Role of Envelopment in the HEV Life Cycle
Abstract
:1. Introduction
Envelopment of HEV: A Novel Mechanism for Non-Cytolytic Virus Release and Immune Evasion
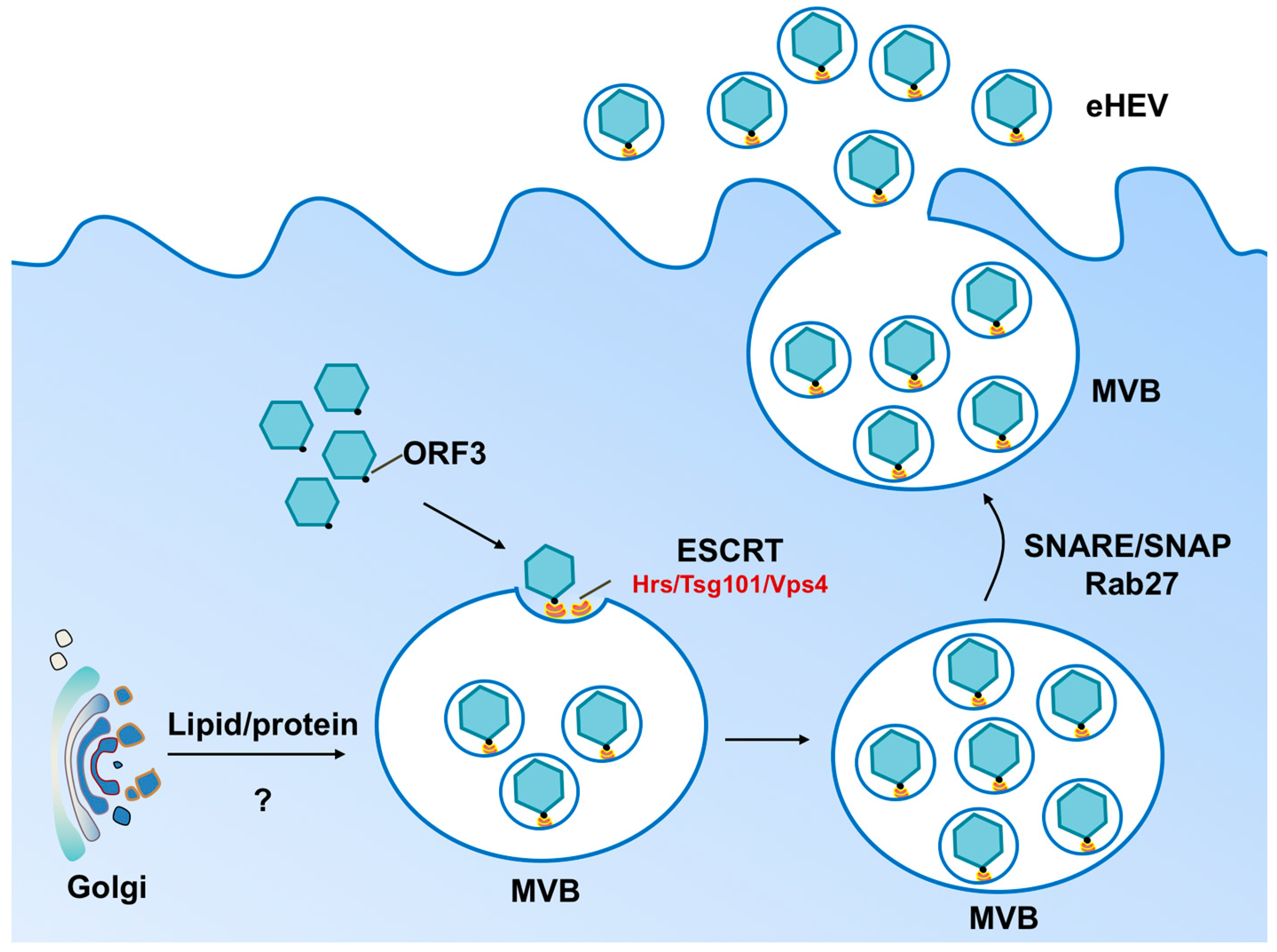
2. Biogenesis of Enveloped HEV Particles
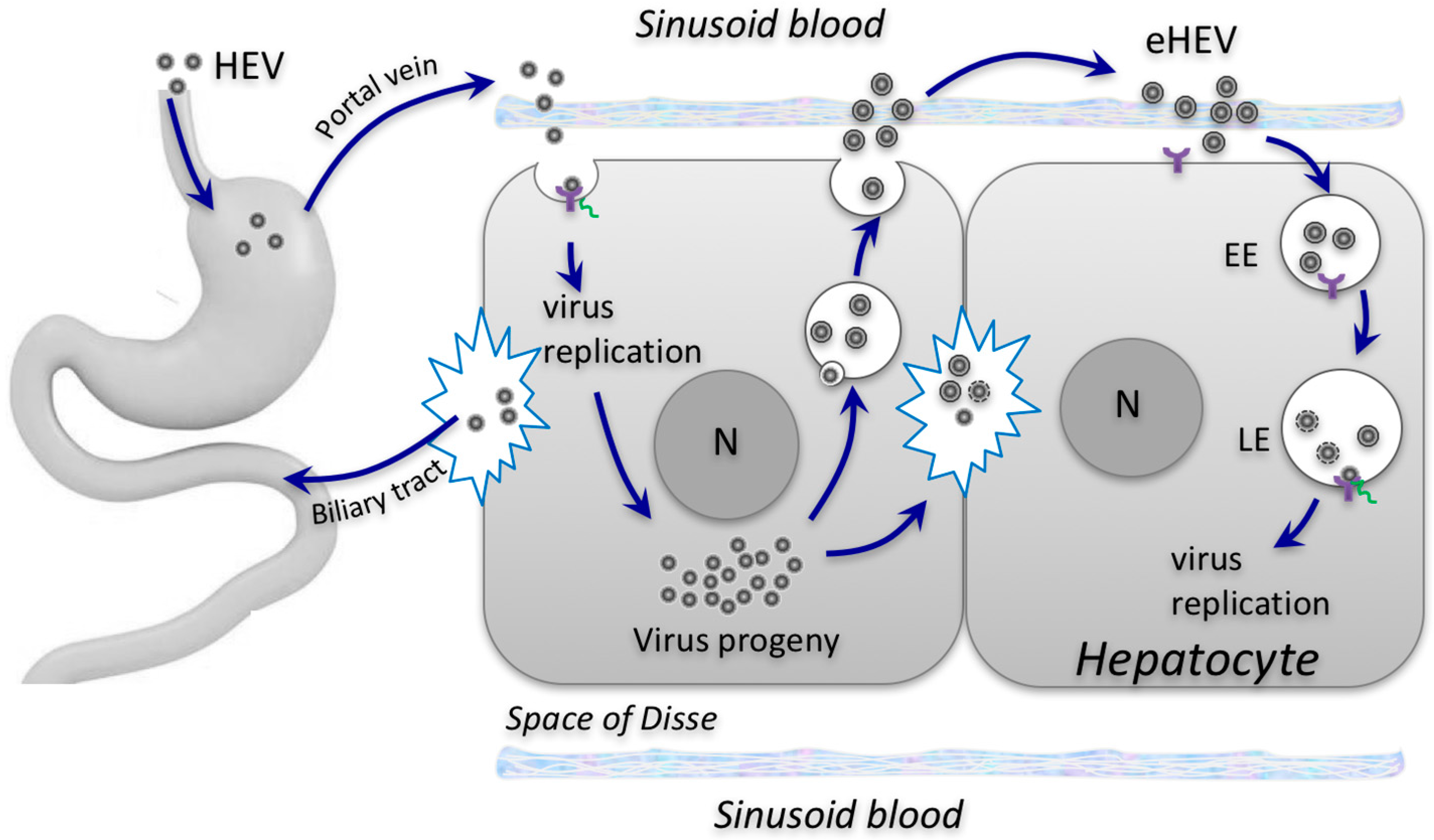
3. Impact of Envelopment on HEV Cell Entry and Spread
4. Impact of Envelopment on HEV Transmission between Hosts
5. Impact of Envelopment on Extrahepatic Manifestation Associated with HEV Infection
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Debing, Y.; Moradpour, D.; Neyts, J.; Gouttenoire, J. Update on hepatitis E virology: Implications for clinical practice. J. Hepatol. 2016, 65, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.R.; Kotwal, V.; Chawla, S. Chronic hepatitis E: A brief review. World J. Hepatol. 2015, 7, 2194–2201. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.Y.; Poovorawan, K.; Intharasongkroh, D.; Sa-Nguanmoo, P.; Vongpunsawad, S.; Chirathaworn, C.; Poovorawan, Y. Hepatitis E virus infection: Epidemiology and treatment implications. World J. Virol. 2015, 4, 343–355. [Google Scholar] [PubMed]
- Ahmad, I.; Holla, R.P.; Jameel, S. Molecular virology of hepatitis E virus. Semin. Liver Dis. 2013, 33, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, J.M.; Bendall, R.; Legrand-Abravanel, F.; Saune, K.; Miedouge, M.; Ellis, V.; Rech, H.; Destruel, F.; Kamar, N.; Dalton, H.R.; et al. Hepatitis E virus antibodies in blood donors, France. Emerg. Infect. Dis. 2011, 17, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, T.; Yoshizumi, T.; Itoh, S.; Harimoto, N.; Harada, N.; Ikegami, T.; Inagaki, Y.; Oshiro, Y.; Ohkohchi, N.; Okamoto, H.; et al. Chronic hepatitis E virus infection after living donor liver transplantation via blood transfusion: A case report. Surg. Case Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Perumpail, R.B.; Tummala, S.; Ahmed, A. Hepatitis E virus infection in the liver transplant recipients: Clinical presentation and management. World J. Hepatol. 2016, 8, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.E. Transmission of hepatitis E virus by transfusion: What is the risk? Transfusion 2014, 54, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.W. Enterically-transmitted non-A, non-B hepatitis. Br. Med. Bull. 1990, 46, 442–461. [Google Scholar] [CrossRef]
- Purcell, R.H.; Emerson, S.U. Hepatitis E: An emerging awareness of an old disease. J. Hepatol. 2008, 48, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Tanaka, T.; Takahashi, H.; Hoshino, Y.; Nagashima, S.; Jirintai; Mizuo, H.; Yazaki, Y.; Takagi, T.; Azuma, M.; et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: Characterization of HEV virions in blood circulation. J. Clin. Microbiol. 2010, 48, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H. Hepatitis E virus cell culture models. Virus Res. 2011, 161, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Reyes, G.R.; Purdy, M.A.; Kim, J.P.; Luk, K.C.; Young, L.M.; Fry, K.E.; Bradley, D.W. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science 1990, 247, 1335–1339. [Google Scholar] [CrossRef] [PubMed]
- Surjit, M.; Jameel, S.; Lal, S.K. The ORF2 protein of hepatitis E virus binds the 5′ region of viral RNA. J. Virol. 2004, 78, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Guu, T.S.; Liu, Z.; Ye, Q.; Mata, D.A.; Li, K.; Yin, C.; Zhang, J.; Tao, Y.J. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. USA 2009, 106, 12992–12997. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Tang, X.; Zhang, X.; Song, C.; Zheng, M.; Wang, K.; Zhang, J.; Ng, M.H.; Hew, C.L.; Li, S.; et al. Structural basis for the neutralization of hepatitis E virus by a cross-genotype antibody. Cell Res. 2015, 25, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.W.; Wu, T.; Zhao, Q.; Ng, M.H.; Xia, N.S. Hepatitis E virus: Neutralizing sites, diagnosis, and protective immunity. Rev. Med. Virol. 2012, 22, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, X.J.; Tang, Z.M.; Yang, F.; Wang, S.L.; Cai, W.; Zhang, K.; Xia, N.S.; Zheng, Z.Z. A comprehensive study of neutralizing antigenic sites on the hepatitis E virus (HEV) capsid by constructing, clustering, and characterizing a tool box. J. Biol. Chem. 2015, 290, 19910–19922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shih, J.W.; Xia, N.S. Long-term efficacy of a hepatitis E vaccine. N. Engl. J. Med. 2015, 372, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yamada, K.; Hoshino, Y.; Takahashi, H.; Ichiyama, K.; Tanaka, T.; Okamoto, H. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 2008, 153, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, F.; Zhang, L.; Harrison, T.J.; Huang, W.; Zhao, C.; Kong, W.; Jiang, C.; Wang, Y. Hepatitis E virus produced from cell culture has a lipid envelope. PLoS ONE 2015, 10, e0132503. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Lemon, S.M. Peek-a-boo: Membrane hijacking and the pathogenesis of viral hepatitis. Trends Microbiol. 2014, 22, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H. Culture systems for hepatitis E virus. J. Gastroenterol. 2013, 48, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.P.; Pudupakam, R.S.; Huang, Y.W.; Pierson, F.W.; LeRoith, T.; Meng, X.J. The PSAP motif within the ORF3 protein of an avian strain of the hepatitis E virus is not critical for viral infectivity in vivo but plays a role in virus release. J. Virol. 2012, 86, 5637–5646. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanaka, T.; Nishizawa, T.; Yasuda, J.; Okamoto, H. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J. Gen. Virol. 2011, 92, 2838–2848. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Jirintai; Tanaka, T.; Yamada, K.; Nishizawa, T.; Okamoto, H. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J. Gen. Virol. 2011, 92, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Babst, M.; Odorizzi, G.; Estepa, E.J.; Emr, S.D. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 2000, 1, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Jirintai, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Kouki, T.; Yashiro, T.; Okamoto, H. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J. Gen. Virol. 2014, 95, 2166–2175. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanggis; Kobayashi, T.; Nishizawa, T.; Okamoto, H. The membrane on the surface of hepatitis E virus particles is derived from the intracellular membrane and contains trans-Golgi network protein 2. Arch. Virol. 2014, 159, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M.; Chandra, V.; Rahman, S.A.; Sehgal, D.; Jameel, S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 2009, 83, 12714–12724. [Google Scholar] [CrossRef] [PubMed]
- Raff, A.B.; Woodham, A.W.; Raff, L.M.; Skeate, J.G.; Yan, L.; da Silva, D.M.; Schelhaas, M.; Kast, W.M. The evolving field of human papillomavirus receptor research: A review of binding and entry. J. Virol. 2013, 87, 6062–6072. [Google Scholar] [CrossRef] [PubMed]
- Urban, S.; Bartenschlager, R.; Kubitz, R.; Zoulim, F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology 2014, 147, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct entry mechanisms for non-enveloped and quasi-enveloped hepatitis E virus. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef] [PubMed]
- Allweiss, L.; Gass, S.; Giersch, K.; Groth, A.; Kah, J.; Volz, T.; Rapp, G.; Schöbel, A.; Lohse, A.W.; Polywka, S.; et al. Polywka S human liver chimeric mice as a new model of chronic hepatitis E virus infection and preclinical drug evaluation. J. Hepatol. 2016, 64, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Van de Garde, M.D.; Pas, S.D.; van der Net, G.; de Man, R.A.; Osterhaus, A.D.M.E.; Haagmans, B.L.; Boonstra, A.; Vanwolleghem, T. Hepatitis E virus (HEV) genotype 3 infection of human liver chimeric mice as a model for chronic HEV Infection. J. Virol. 2016, 90, 4394–4401. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Kato, K.; Li, T.; Takeda, N.; Miyamura, T.; Hammar, L.; Cheng, R.H. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology 1999, 265, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Kapur, N.; Thakral, D.; Durgapal, H.; Panda, S.K. Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J. Viral. Hepat. 2012, 19, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Emerson, S.U.; Nguyen, H.T.; Torian, U.; Burke, D.; Engle, R.; Purcell, R.H. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J. Virol. 2010, 84, 9059–9069. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Feng, Z.; (The Research Institute at Nationwide Children’s Hospital, Columbus, OH, USA). Manuscript in preparation. 2016. [Google Scholar]
- German Advisory Committee Blood (Arbeitskreis Blut), S.A.O.P.; Blood, T.B. Hepatitis E virus. Transfus. Med. Hemother. 2015, 42, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, P.E.; Ijaz, S.; Brailsford, S.R.; Brett, R.; Dicks, S.; Haywood, B.; Kennedy, I.T.R.; Kitchen, A.; Patel, P.; Poh, J.; et al. Hepatitis E virus in blood components: A prevalence and transmission study in southeast England. Lancet 2014, 384, 1766–1773. [Google Scholar] [CrossRef]
- Gallian, P.; Lhomme, S.; Piquet, Y.; Sauné, K.; Abravanel, F.; Assal, A.; Tiberghien, P.; Izopet, J. Hepatitis E virus infections in blood donors, France. Emerg. Infect. Dis. 2014, 20, 1914–1917. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M.; Verhoye, L.; Cocquerel, L.; Abravanel, F.; Foquet, L.; Montpellier, C.; Debing, Y.; Farhoudi, A.; Wychowski, C.; Dubuisson, J.; et al. Study of hepatitis E virus infection of genotype 1 and 3 in mice with humanised liver. Gut 2016. [Google Scholar] [CrossRef] [PubMed]
- Drave, S.A.; Debing, Y.; Walter, S.; Todt, D.; Engelmann, M.; Friesland, M.; Wedemeyer, H.; Neyts, J.; Behrendt, P.; Steinmann, E. Extra-hepatic replication and infection of hepatitis E virus in neuronal-derived cells. J. Viral. Hepat. 2016, 23, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Zhao, C.; Huang, W.; Harrison, T.J.; Zhang, H.; Geng, K.; Wang, Y. Detection and assessment of infectivity of hepatitis E virus in urine. J. Hepatol. 2016, 64, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Pischke, S.; Behrendt, P.; Manns, M.P.; Wedemeyer, H. HEV-associated cryoglobulinaemia and extrahepatic manifestations of hepatitis E. Lancet Infect. Dis. 2014, 14, 678–679. [Google Scholar] [CrossRef]
- Dalton, H.R.; Kamar, N.; van Eijk, J.J.; Mclean, B.N.; Cintas, P.; Bendall, R.P.; Jacobs, B.C. Hepatitis E virus and neurological injury. Nat. Rev. Neurol. 2016, 12, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.D.; Das, B.C.; Hazam, R.K.; Kumar, A.; Medhi, S.; Kar, P. Evidence of extrahepatic replication of hepatitis E virus in human placenta. J. Gen. Virol. 2014, 95, 1266–1271. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Lakhal, S.; Mäger, I.; Wood, M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Li, X.; Feng, Z. Role of Envelopment in the HEV Life Cycle. Viruses 2016, 8, 229. https://doi.org/10.3390/v8080229
Yin X, Li X, Feng Z. Role of Envelopment in the HEV Life Cycle. Viruses. 2016; 8(8):229. https://doi.org/10.3390/v8080229
Chicago/Turabian StyleYin, Xin, Xinlei Li, and Zongdi Feng. 2016. "Role of Envelopment in the HEV Life Cycle" Viruses 8, no. 8: 229. https://doi.org/10.3390/v8080229
APA StyleYin, X., Li, X., & Feng, Z. (2016). Role of Envelopment in the HEV Life Cycle. Viruses, 8(8), 229. https://doi.org/10.3390/v8080229







