Discovery of Novel Alphacoronaviruses in European Rodents and Shrews
Abstract
:1. Introduction
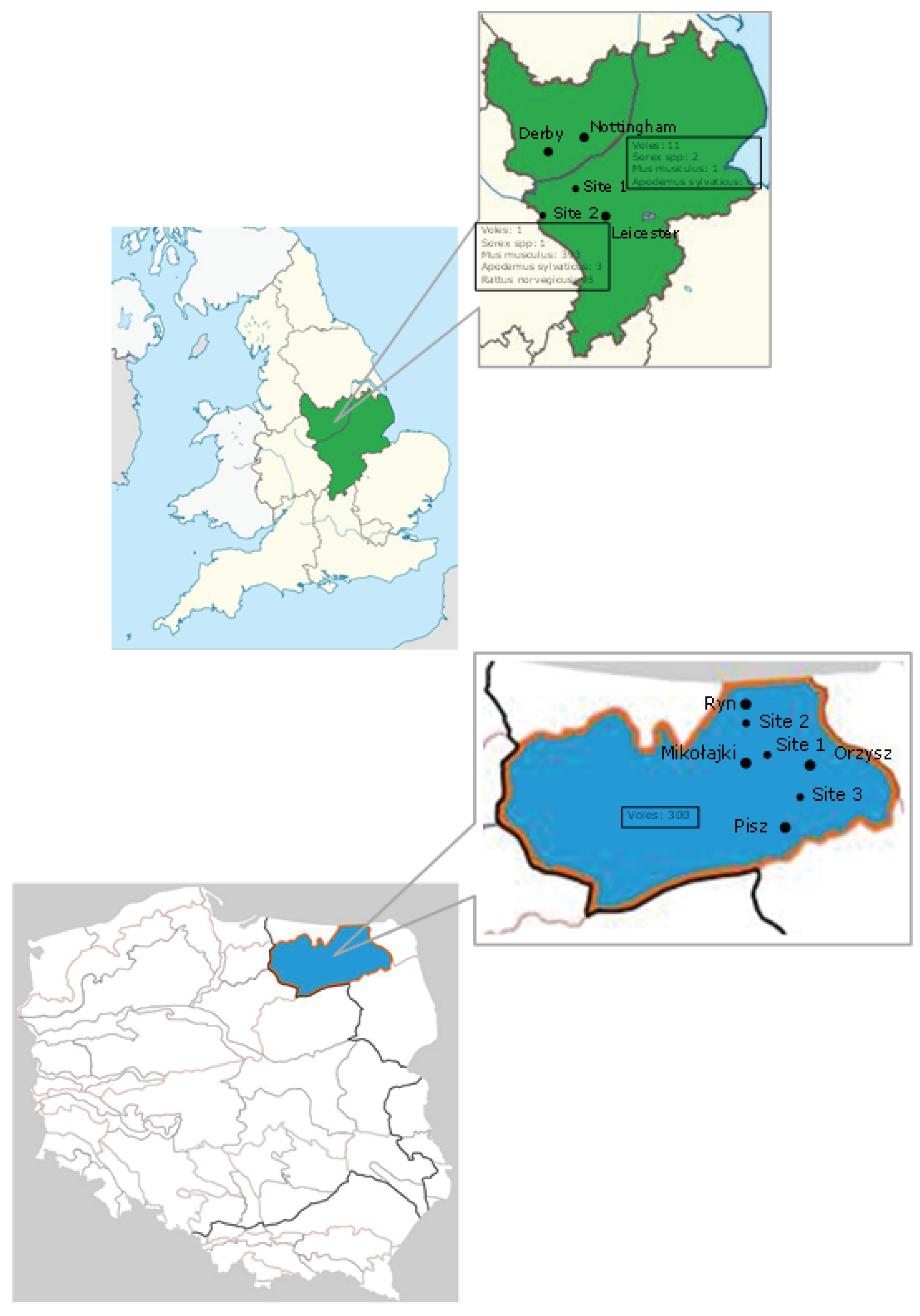
2. Materials and Methods
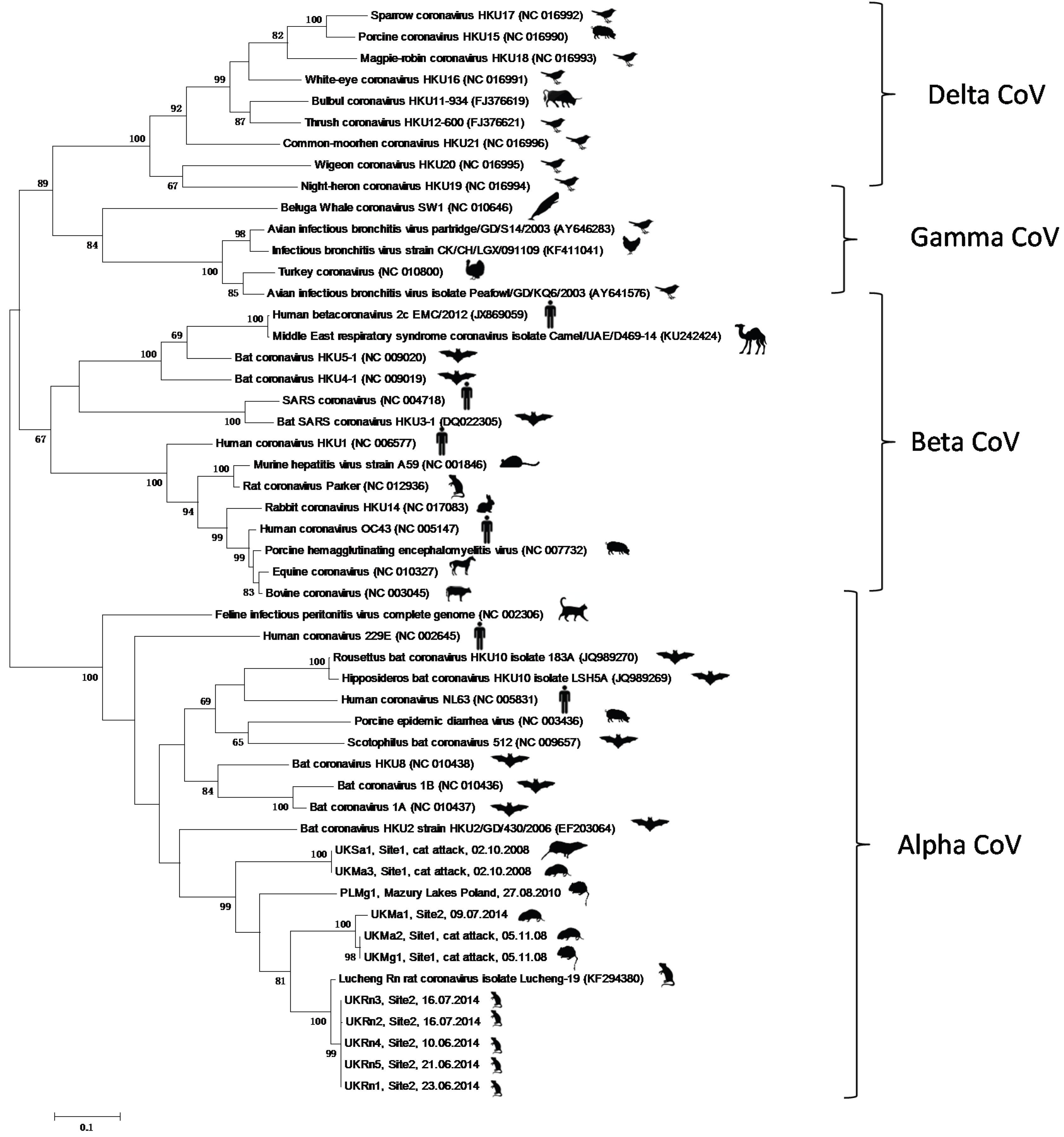
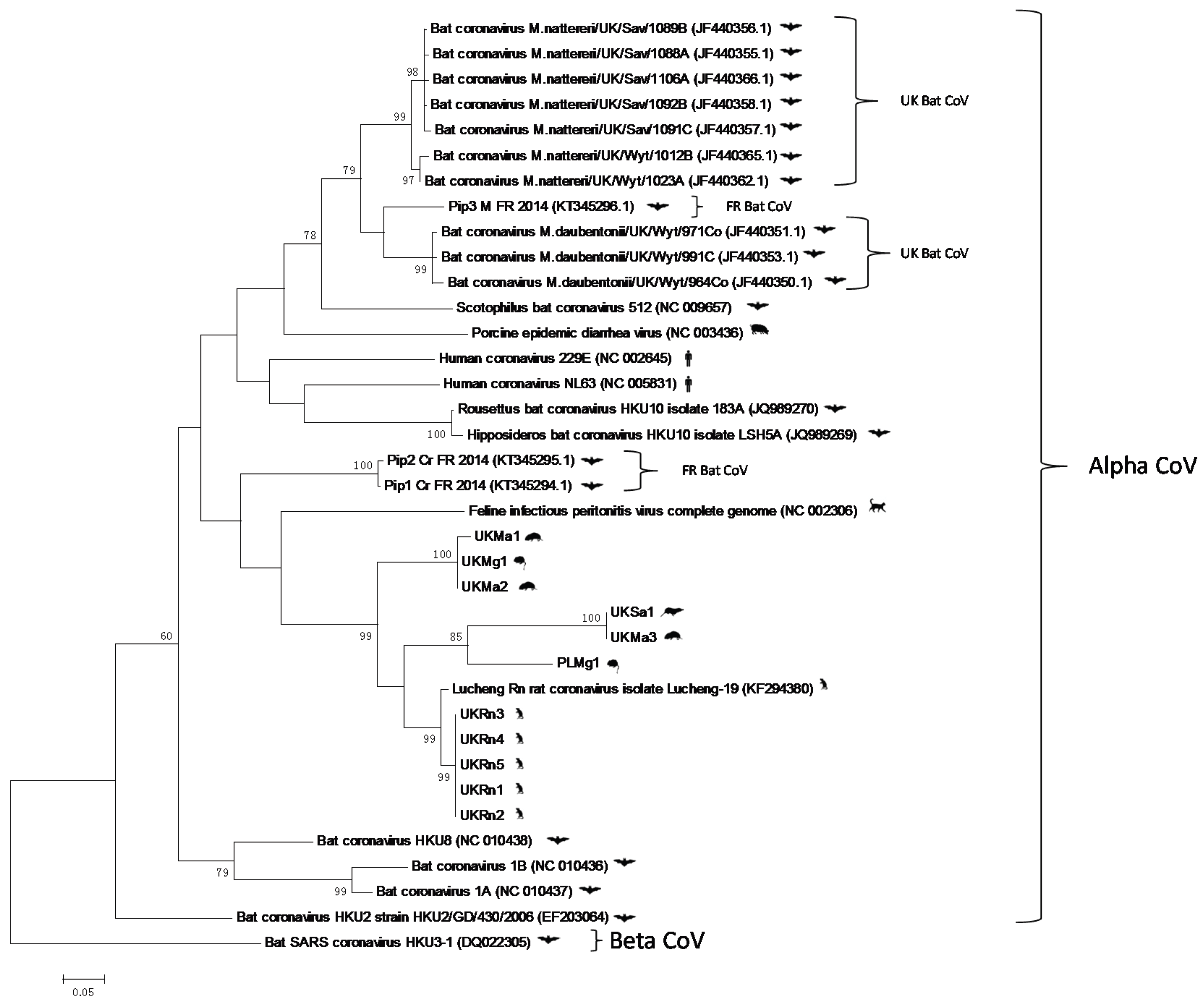
3. Results and Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hudson, C.; Beaudette, F.R. Infection of the cloaca with the virus of infectious bronchitis. Science 1932, 76, 34. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Navas-Martin, S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Frieman, M.B. Coronaviruses: Important Emerging Human Pathogens. J. Virol. 2014, 88, 5209–5212. [Google Scholar] [CrossRef] [PubMed]
- Bolles, M.; Donaldson, E.; Baric, R. SARS-CoV and Emergent Coronaviruses: Viral Determinants of Interspecies Transmission. Cur. Opin. Virol. 2011, 1, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, X.D.; Guo, W.P.; Zhou, R.H.; Wang, M.R.; Wang, C.Q.; Ge, S.; Mei, S.H.; Li, M.H.; Shi, M.; et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology 2015, 474, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Tsang, A.K.; Fan, R.Y.; Luk, H.K.; Cai, J.P.; Chan, K.H.; Zheng, B.J.; Wang, M.; et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J. Virol. 2015, 89, 3076–3092. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Kapusinszky, B.; Wang, C.; Rose, R.K.; Lipton, H.L.; Delwart, E.L. The fecal viral flora of wild rodents. PLoS Pathogens 2011, 7, e1002218. [Google Scholar] [CrossRef] [PubMed]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.N.; Childs, J.E. Ecologic studies of rodent reservoirs: Their relevance for human health. Emerg. Infect. Dis. 1998, 4, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Backhans, A.; Fellström, C. Rodents on pig and chicken farms—A potential threat to human and animal health. Infect. Ecol. Epidemiol. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Bajer, A.; Welc-Falęciak, R.; Bednarska, M.; Alsarraf, M.; Behnke-Borowczyk, J.; Siński, E.; Behnke, J.M. Long-Term Spatiotemporal Stability and Dynamic Changes in the Haemoparasite Community of Bank Voles (Myodes glareolus) in NE Poland. Microb. Ecol. 2014, 68, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Epstein, J.H.; Murray, K.A.; Navarrete-Macias, I.; Zambrana-Torrelio, C.M.; Solovyov, A.; Ojeda-Flores, R.; Arrigo, N.C.; Islam, A.; Ali Khan, S.; et al. A Strategy To Estimate Unknown Viral Diversity in Mammals. MBio 2013, 4, e00598-13. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Chu, C.M.; Chan, K.H.; Tsoi, H.W.; Huang, Y.; Wong, B.H.; Poon, R.W.; Cai, J.J.; Luk, W.K.; et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Halpin, R.; Wang, S.; Ghedin, E.; Spiro, D.J.; Saif, L.J. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J. Gen. Virol. 2011, 92, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Nollens, H.H.; Wellehan, J.F.; Archer, L.; Lowenstine, L.J.; Gulland, F.M. Detection of a respiratory coronavirus from tissues archived during a pneumonia epizootic in free-ranging Pacific harbor seals Phoca vitulina richardsii. Dis. Aquat. Organ 2010, 90, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.A.; Wang, P.; Gomaa, M.R.; El-Shesheny, R.; Kandeil, A.; Bagato, O.; Siu, L.Y.; Shehata, M.M.; Kayed, A.S.; Moatasim, Y.; et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013, 18, 574. [Google Scholar] [CrossRef]
- Reusken, C.B.; Haagmans, B.L.; Müller, M.A.; Gutierrez, C.; Godeke, G.J.; Meyer, B.; Muth, D.; Raj, V.S.; Smits-De Vries, L.; Corman, V.M.; et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infect. Dis. 2013, 13, 859–866. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Duengkae, P.; Rodpan, A.; Kaewpom, T.; Maneeorn, P.; Kanchanasaka, B.; Yingsakmongkon, S.; Sittidetboripat, N.; Chareesaen, C.; Khlangsap, N.; et al. Diversity of coronavirus in bats from Eastern Thailand. Virology J. 2015, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Goffard, A.; Demanche, C.; Arthur, L.; Pinçon, C.; Michaux, J.; Dubuisson, J. Alphacoronaviruses Detected in French Bats Are Phylogeographically Linked to Coronaviruses of European Bats. Viruses 2015, 7, 2937. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Gloza-Rausch, F.; Glende, J.; Corman, V.M.; Muth, D.; Goettsche, M.; Seebens, A.; Niedrig, M.; Pfefferle, S.; Yordanov, S.; et al. Genomic Characterization of Severe Acute Respiratory Syndrome-Related Coronavirus in European Bats and Classification of Coronaviruses Based on Partial RNA-Dependent RNA Polymerase Gene Sequences. J. Virol. 2010, 84, 11336–11349. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Ithete, N.L.; Richards, L.R.; Schoeman, M.C.; Preiser, W.; Drosten, C.; Drexler, J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014, 88, 11297–11303. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Hu, B.; Wang, B.; Wang, M.N.; Zhang, Q.; Zhang, W.; Wu, L.J.; Ge, X.Y.; Zhang, Y.Z.; Daszak, P.; et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of SARS coronavirus. J. Virol. 2015, 90, 3253–3256. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Dickerman, A.W.; Piñeyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 2013, 4, e00737-13. [Google Scholar] [CrossRef] [PubMed]
- August, T.A.; Mathews, F.; Nunn, M.A. Alphacoronavirus detected in bats in the United Kingdom. Vector Borne Zoonot. Dis. 2012, 12, 530–533. [Google Scholar] [CrossRef] [PubMed]

| Species | Location | Organ | Number | CoV Positives |
|---|---|---|---|---|
| Mus musculus | United Kingdom | Liver | 394 | 0 |
| Gut | 58 | |||
| Rattus norvegicus | United Kingdom | Liver | 95 | 5 ‡ |
| Gut | 28 | |||
| Microtus agrestis | United Kingdom | Liver | 11 | 3 |
| Myodes glareolus | United Kingdom | Liver | 1 | 1 |
| Poland | Liver | 300 | 1 | |
| Sorex araneus | United Kingdom | Liver | 3 | 1 |
| Apodemus sylvaticus | United Kingdom | Liver | 9 | 0 |
| Total animals | 813 | |||
| Total samples | 899 | |||
| Pairwise Percentage Similarity | ||||||
|---|---|---|---|---|---|---|
| Lucheng_Rn | UKRn1-5 | UK Ma1 | UKMa2/UKMg1 | UKSa1/UKMa3 | PLMg1 | |
| Lucheng_Rn | 97.6 | 83.8 | 84.6 | 78.4 | 81.8 | |
| UKRn1-5 | 98.6 | 83.7 | 84.6 | 77.8 | 81.3 | |
| UKMa1 | 96.2 | 95.7 | 97.6 | 76.7 | 79.4 | |
| UKMa2/UKMg1 | 96.2 | 95.7 | 100 | 76.2 | 80.3 | |
| UKSa1/UKMa3 | 91.9 | 91.9 | 90.5 | 90.5 | 79 | |
| PLMg1 | 94.3 | 93.8 | 93.8 | 93.8 | 89 | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsoleridis, T.; Onianwa, O.; Horncastle, E.; Dayman, E.; Zhu, M.; Danjittrong, T.; Wachtl, M.; Behnke, J.M.; Chapman, S.; Strong, V.; et al. Discovery of Novel Alphacoronaviruses in European Rodents and Shrews. Viruses 2016, 8, 84. https://doi.org/10.3390/v8030084
Tsoleridis T, Onianwa O, Horncastle E, Dayman E, Zhu M, Danjittrong T, Wachtl M, Behnke JM, Chapman S, Strong V, et al. Discovery of Novel Alphacoronaviruses in European Rodents and Shrews. Viruses. 2016; 8(3):84. https://doi.org/10.3390/v8030084
Chicago/Turabian StyleTsoleridis, Theocharis, Okechukwu Onianwa, Emma Horncastle, Emma Dayman, Miaoran Zhu, Taechasit Danjittrong, Marta Wachtl, Jerzy M. Behnke, Sarah Chapman, Victoria Strong, and et al. 2016. "Discovery of Novel Alphacoronaviruses in European Rodents and Shrews" Viruses 8, no. 3: 84. https://doi.org/10.3390/v8030084
APA StyleTsoleridis, T., Onianwa, O., Horncastle, E., Dayman, E., Zhu, M., Danjittrong, T., Wachtl, M., Behnke, J. M., Chapman, S., Strong, V., Dobbs, P., Ball, J. K., Tarlinton, R. E., & McClure, C. P. (2016). Discovery of Novel Alphacoronaviruses in European Rodents and Shrews. Viruses, 8(3), 84. https://doi.org/10.3390/v8030084






