oxidation in catecholases and tyrosinases [30]. Moreover, sodium azide could also act as a singlet oxygen scavenger. However, the presence of singlet oxygen was highly unlikely in this reaction, as it did not involve photo-irradiation. The presence of reducing agents was necessary for the nuclease activity, and sodium azide appeared to act as a copper chelator but not a singlet oxygen scavenger in this reaction. Nuclease inhibition by sodium formate implied that the hydroxyl radical was the ROS involved in DNA cleavage. Nevertheless, the failure of neocuproine to remove the copper(I) ions and hence stop the reaction may have been due to the strong binding of Cu(I) ions to the bis(2-pyridyl)amino moieties, indicating that neocuproine could not remove the copper(I) ions or stop the reaction.
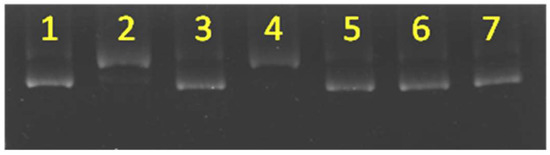
Figure 4.
Identification of the active species. Lane1, plasmid alone; Lane 2, plasmid with 5e (33 nM) + DTT (0.66 mM); Lane 3, plasmid with 5e (33 nM) + DTT (0.66 mM) + sodium formate (53 mM); Lane 4, plasmid with 5e (33 nM) + DTT (0.66 mM) + neocuproine (0.53 mM); Lane 5, plasmid with 5e (33 nM) + DTT (0.66 mM) + EDTA (13 mM); Lane 6, plasmid with 5e (33 nM) + DTT (0.66 mM) + sodium azide (53 mM); Lane 7, plasmid with 5e (33 nM).
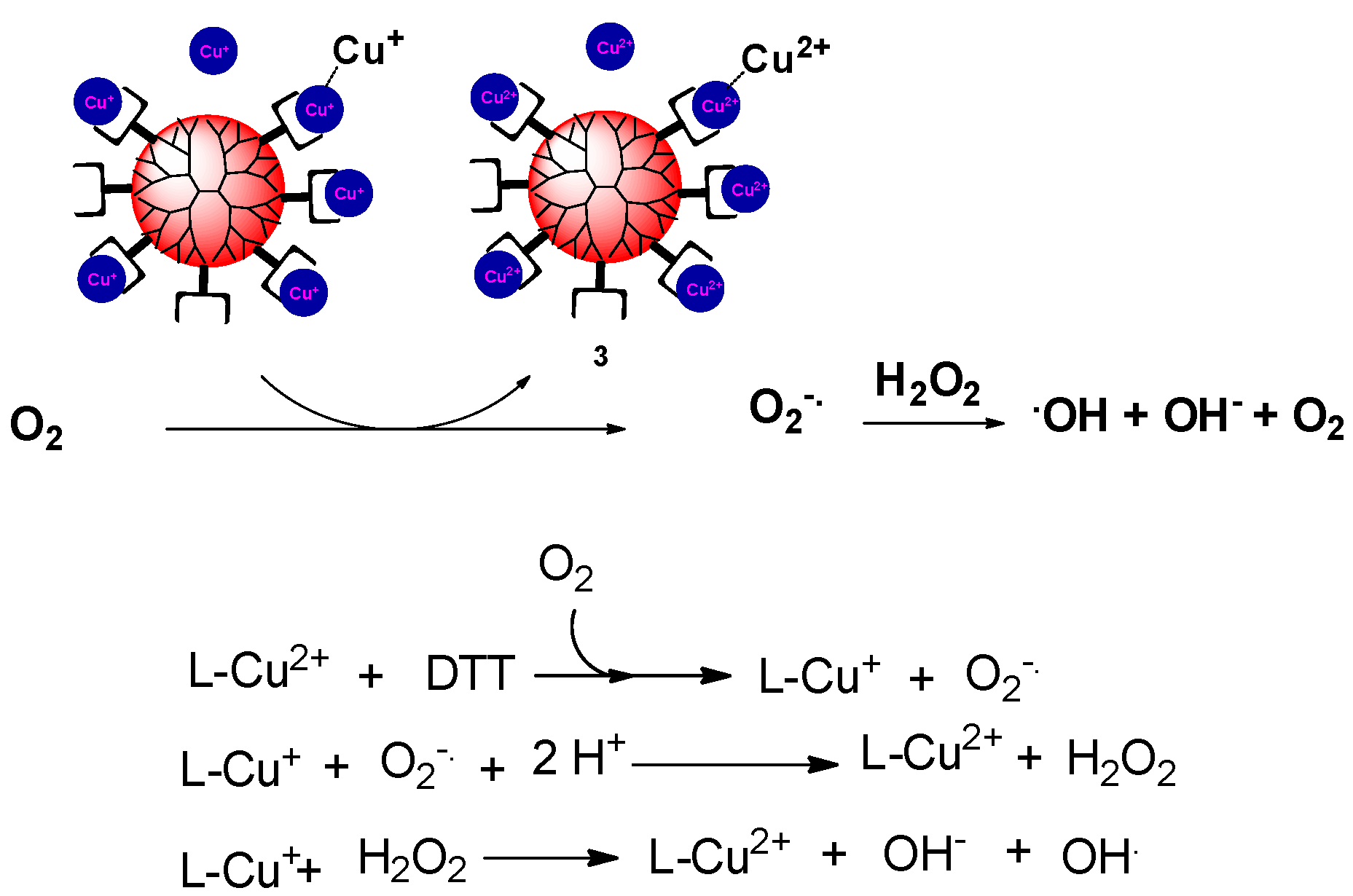
In view of previous studies [31], the Cu dendrimer complexes apparently generated superoxide anion radicals in the presence of reducing agents. The superoxide anion radicals could be then further reduced and protonated to form hydrogen peroxide. The hydroxyl radical could have been obtained from either one of two routes. With excess reducing agent, Cu+ ion conjugated to hydrogen peroxide may have produced a highly reactive hydroxyl radical through the copper Fenton reaction. Otherwise, the Haber–Weiss reaction might have been involved while reductant was being consumed. Under this circumstance, Cu2+ was reduced by superoxide anion radicals to give the Cu+ ion, which could lead to the formation of the hydroxyl radical (Figure 5).
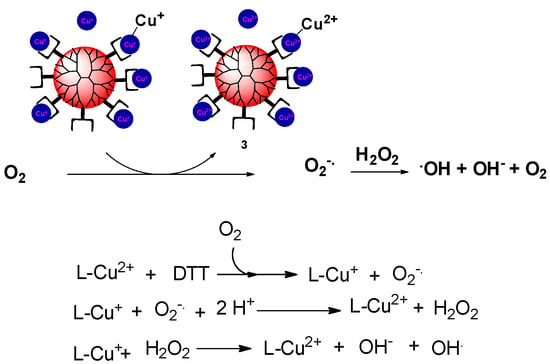
Figure 5.
Hypothetical mechanism for the dendrimer-induced production of hydroxyl radicals.
4. Conclusions
The most significant discovery of this investigation was the establishment of a correlation between the binding affinity of copper dendrimer-based nucleases and their nuclease activity. For the first time, a linear correlation between Kb and chemical nuclease activity was found among the complexes tested. The higher-generation dendrimers displayed higher binding affinities for the DNA plasmid, and the nuclease activity of the dendrimer complex was found to depend on the size of the dendrimer. These results revealed the importance of binding affinity for macromolecular nuclease activity and could be used for the development of new macromolecular nucleases. We also identified the formation of hydroxyl radicals to be the major working mechanism for this cleavage reaction. Our observations here have provided a fundamental understanding of how macromolecules can serve as nucleases and pave the way for the design of efficient chemical nucleases.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4923/10/4/258/s1, Procedures used to synthesize compounds 2 and 4, complexes 3a and 5; H-NMR spectra of compounds 2. Figure S1. Structure of compound 4; Figure S2. Nuclease activity of hexaCu complexes 5; Figure S3. Fluorescence spectra of HPF in the presence and absence of 5e; Table S1. Relative fluorescence intensity of II.
Author Contributions
Conceptualization, Y.-H.T. and C.-L.K.; Data curation: Y.-H.T.; S.-t.L., and C.H.-Y.W.; Methodology, Y.-H.T.; C.H.-Y.W.; S.C.N.H.; H.-T.C. and P.-Y.C.; Validation, Y.-H.T. and C.-L.K.; Formal Analysis, Y.-H.T. and S.-t.L.; Investigation, Y.-H.T.; Data Curation, Y.-H.T. and S.-t.L.; Writing—Original Draft Preparation, Y.-H.T.; Writing—Review and Editing, C.-L.K. Supervision, C.-L.K.; Funding Acquisition, C.-L.K.
Funding
This work was supported by the Ministry of Science and Technology (MoST), Taiwan (MOST 106-2632-M-037-001- and 106-2113-M-037-008-) Kaohsiung Medical University (KMU-DK108009) and Kaohsiung Medical University Hospital (KMUH106-M625).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Astruc, D.; Ornelas, C.; Ruiz, J. Metallocenyl Dendrimers and Their Applications in Molecular Electronics, Sensing, and Catalysis. Acc. Chem. Res. 2008, 41, 841–856. [Google Scholar] [CrossRef] [PubMed]
- Newkome, G.R.; He, E.; Moorefield, C.N. Suprasupermolecules with Novel Properties: Metallodendrimers. Chem. Rev. 1999, 99, 1689–1746. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-H.; Huang, A.Y.; Chen, P.-Y.; Chen, H.-T.; Kao, C.-L. Metallodendrimers and Dendrimer Nanocomposites. Curr. Pharm. Des. 2011, 17, 2308–2330. [Google Scholar] [CrossRef] [PubMed]
- Corbin, D.A.; Shircliff, D.M.; Reeves, B.J.; Boardman, B.M. Metallopolymers from direct polymerization of functionalized cobalt chalcogenide clusters and thiophene comonomers. Polym. Chem. 2017, 8, 3801–3809. [Google Scholar] [CrossRef]
- Greenfield, J.L.; Rizzuto, F.J.; Goldberga, I.; Nitschke, J.R. Self-Assembly of Conjugated Metallopolymers with Tunable Length and Controlled Regiochemistry. Angew. Chem. Int. Ed. 2017, 56, 7541–7545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Liu, S.; Zhao, Q.; Huang, W. Stimuli–responsive metallopolymers. Coord. Chem. Rev. 2016, 319, 180–195. [Google Scholar] [CrossRef]
- Vishwanath, R.S.; Kandaiah, S. Metal ion-containing C3N3S3 coordination polymers chemisorbed to a copper surface as acid stable hydrogen evolution electrocatalysts. J. Mater. Chem. A 2017, 5, 2052–2065. [Google Scholar]
- Liang, Y.; Strohecker, D.; Lynch, V.; Holliday, B.J.; Jones, R.A. A Thiophene-Containing Conductive Metallopolymer Using an Fe(II) Bis(terpyridine) Core for Electrochromic Materials. ACS Appl. Mater. Interfaces 2016, 8, 34568–34580. [Google Scholar] [CrossRef]
- Caminade, A.-M.; Ouali, A.; Laurent, R.; Turrin, C.-O.; Majoral, J.-P. Coordination chemistry with phosphorus dendrimers. Applications as catalysts, for materials, and in biology. Coord. Chem. Rev. 2016, 308, 478–497. [Google Scholar] [CrossRef]
- Verma, S.; Srivatsan, S.G.; Claussen, C.A.; Long, E.C. DNA strand scission by a Cu(I)·adenylated polymeric template: Preliminary mechanistic and recycling studies. Bioorg. Med. Chem. Lett. 2003, 13, 2501–2504. [Google Scholar] [CrossRef]
- Melvin, M.S.; Tomlinson, J.T.; Saluta, G.R.; Kucera, G.L.; Lindquist, N.; Manderville, R.A. Double-Strand DNA Cleavage by Copper·Prodigiosin. J. Am. Chem. Soc. 2000, 122, 6333–6334. [Google Scholar] [CrossRef]
- Zandarashvili, L.; Esadze, A.; Vuzman, D.; Kemme, C.A.; Levy, Y.; Iwahara, J. Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc. Natl. Acad. Sci. USA 2015, 112, E5142–E5149. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-L.; Chao, H.; Li, H.; Hong, X.-L.; Ji, L.-N.; Li, X.-Y. Synthesis, crystal structure and DNA cleavage activities of copper(II) complexes with asymmetric tridentate ligands. J. Inorg. Biochem. 2004, 98, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Xiao, N.; Shi, P.; Zhu, Y.; Guo, Z. Design of artificial metallonucleases with oxidative mechanism. Coord. Chem. Rev. 2007, 251, 1951–1972. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Zhao, J.; Yang, P. DNA-binding and cleavage studies of novel binuclear copper(II) complex with 1,1′-dimethyl-2,2′-biimidazole ligand. J. Inorg. Biochem. 2007, 101, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, M.; Zhang, T.; Sun, H. DNA hydrolysis promoted by di- and multi-nuclear metal complexes. Coord. Chem. Rev. 2004, 248, 147–168. [Google Scholar] [CrossRef]
- Hurley, A.L.; Mohler, D.L. Organometallic Photonucleases: Synthesis and DNA-Cleavage Studies of Cyclopentadienyl Metal-Substituted Dendrimers Designed To Increase Double-Strand Scission. Org. Lett. 2000, 2, 2745–2748. [Google Scholar] [CrossRef]
- Teyssot, M.-L.; Jarrousse, A.-S.; Chevry, A.; De Haze, A.; Beaudoin, C.; Manin, M.; Nolan, S.P.; Díez-González, S.; Morel, L.; Gautier, A. Toxicity of Copper(I)–NHC Complexes Against Human Tumor Cells: Induction of Cell Cycle Arrest, Apoptosis, and DNA Cleavage. Chemistry 2009, 15, 314–318. [Google Scholar] [CrossRef]
- Mack, D.P.; Iverson, B.L.; Dervan, P.B. Design and chemical synthesis of a sequence-specific DNA-cleaving protein. J. Am. Chem. Soc. 1988, 110, 7572–7574. [Google Scholar] [CrossRef]
- Suntharalingam, K.; Hunt, D.J.; Duarte, A.A.; White, A.J.P.; Mann, D.J.; Vilar, R. A Tri-copper(II) Complex Displaying DNA-Cleaving Properties and Antiproliferative Activity against Cancer Cells. Chemistry 2012, 18, 15133–15141. [Google Scholar] [CrossRef]
- Mancin, F.; Scrimin, P.; Tecilla, P.; Tonellato, U. Artificial metallonucleases. Chem. Commun. 2005, 20, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.D.; Hudson, S.E.; Brown, S.J.; Olmstead, M.M.; Mascharak, P.K. Syntheses, structures, and reactivities of synthetic analogs of the three forms of cobalt(III)-bleomycin: Proposed mode of light-induced DNA damage by the cobalt(III) chelate of the drug. J. Am. Chem. Soc. 1992, 114, 3841–3853. [Google Scholar] [CrossRef]
- El Amrani, F.B.A.; Perelló, L.; Real, J.A.; González-Alvarez, M.; Alzuet, G.; Borrás, J.; García-Granda, S.; Montejo-Bernardo, J. Oxidative DNA cleavage induced by an iron(III) flavonoid complex: Synthesis, crystal structure and characterization of chlorobis(flavonolato)(methanol) iron(III) complex. J. Inorg. Biochem. 2006, 100, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Wende, C.; Lüdtke, C.; Kulak, N. Copper Complexes of N-Donor Ligands as Artificial Nucleases. Eur. J. Inorg. Chem. 2014, 16, 2597–2612. [Google Scholar] [CrossRef]
- Soler, M.; Figueras, E.; Serrano-Plana, J.; González-Bártulos, M.; Massaguer, A.; Company, A.; Martínez, M.Á.; Malina, J.; Brabec, V.; Feliu, L.; Planas, M.; et al. Design, Preparation, and Characterization of Zn and Cu Metallopeptides Based On Tetradentate Aminopyridine Ligands Showing Enhanced DNA Cleavage Activity. Inorg. Chem. 2015, 54, 10542–10558. [Google Scholar] [CrossRef] [PubMed]
- Zhiryakova, M.V.; Izumrudov, V.A. Interaction of Astramol Poly(propyleneimine) Dendrimers with DNA and Poly(methacrylate) Anion in Water and Water–Salt Solutions. J. Phys. Chem. B 2014, 118, 8819–8826. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Cowan, J.A. DNA Cleavage by Copper−ATCUN Complexes. Factors Influencing Cleavage Mechanism and Linearization of dsDNA. J. Am. Chem. Soc. 2005, 127, 8408–8415. [Google Scholar] [CrossRef]
- Kao, C.-L.; Tang, Y.-H.; Lin, Y.C.; Chiu, L.-T.; Chen, H.-T.; Hsu, S.C.N.; Hsieh, K.-C.; Lu, C.-Y.; Chen, Y.-L. Copper complex of a pyridine-modified poly(amidoamine) dendrimer as a chemical nuclease: Synthetic and catalytic study. Nanomedicine 2011, 7, 273–276. [Google Scholar] [CrossRef]
- Tang, Y.-H.; Lin, Y.C.; Hsu, S.C.N.; Liou, S.-T.; Chen, H.-Y.; Hsieh, K.-C.; Chuang, W.-J.; Chiu, L.-T.; Chen, Y.-L.; Kao, C.-L. Cooperative Effects in Copper Polyamidoamine Dendrimer Complexes Catalyzing the Reduction of Molecular Oxygen. Eur. J. Inorg. Chem. 2015, 29, 4839–4847. [Google Scholar] [CrossRef]
- Healey, D.F.; Strothkamp, K.G. Inhibition of the catecholase and cresolase activity of mushroom tyrosinase by azide. Arch. Biochem. Biophys. 1981, 211, 86–91. [Google Scholar] [CrossRef]
- Tang, Y.-H.; Cangiotti, M.; Kao, C.-L.; Ottaviani, M.F. EPR Characterization of Copper(II) Complexes of PAMAM-Py Dendrimers for Biocatalysis in the Absence and Presence of Reducing Agents and a Spin Trap. J. Phys. Chem. B 2017, 121, 10498–10507. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).