LEDGF/p75 Deficiency Increases Deletions at the HIV-1 cDNA Ends
Abstract
:1. Introduction
2. Materials and Methods
2.1. LEDGF/p75-Deficient Cell Lines
2.2. Generation of Retroviral Vectors
2.3. Single-Round Viral Infectivity Assay
2.4. Analysis of HIV-1 2-LTR Circles
2.5. HIV-1 cDNA Analysis
2.6. Immunoblotting
2.7. Statistical Analysis
3. Results
3.1. Susceptibility of TL3 and TC3 Cells to HIV-1 Infection
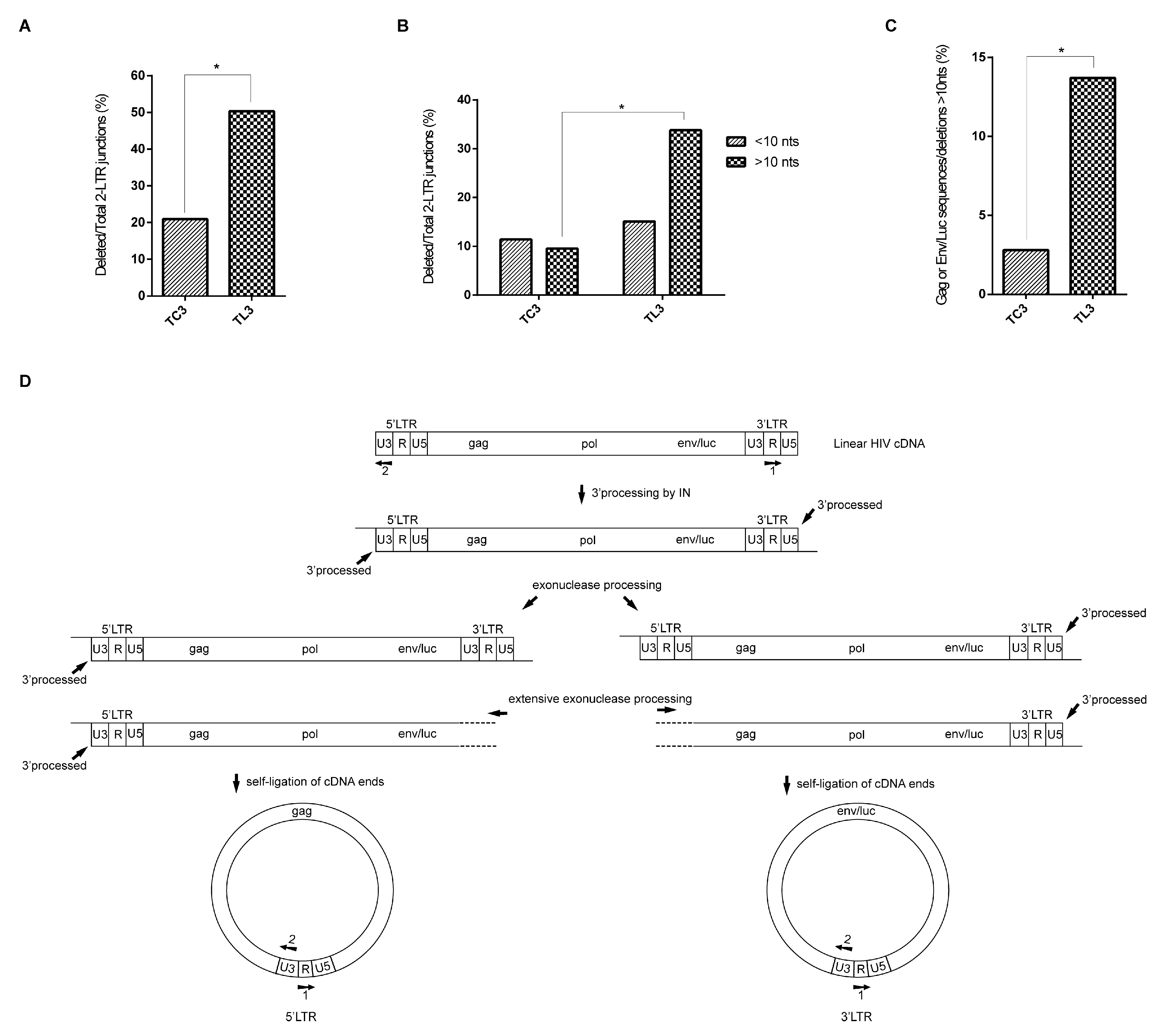
3.2. LEDGF/p75 Deficiency Increases the Frequency of Deletions at HIV-1 LTR Termini
3.3. HIV-1 Reverse Transcription Activity Is Not Affected by LEDGF/p75 Depletion
3.4. Integrase-Mediated 3′ Processing Is a Pre-Requisite for the Increased Frequency of Deletions Observed at the 2-LTR Junctions Isolated from LEDGF/p75-Deficient Cells
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sloan, R.D.; Wainberg, M.A. The role of unintegrated DNA in HIV infection. Retrovirology 2011, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Yoder, K.; Sarasin, A.; Kraemer, K.; McIlhatton, M.; Bushman, F.; Fishel, R. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc. Natl. Acad. Sci. USA 2006, 103, 4622–4627. [Google Scholar] [CrossRef] [PubMed]
- Yoder, K.E.; Roddick, W.; Hoellerbauer, P.; Fishel, R. XPB mediated retroviral cDNA degradation coincides with entry to the nucleus. Virology 2011, 410, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Weil, A.F.; Ghosh, D.; Zhou, Y.; Seiple, L.; McMahon, M.A.; Spivak, A.M.; Siliciano, R.F.; Stivers, J.T. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl. Acad. Sci. USA 2013, 110, E448–457. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Kanaar, R.; Jackson, S.P.; O’Connor, M.J. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004, 23, 3421–3429. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Olvera, J.M.; Yoder, K.E.; Mitchell, R.S.; Butler, S.L.; Lieber, M.; Martin, S.L.; Bushman, F.D. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. Embo J. 2001, 20, 3272–3281. [Google Scholar] [CrossRef] [PubMed]
- Jeanson, L.; Subra, F.; Vaganay, S.; Hervy, M.; Marangoni, E.; Bourhis, J.; Mouscadet, J.F. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 2002, 300, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ao, Z.; Wang, B.; Jayappa, K.D.; Yao, X. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J. Biol. Chem. 2011, 286, 17722–17735. [Google Scholar] [CrossRef] [PubMed]
- Butler, S.L.; Hansen, M.S.; Bushman, F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001, 7, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Hazuda, D.J.; Felock, P.; Witmer, M.; Wolfe, A.; Stillmock, K.; Grobler, J.A.; Espeseth, A.; Gabryelski, L.; Schleif, W.; Blau, C.; et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 2000, 287, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Reigadas, S.; Andreola, M.L.; Wittkop, L.; Cosnefroy, O.; Anies, G.; Recordon-Pinson, P.; Thiebaut, R.; Masquelier, B.; Fleury, H. Evolution of 2-long terminal repeat (2-LTR) episomal HIV-1 DNA in raltegravir-treated patients and in in vitro infected cells. J. Antimicrob. Chemother. 2010, 65, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Svarovskaia, E.S.; Barr, R.; Zhang, X.; Pais, G.C.; Marchand, C.; Pommier, Y.; Burke, T.R., Jr.; Pathak, V.K. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J. Virol. 2004, 78, 3210–3222. [Google Scholar] [CrossRef] [PubMed]
- Buckman, J.S.; Bosche, W.J.; Gorelick, R.J. Human immunodeficiency virus type 1 nucleocapsid Zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003, 77, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, S.; Mousnier, A.; Busschots, K.; Maroun, M.; Van Maele, B.; Tempe, D.; Vandekerckhove, L.; Moisant, F.; Ben-Slama, L.; Witvrouw, M.; et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005, 280, 25517–25523. [Google Scholar] [CrossRef] [PubMed]
- Thierry, S.; Munir, S.; Thierry, E.; Subra, F.; Leh, H.; Zamborlini, A.; Saenz, D.; Levy, D.N.; Lesbats, P.; Saib, A.; et al. Integrase inhibitor reversal dynamics indicate unintegrated HIV-1 DNA initiate de novo integration. Retrovirology 2015, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Saenz, D.T.; Meehan, A.; Wongthida, P.; Peretz, M.; Walker, W.H.; Teo, W.; Poeschla, E.M. An essential role for LEDGF/p75 in HIV integration. Science 2006, 314, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Shun, M.C.; Raghavendra, N.K.; Vandegraaff, N.; Daigle, J.E.; Hughes, S.; Kellam, P.; Cherepanov, P.; Engelman, A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007, 21, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, L.; Christ, F.; van Maele, B.; de Rijck, J.; Gijsbers, R.; van den Haute, C.; Witvrouw, M.; Debyser, Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006, 80, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rivera, J.A.; Bueno, M.T.; Morales, E.; Kugelman, J.R.; Rodriguez, D.F.; Llano, M. Implication of serine residues 271, 273 and 275 in the HIV-1 cofactor activity of LEDGF/p75. J. Virol 2009, 84, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Cohn, L.B.; Silva, I.T.; Oliveira, T.Y.; Rosales, R.A.; Parrish, E.H.; Learn, G.H.; Hahn, B.H.; Czartoski, J.L.; McElrath, M.J.; Lehmann, C.; et al. HIV-1 integration landscape during latent and active infection. Cell 2015, 160, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Meehan, A.M.; Saenz, D.T.; Morrison, J.H.; Garcia-Rivera, J.A.; Peretz, M.; Llano, M.; Poeschla, E.M. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009, 5, e1000522. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Cherepanov, P.; Daigle, J.E.; Engelman, A.; Lieberman, J. The set complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 2009, 5, e1000327. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, J.M.; Hughes, S.H. Retroviral reverse transcription and integration: Progress and problems. Annu. Rev. Cell Biol. 1992, 8, 275–306. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, J.M.; Kumar, R.; Hughes, S.H. Sequence of the circle junction of human immunodeficiency virus type 1: Implications for reverse transcription and integration. J. Virol. 1990, 64, 4903–4906. [Google Scholar] [PubMed]
- Furfine, E.S.; Reardon, J.E. Human immunodeficiency virus reverse transcriptase ribonuclease H: Specificity of tRNA(LYS3)-primer excision. Biochemistry 1991, 30, 7041–7046. [Google Scholar] [CrossRef] [PubMed]
- Pullen, K.A.; Ishimoto, L.K.; Champoux, J.J. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 1992, 66, 367–373. [Google Scholar] [PubMed]
- Smith, J.S.; Roth, M.J. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(LYS3). J. Biol. Chem. 1992, 267, 15071–15079. [Google Scholar] [PubMed]
- Smith, J.S.; Kim, S.Y.; Roth, M.J. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J. Virol. 1990, 64, 6286–6290. [Google Scholar] [PubMed]
- Julias, J.G.; McWilliams, M.J.; Sarafianos, S.G.; Arnold, E.; Hughes, S.H. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 9515–9520. [Google Scholar] [CrossRef] [PubMed]
- Jurriaans, S.; de Ronde, A.; Dekker, J.; Goudsmit, J.; Cornelissen, M. Analysis of human immunodeficiency virus type 1 LTR-LTR junctions in peripheral blood mononuclear cells of infected individuals. J. Gen. Virol. 1992, 73, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.; Drlica, K.; Pinter, A.; Murphy, E. Circular DNA of human immunodeficiency virus: Analysis of circle junction nucleotide sequences. J. Virol. 1991, 65, 551–555. [Google Scholar] [PubMed]
- Gorelick, R.J.; Fu, W.; Gagliardi, T.D.; Bosche, W.J.; Rein, A.; Henderson, L.E.; Arthur, L.O. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from moloney murine leukemia virus. J. Virol. 1999, 73, 8185–8195. [Google Scholar] [PubMed]
- McWilliams, M.J.; Julias, J.G.; Sarafianos, S.G.; Alvord, W.G.; Arnold, E.; Hughes, S.H. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect rnase h cleavage specificity and virus titer. J. Virol. 2003, 77, 11150–11157. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Dash, C.; Le Grice, S.F.; Prasad, V.R. Analysis of HIV-1 replication block due to substitutions at F61 residue of reverse transcriptase reveals additional defects involving the RNase H function. Nucleic Acids Res. 2006, 34, 2853–2863. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.D.; Bebenek, K.; Kunkel, T.A. The accuracy of reverse transcriptase from HIV-1. Science 1988, 242, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Vranckx, L.S.; Demeulemeester, J.; Saleh, S.; Boll, A.; Vansant, G.; Schrijvers, R.; Weydert, C.; Battivelli, E.; Verdin, E.; Cereseto, A.; et al. Ledgin-mediated inhibition of integrase-LEDGF/p75 interaction reduces reactivation of residual latent HIV. EBioMedicine 2016, 8, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Trinite, B.; Ohlson, E.C.; Voznesensky, I.; Rana, S.P.; Chan, C.N.; Mahajan, S.; Alster, J.; Burke, S.A.; Wodarz, D.; Levy, D.N. An HIV-1 replication pathway utilizing reverse transcription products that fail to integrate. J. Virol. 2013, 87, 12701–12720. [Google Scholar] [CrossRef] [PubMed]
- Gelderblom, H.C.; Vatakis, D.N.; Burke, S.A.; Lawrie, S.D.; Bristol, G.C.; Levy, D.N. Viral complementation allows HIV-1 replication without integration. Retrovirology 2008, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Hamid, F.B.; Kim, J.; Shin, C.G. Distribution and fate of HIV-1 unintegrated DNA species: A comprehensive update. AIDS Res. Ther. 2017, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Munir, S.; Thierry, S.; Subra, F.; Deprez, E.; Delelis, O. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology 2013, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Malet, I.; Calvez, V.; Marcelin, A.G. The future of integrase inhibitors of HIV-1. Curr. Opin. Virol. 2012, 2, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Tsiang, M.; Jones, G.S.; Niedziela-Majka, A.; Kan, E.; Lansdon, E.B.; Huang, W.; Hung, M.; Samuel, D.; Novikov, N.; Xu, Y.; et al. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J. Biol. Chem. 2012, 287, 21189–21203. [Google Scholar] [CrossRef] [PubMed]
- Schrijvers, R.; Vets, S.; de Rijck, J.; Malani, N.; Bushman, F.D.; Debyser, Z.; Gijsbers, R. HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology 2012, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jurado, K.A.; Wu, X.; Shun, M.C.; Li, X.; Ferris, A.L.; Smith, S.J.; Patel, P.A.; Fuchs, J.R.; Cherepanov, P.; et al. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012, 40, 11518–11530. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bueno, M.T.D.; Reyes, D.; Llano, M. LEDGF/p75 Deficiency Increases Deletions at the HIV-1 cDNA Ends. Viruses 2017, 9, 259. https://doi.org/10.3390/v9090259
Bueno MTD, Reyes D, Llano M. LEDGF/p75 Deficiency Increases Deletions at the HIV-1 cDNA Ends. Viruses. 2017; 9(9):259. https://doi.org/10.3390/v9090259
Chicago/Turabian StyleBueno, Murilo T. D., Daniel Reyes, and Manuel Llano. 2017. "LEDGF/p75 Deficiency Increases Deletions at the HIV-1 cDNA Ends" Viruses 9, no. 9: 259. https://doi.org/10.3390/v9090259
APA StyleBueno, M. T. D., Reyes, D., & Llano, M. (2017). LEDGF/p75 Deficiency Increases Deletions at the HIV-1 cDNA Ends. Viruses, 9(9), 259. https://doi.org/10.3390/v9090259




