Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis
Abstract
:1. Introduction
2. Marek’s Disease Virus Integration
3. MDV Telomeric Repeats
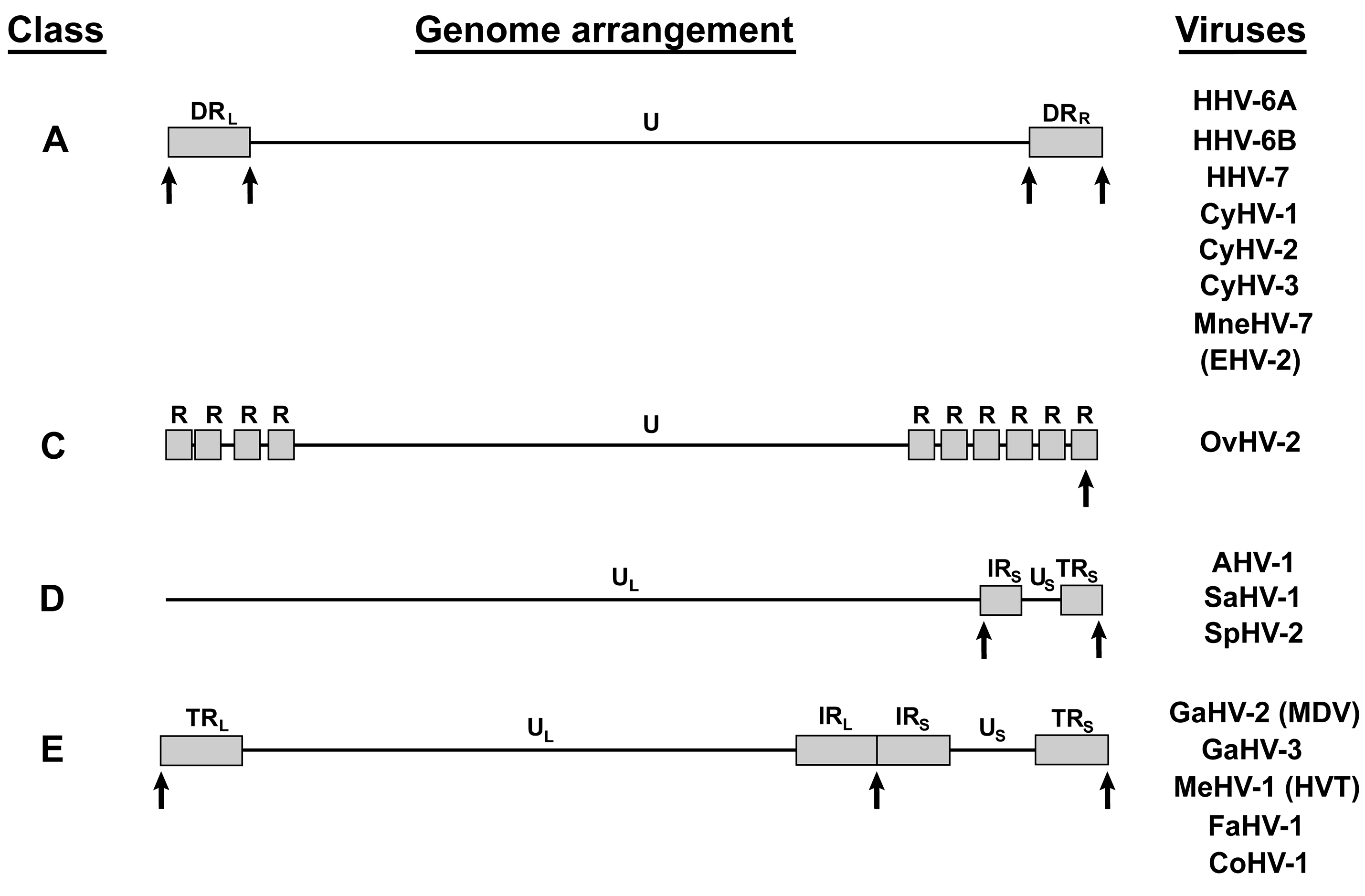
4. Other Telomere Herpesviruses
5. Factors Involved in Telomere Herpesvirus Integration
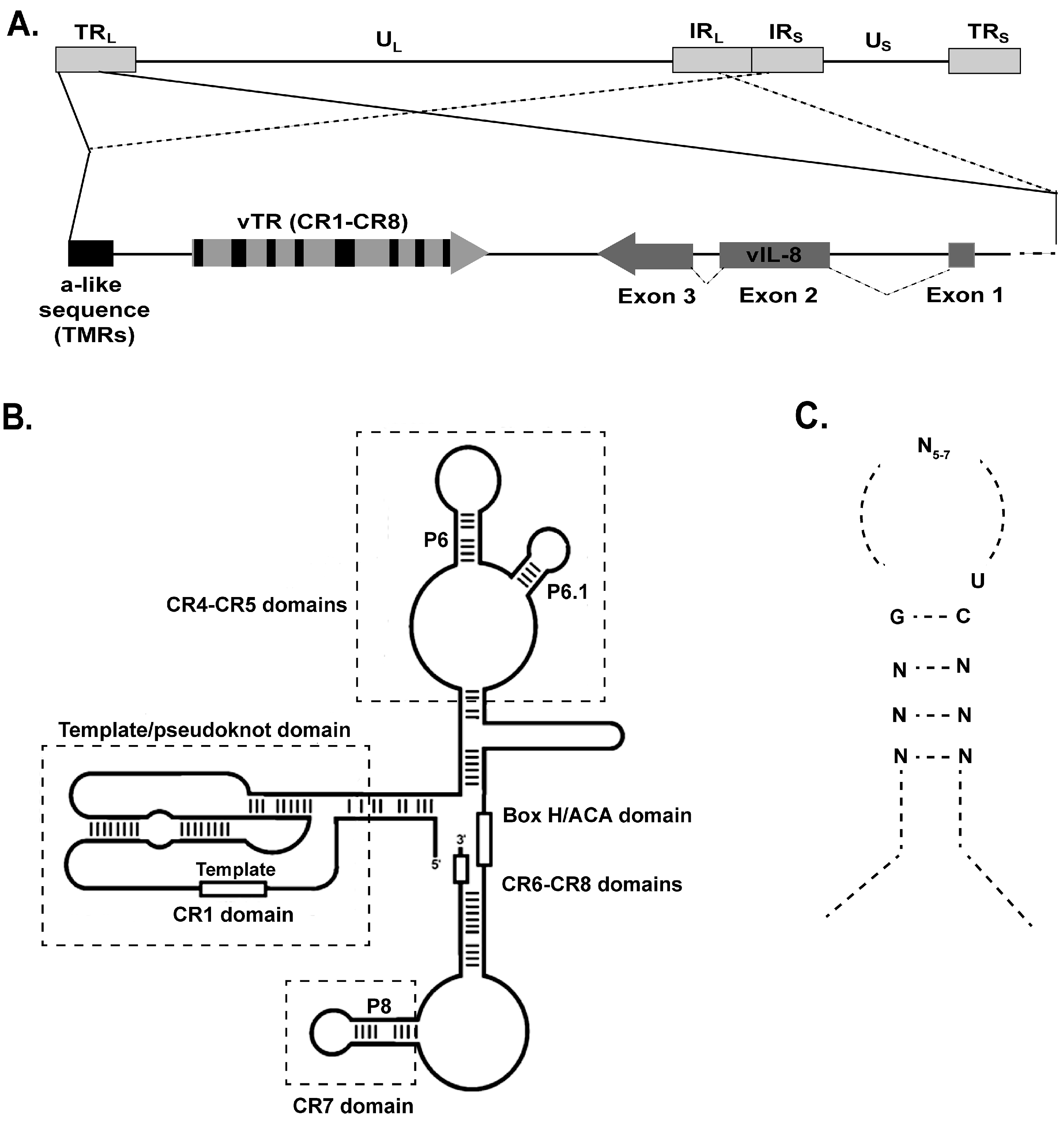
6. MDV Telomerase RNA (vTR)
7. Role of vTR in Tumorigenesis
8. Telomerase Independent Functions of vTR
9. vTR Expression Levels
10. vTR-Based Vaccines
11. Conclusions
Author Contributions
Conflicts of Interest
References
- Davison, T.F.; Nair, V. Marek’s Disease: An Evolving Problem; Elsevier: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Parcells, M.S.; Burnside, J.; Morgan, R.W. Marek’s disease virus-induced t-cell lymphomas. In Cancer Associated Viruses; Robertson, E.S., Ed.; Springer: New York, NY, USA, 2012; pp. 307–335. [Google Scholar]
- Witter, R.L. Increased virulence of Marek’s disease virus field isolates. Avian Dis. 1997, 41, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Lee, L.; Liu, J.L.; Kung, H.J.; Tillotson, J.K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors (published erratum appears in Proc. Natl. Acad. Sci. USA 1993, 90, 2556). Proc. Natl. Acad. Sci. USA 1992, 89, 4042–4046. [Google Scholar] [CrossRef] [PubMed]
- Parcells, M.S.; Lin, S.F.; Dienglewicz, R.L.; Majerciak, V.; Robinson, D.R.; Chen, H.C.; Wu, Z.; Dubyak, G.R.; Brunovskis, P.; Hunt, H.D.; et al. Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): Characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 2001, 75, 5159–5173. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.T.; Selvaraj, R.K.; Kamil, J.P.; Osterrieder, N.; Kaufer, B.B. Marek’s disease viral interleukin-8 (vIL-8) promotes lymphoma formation through targeted recruitment of B-cells and CD4+ CD25+ T-cells. J. Virol. 2012, 86, 8536–8545. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Lawrie, C.H.; Saunders, N.J.; Watson, M.; Nair, V. Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J. Gen. Virol. 2009, 90, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.W.; Osterrieder, N.; Nair, V.K.; Schat, K.A. Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 2005, 79, 11647–11659. [Google Scholar] [CrossRef] [PubMed]
- Jarosinski, K.W.; Schat, K.A. Multiple alternative splicing to exons II and III of viral interleukin-8 (vIL-8) in the Marek’s disease virus genome: The importance of vIL-8 exon I. Virus Genes. 2007, 34, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Kishi, M.; Harada, H.; Takahashi, M.; Tanaka, A.; Hayashi, M.; Nonoyama, M.; Josephs, S.F.; Buchbinder, A.; Schachter, F.; Ablashi, D.V. A repeat sequence, GGGTTA, is shared by DNA of human herpesvirus 6 and Marek’s disease virus. J. Virol. 1988, 62, 4824–4827. [Google Scholar] [PubMed]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus genome integration into telomeric repeats of host cell chromosomes. Annu. Rev. Virol. 2014, 1, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Fragnet, L.; Blasco, M.A.; Klapper, W.; Rasschaert, D. The RNA subunit of telomerase is encoded by Marek’s disease virus. J. Virol. 2003, 77, 5985–5996. [Google Scholar] [CrossRef] [PubMed]
- Trapp, S.; Parcells, M.S.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Kumar, P.M.; Nair, V.K.; Osterrieder, N. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2006, 203, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, B.B.; Trapp, S.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomerase RNA(vTR)-dependent lymphoma formation does not require interaction of vTR with telomerase reverse transcriptase (TERT). PLoS Pathog. 2010, 6, e1001073. [Google Scholar] [CrossRef] [PubMed]
- Schermuly, J.; Greco, A.; Hartle, S.; Osterrieder, N.; Kaufer, B.B.; Kaspers, B. In vitro model for lytic replication, latency, and transformation of an oncogenic alphaherpesvirus. Proc. Natl. Acad. Sci. USA 2015, 112, 7279–7284. [Google Scholar] [CrossRef] [PubMed]
- Delecluse, H.J.; Hammerschmidt, W. Status of Marek’s disease virus in established lymphoma cell lines: Herpesvirus integration is common. J. Virol. 1993, 67, 82–92. [Google Scholar] [PubMed]
- Kaufer, B.B.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 2011, 208, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Delecluse, H.J.; Schuller, S.; Hammerschmidt, W. Latent Marek’s disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993, 12, 3277–3286. [Google Scholar] [PubMed]
- Robinson, C.M.; Cheng, H.H.; Delany, M.E. Temporal kinetics of Marek’s disease herpesvirus: Integration occurs early after infection in both B and T cells. Cytogenet. Genome Res. 2014, 144, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.M.; Hunt, H.D.; Cheng, H.H.; Delany, M.E. Chromosomal integration of an avian oncogenic herpesvirus reveals telomeric preferences and evidence for lymphoma clonality. Herpesviridae 2010, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Fester, N.; Engel, A.T.; Kaufer, B.B. Role of the short telomeric repeat region in Marek’s disease virus replication, genomic integration, and lymphomagenesis. J. Virol. 2014, 88, 14138–14147. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, B.B. Detection of integrated herpesvirus genomes by fluorescence in situ hybridization (FISH). Methods Mol. Biol. 2013, 1064, 141–152. [Google Scholar] [PubMed]
- McPherson, M.C.; Cheng, H.H.; Delany, M.E. Marek’s disease herpesvirus vaccines integrate into chicken host chromosomes yet lack a virus-host phenotype associated with oncogenic transformation. Vaccine 2016, 34, 5554–5561. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Sanyal, A.; Pirzer, F.; Gravel, A.; Mori, Y.; Flamand, L.; Kaufer, B.B. The telomeric repeats of human herpesvirus 6A (HHV-6A) are required for efficient virus integration. PLoS Pathog. 2016, 12, e1005666. [Google Scholar] [CrossRef] [PubMed]
- Cebrian, J.; Kaschka-Dierich, C.; Berthelot, N.; Sheldrick, P. Inverted repeat nucleotide sequences in the genomes of Marek disease virus and the herpesvirus of the turkey. Proc. Natl. Acad. Sci. USA 1982, 79, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Nixon, D.E.; McVoy, M.A. Terminally repeated sequences on a herpesvirus genome are deleted following circularization but are reconstituted by duplication during cleavage and packaging of concatemeric DNA. J. Virol. 2002, 76, 2009–2013. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Dewhurst, S. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J. Virol. 1998, 72, 320–329. [Google Scholar] [PubMed]
- Arbuckle, J.H.; Medveczky, M.M.; Luka, J.; Hadley, S.H.; Luegmayr, A.; Ablashi, D.; Lund, T.C.; Tolar, J.; De Meirleir, K.; Montoya, J.G.; et al. The latent human herpesvirus-6a genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 5563–5568. [Google Scholar] [CrossRef] [PubMed]
- Prusty, B.K.; Gulve, N.; Rasa, S.; Murovska, M.; Hernandez, P.C.; Ablashi, D.V. Possible chromosomal and germline integration of human herpesvirus 7. J. General. Virol. 2017, 98, 266–274. [Google Scholar]
- Kaufer, B.B.; Flamand, L. Chromosomally integrated HHV-6: Impact on virus, cell and organismal biology. Curr. Opin. Virol. 2014, 9, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Ablashi, D.; Agut, H.; Alvarez-Lafuente, R.; Clark, D.A.; Dewhurst, S.; DiLuca, D.; Flamand, L.; Frenkel, N.; Gallo, R.; Gompels, U.A.; et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch. Virol. 2014, 159, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Ablashi, D.V.; Balachandran, N.; Josephs, S.F.; Hung, C.L.; Krueger, G.R.; Kramarsky, B.; Salahuddin, S.Z.; Gallo, R.C. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 1991, 184, 545–552. [Google Scholar] [CrossRef]
- Adams, M.J.; Carstens, E.B. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2012). Arch. Virol. 2012, 157, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Luppi, M.; Marasca, R.; Barozzi, P.; Ferrari, S.; Ceccherini-Nelli, L.; Batoni, G.; Merelli, E.; Torelli, G. Three cases of human herpesvirus-6 latent infection: Integration of viral genome in peripheral blood mononuclear cell DNA. J. Med. Virol. 1993, 40, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Torelli, G.; Barozzi, P.; Marasca, R.; Cocconcelli, P.; Merelli, E.; Ceccherini-Nelli, L.; Ferrari, S.; Luppi, M. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J. Med. Virol. 1995, 46, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Daibata, M.; Taguchi, T.; Taguchi, H.; Miyoshi, I. Integration of human herpesvirus 6 in a Burkitt’s lymphoma cell line. Br. J. Haematol. 1998, 102, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Nacheva, E.P.; Ward, K.N.; Brazma, D.; Virgili, A.; Howard, J.; Leong, H.N.; Clark, D.A. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J. Med. Virol. 2008, 80, 1952–1958. [Google Scholar] [CrossRef] [PubMed]
- Leong, H.N.; Tuke, P.W.; Tedder, R.S.; Khanom, A.B.; Eglin, R.P.; Atkinson, C.E.; Ward, K.N.; Griffiths, P.D.; Clark, D.A. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J. Med. Virol. 2007, 79, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.N.; Leong, H.N.; Thiruchelvam, A.D.; Atkinson, C.E.; Clark, D.A. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J. Clin. Microbiol. 2007, 45, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Potenza, L.; Barozzi, P.; Masetti, M.; Pecorari, M.; Bresciani, P.; Gautheret-Dejean, A.; Riva, G.; Vallerini, D.; Tagliazucchi, S.; Codeluppi, M.; et al. Prevalence of human herpesvirus-6 chromosomal integration (ciHHV-6) in italian solid organ and allogeneic stem cell transplant patients. Am. J. Transplant. 2009, 9, 1690–1697. [Google Scholar] [CrossRef] [PubMed]
- Hubacek, P.; Muzikova, K.; Hrdlickova, A.; Cinek, O.; Hyncicova, K.; Hrstkova, H.; Sedlacek, P.; Stary, J. Prevalence of HHV-6 integrated chromosomally among children treated for acute lymphoblastic or myeloid leukemia in the Czech Republic. J. Med. Virol. 2009, 81, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, U.; Lassner, D.; Wallaschek, N.; Gross, U.M.; Krueger, G.R.; Seeberg, B.; Kaufer, B.B.; Escher, F.; Poller, W.; Schultheiss, H.P. Chromosomally integrated human herpesvirus 6 in heart failure: Prevalence and treatment. Eur. J. Heart Fail. 2015, 17, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Brown, R.A.; Eid, A.J.; Razonable, R.R. Chromosomally integrated human herpesvirus-6 in kidney transplant recipients. Nephrol. Dial. Transplant. 2011, 26, 2391–2393. [Google Scholar] [CrossRef] [PubMed]
- Tanaka-Taya, K.; Sashihara, J.; Kurahashi, H.; Amo, K.; Miyagawa, H.; Kondo, K.; Okada, S.; Yamanishi, K. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J. Med. Virol. 2004, 73, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Pellett, P.E.; Ablashi, D.V.; Ambros, P.F.; Agut, H.; Caserta, M.T.; Descamps, V.; Flamand, L.; Gautheret-Dejean, A.; Hall, C.B.; Kamble, R.T.; et al. Chromosomally integrated human herpesvirus 6: Questions and answers. Rev. Med. Virol. 2012, 22, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Di Luca, D. Molecular biology and clinical associations of roseoloviruses human herpesvirus 6 and human herpesvirus 7. New Microbiol. 2007, 30, 173–187. [Google Scholar] [PubMed]
- De Bolle, L.; Naesens, L.; De Clercq, E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 2005, 18, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Gompels, U.A.; Macaulay, H.A. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J. Gen. Virol. 1995, 76, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Achour, A.; Malet, I.; Deback, C.; Bonnafous, P.; Boutolleau, D.; Gautheret-Dejean, A.; Agut, H. Length variability of telomeric repeat sequences of human herpesvirus 6 DNA. J. Virol. Meth. 2009, 159, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Liu, X.; Witter, R.L. Monoclonal antibodies with specificity for three different serotypes of Marek’s disease viruses in chickens. J. Immunol. (Baltimore MD 1950) 1983, 130, 1003–1006. [Google Scholar]
- Schumacher, A.J.; Mohni, K.N.; Kan, Y.; Hendrickson, E.A.; Stark, J.M.; Weller, S.K. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012, 8, e1002862. [Google Scholar] [CrossRef] [PubMed]
- Reuven, N.B.; Staire, A.E.; Myers, R.S.; Weller, S.K. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 2003, 77, 7425–7433. [Google Scholar] [CrossRef] [PubMed]
- Reuven, N.B.; Antoku, S.; Weller, S.K. The ul12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 2004, 78, 4599–4608. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.J.; Weindler, F.W.; Gray, D.; Schwaab, V.; Heilbronn, R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology 1994, 204, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.J.; Efstathiou, S.; Honess, R.W. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature 1991, 351, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Trempe, F.; Gravel, A.; Dubuc, I.; Wallaschek, N.; Collin, V.; Gilbert-Girard, S.; Morissette, G.; Kaufer, B.B.; Flamand, L. Characterization of human herpesvirus 6a/b U94 as atpase, helicase, exonuclease and DNA-binding proteins. Nucleic Acids Res. 2015, 43, 6084–6098. [Google Scholar] [CrossRef] [PubMed]
- Dhepakson, P.; Mori, Y.; Jiang, Y.B.; Huang, H.L.; Akkapaiboon, P.; Okuno, T.; Yamanishi, K. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J. Gen. Virol. 2002, 83, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Gravel, A.; Flamand, L.; Kaufer, B.B. The putative U94 integrase is dispensable for human herpesvirus 6 (HHV-6) chromosomal integration. J. Gen. Virol. 2016, 97, 1899–1903. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.K.; Park, D.; Xu, L.; Kleckner, N. DMC1: A meiosis-specific yeast homolog of E. coli reca required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 1992, 69, 439–456. [Google Scholar] [CrossRef]
- Tsuzuki, T.; Fujii, Y.; Sakumi, K.; Tominaga, Y.; Nakao, K.; Sekiguchi, M.; Matsushiro, A.; Yoshimura, Y.; Morita, T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 1996, 93, 6236–6240. [Google Scholar] [CrossRef] [PubMed]
- Arnaudeau, C.; Helleday, T.; Jenssen, D. The Rad51 protein supports homologous recombination by an exchange mechanism in mammalian cells. J. Mol. Biol. 1999, 289, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; Benson, F.E.; West, S.C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 1996, 87, 757–766. [Google Scholar] [CrossRef]
- Stark, J.M.; Pierce, A.J.; Oh, J.; Pastink, A.; Jasin, M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004, 24, 9305–9316. [Google Scholar] [CrossRef] [PubMed]
- McEachern, M.J.; Krauskopf, A.; Blackburn, E.H. Telomeres and their control. Annu. Rev. Genet. 2000, 34, 331–358. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.S.; Wright, W.E.; Shay, J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. MMBR 2002, 66, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, B.B.; Arndt, S.; Trapp, S.; Osterrieder, N.; Jarosinski, K.W. Herpesvirus telomerase RNA (vTR) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog. 2011, 7, e1002333. [Google Scholar] [CrossRef] [PubMed]
- Fragnet, L.; Kut, E.; Rasschaert, D. Comparative functional study of the viral telomerase RNA based on natural mutations. J. Biol. Chem. 2005, 280, 23502–23515. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Opperman, K.K.; Greider, C.W. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002, 30, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Toczyski, D.P.; Matera, A.G.; Ward, D.C.; Steitz, J.A. The epstein-barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein l22 in EBV-infected human B lymphocytes. Proc. Natl. Acad. Sci. USA 1994, 91, 3463–3467. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Cai, K.Q.; Stadanlick, J.E.; Greenberg-Kushnir, N.; Solanki-Patel, N.; Lee, S.Y.; Fahl, S.P.; Testa, J.R.; Wiest, D.L. Ribosomal protein Rpl22 controls the dissemination of T-cell lymphoma. Cancer Res. 2016, 76, 3387–3396. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Lee, S.Y.; Gutierrez, A.; Perrigoue, J.; Thapa, R.J.; Tu, Z.; Jeffers, J.R.; Rhodes, M.; Anderson, S.; Oravecz, T.; et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, lin28b. Blood 2012, 120, 3764–3773. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, M.N.; Schreiber, K.H.; Zhang, Y.; Duc, A.C.; Rao, S.; Hale, J.S.; Academia, E.C.; Shah, S.R.; Morton, J.F.; Holstein, C.A.; et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 2013, 9, e1003708. [Google Scholar] [CrossRef] [PubMed]
- Dobbelstein, M.; Shenk, T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with epstein-barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J. Virol. 1995, 69, 8027–8034. [Google Scholar] [PubMed]
- Elia, A.; Vyas, J.; Laing, K.G.; Clemens, M.J. Ribosomal protein L22 inhibits regulation of cellular activities by the epstein-barr virus small RNA EBER-1. Eur. J. Biochem. 2004, 271, 1895–1905. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, F.S.; Blackburn, E.H. An antiapoptotic role for telomerase RNA in human immune cells independent of telomere integrity or telomerase enzymatic activity. Blood 2014, 124, 3675–3684. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Nair, V.; Allday, M.J. Epigenetic regulation of the latency-associated region of Marek’s disease virus in tumor-derived T-cell lines and primary lymphoma. J. Virol. 2012, 86, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Chbab, N.; Egerer, A.; Veiga, I.; Jarosinski, K.W.; Osterrieder, N. Viral control of vTR expression is critical for efficient formation and dissemination of lymphoma induced by Marek’s disease virus (MDV). Vet. Res. 2010, 41, 56. [Google Scholar] [CrossRef] [PubMed]
- Shkreli, M.; Dambrine, G.; Soubieux, D.; Kut, E.; Rasschaert, D. Involvement of the oncoprotein c-myc in viral telomerase RNA gene regulation during Marek’s disease virus-induced lymphomagenesis. J. Virol. 2007, 81, 4848–4857. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.M.; Rivera, M.A.; Botchkina, I.L.; Shalaby, R.; Thor, A.D.; Blackburn, E.H. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 7982–7987. [Google Scholar] [CrossRef] [PubMed]
- Morissette, G.; Flamand, L. Herpesviruses and chromosomal integration. J. Virol. 2010, 84, 12100–12109. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Zou, Y.; Shay, J.W.; Wright, W.E. Telomere position effect in human cells. Science 2001, 292, 2075–2077. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kheimar, A.; Previdelli, R.L.; Wight, D.J.; Kaufer, B.B. Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis. Viruses 2017, 9, 173. https://doi.org/10.3390/v9070173
Kheimar A, Previdelli RL, Wight DJ, Kaufer BB. Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis. Viruses. 2017; 9(7):173. https://doi.org/10.3390/v9070173
Chicago/Turabian StyleKheimar, Ahmed, Renato L. Previdelli, Darren J. Wight, and Benedikt B. Kaufer. 2017. "Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis" Viruses 9, no. 7: 173. https://doi.org/10.3390/v9070173
APA StyleKheimar, A., Previdelli, R. L., Wight, D. J., & Kaufer, B. B. (2017). Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis. Viruses, 9(7), 173. https://doi.org/10.3390/v9070173





