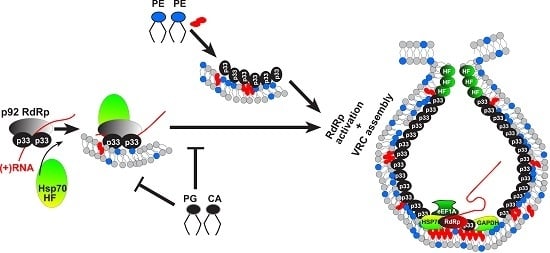
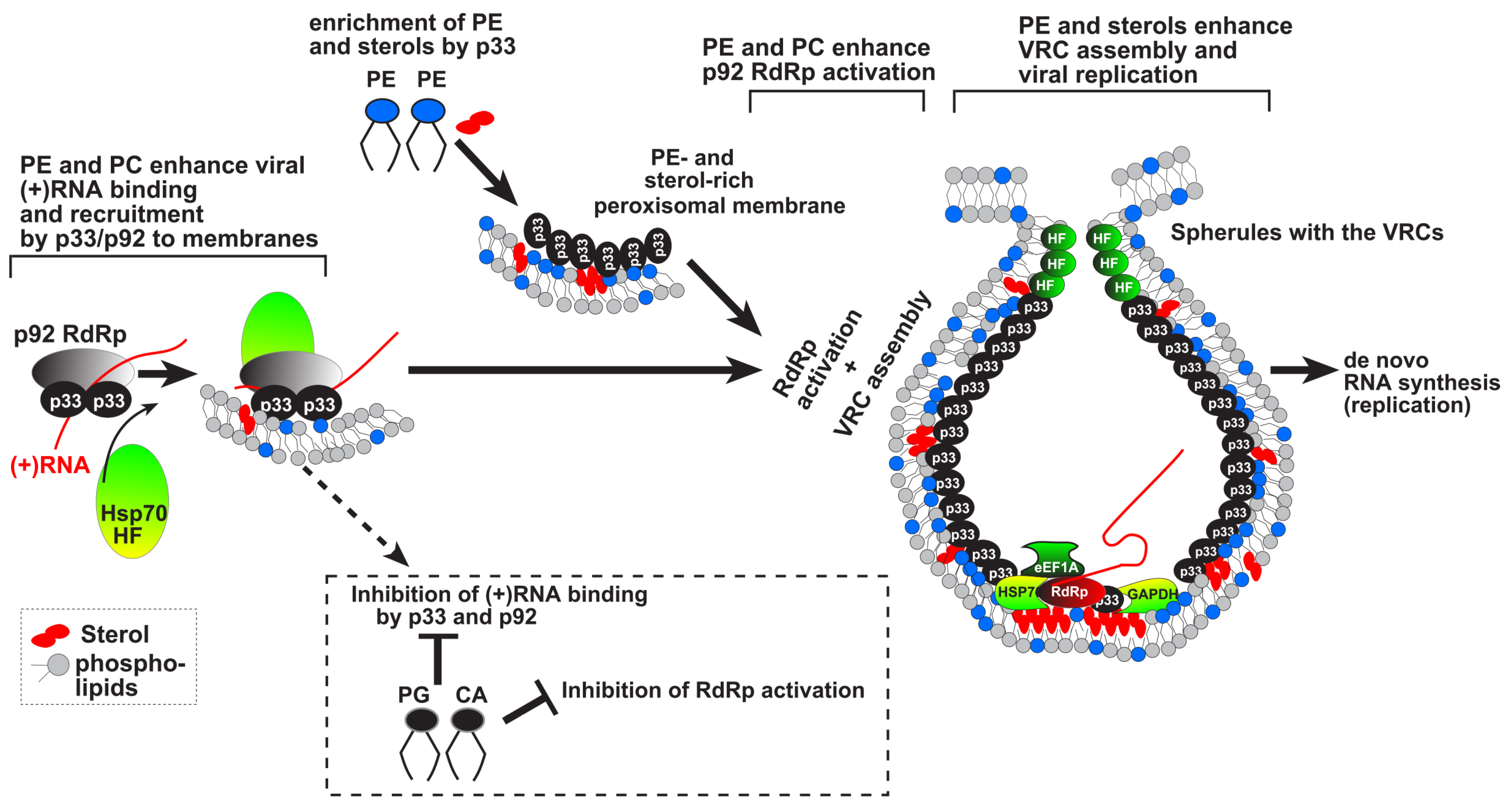
Cell-Free and Cell-Based Approaches to Explore the Roles of Host Membranes and Lipids in the Formation of Viral Replication Compartment Induced by Tombusviruses
Abstract
:1. Introduction
2. Formation of Membranous Viral Replication Compartments
3. Approaches to Identify and Characterize Co-Opted Lipids and Membranes Required for Replication of Plant Positive-Sense RNA Viruses
3.1. Genome-Wide and Proteome-Wide Approaches
3.2. Lipidomics-Based Approaches
3.3. Transcriptomic Analysis
3.4. Cell Biology-Based Approaches
3.5. Cell-Free Studies
4. Additional Plant (+)RNA Viruses
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous replication factories induced by plus-strand RNA viruses. Viruses 2014, 6, 2826–2857. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Hu, J.; Kozlov, M.M.; Rapoport, T.A. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 2009, 25, 329–354. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Burgyan, J.; Martelli, G.P. Molecular biology of tombusviridae. Adv. Virus Res. 1994, 44, 381–428. [Google Scholar] [PubMed]
- Barajas, D.; Jiang, Y.; Nagy, P.D. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009, 5, e1000705. [Google Scholar] [CrossRef] [PubMed]
- McCartney, A.W.; Greenwood, J.S.; Fabian, M.R.; White, K.A.; Mullen, R.T. Localization of the Tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 2005, 17, 3513–3531. [Google Scholar] [CrossRef] [PubMed]
- Pathak, K.B.; Sasvari, Z.; Nagy, P.D. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 2008, 379, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Panavas, T.; Hawkins, C.M.; Panaviene, Z.; Nagy, P.D. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 2005, 338, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Russo, M.; Pantaleo, V.; Rubino, L. Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication. J. Gen. Virol. 2006, 87, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Rochon, D.; Singh, B.; Reade, R.; Theilmann, J.; Ghoshal, K.; Alam, S.B.; Maghodia, A. The p33 auxiliary replicase protein of Cucumber necrosis virus targets peroxisomes and infection induces de novo peroxisome formation from the endoplasmic reticulum. Virology 2014, 452, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Huang, T.S.; Nagy, P.D. Authentic in vitro replication of two tombusviruses in isolated mitochondrial and endoplasmic reticulum membranes. J. Virol. 2012, 86, 12779–12794. [Google Scholar] [CrossRef] [PubMed]
- Weber-Lotfi, F.; Dietrich, A.; Russo, M.; Rubino, L. Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J. Virol. 2002, 76, 10485–10496. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.G.; Clendening, E.A.; Sheen, H.; Gidda, S.K.; White, K.A.; Mullen, R.T. A unique N-terminal sequence in the Carnation Italian ringspot virus p36 replicase-associated protein interacts with the host cell ESCRT-I component Vps23. J. Virol. 2014, 88, 6329–6344. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Barajas, D.; Qin, J.; Nagy, P.D. Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog. 2014, 10, e1003944. [Google Scholar] [CrossRef] [PubMed]
- Jonczyk, M.; Pathak, K.B.; Sharma, M.; Nagy, P.D. Exploiting alternative subcellular location for replication: Tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 2007, 362, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Laliberte, J.F.; Sanfacon, H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef] [PubMed]
- Hyodo, K.; Okuno, T. Pathogenesis mediated by proviral host factors involved in translation and replication of plant positive-strand RNA viruses. Curr. Opin. Virol. 2015, 17, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Den Boon, J.A.; Ahlquist, P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jin, X.; Zhang, X.; Li, Y.; Wang, C.; Wang, X.; Hong, J.; Li, D.; Zhang, Y. Morphogenesis of endoplasmic reticulum membrane-invaginated vesicles during Beet black scorch virus infection: Role of auxiliary replication protein and new implications of three-dimensional architecture. J. Virol. 2015, 89, 6184–6195. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Aix, C.; Garcia-Garcia, M.; Aranda, M.A.; Sanchez-Pina, M.A. Melon necrotic spot virus replication occurs in association with altered mitochondria. Mol. Plant Microbe Interact. 2015, 28, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Martin, I.F.; Pogany, J.; Risco, C.; Nagy, P.D. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of Tomato bushy stunt virus replicase. PLoS Pathog. 2014, 10, e1004087. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Zhang, J.; Ollwerther, A.; Wang, X.; Ahlquist, P. Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 2015, 11, e1004742. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Martin, I.F.; Pogany, J.; Barajas, D.; Pathak, K.; Risco, C.; Nagy, P.D. The role of viral RNA and co-opted cellular ESCRT-I and ESCRT-III factors in formation of tombusvirus spherules harboring the tombusvirus replicase. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Wang, X.; Ahlquist, P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. PNAS 2010, 107, 16291–16296. [Google Scholar] [CrossRef] [PubMed]
- Noueiry, A.O.; Ahlquist, P. Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 2003, 41, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J.; Lin, J.Y. How yeast can be used as a genetic platform to explore virus-host interactions: From “omics” to functional studies. Trends Microbiol. 2014, 22, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Panavas, T.; Serviene, E.; Nawaz-Ul-Rehman, M.S.; Nagy, P.D. A high-throughput approach for studying virus replication in yeast. Curr. Protoc. Microbiol. 2010, 19. [Google Scholar] [CrossRef]
- Nagy, P.D.; Pogany, J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 2006, 344, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Serviene, E.; Shapka, N.; Cheng, C.P.; Panavas, T.; Phuangrat, B.; Baker, J.; Nagy, P.D. Genome-wide screen identifies host genes affecting viral RNA recombination. PNAS 2005, 102, 10545–10550. [Google Scholar] [CrossRef] [PubMed]
- Panavas, T.; Serviene, E.; Brasher, J.; Nagy, P.D. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. PNAS 2005, 102, 7326–7331. [Google Scholar] [CrossRef] [PubMed]
- Kushner, D.B.; Lindenbach, B.D.; Grdzelishvili, V.Z.; Noueiry, A.O.; Paul, S.M.; Ahlquist, P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. PNAS 2003, 100, 15764–15769. [Google Scholar] [CrossRef] [PubMed]
- Gancarz, B.L.; Hao, L.; He, Q.; Newton, M.A.; Ahlquist, P. Systematic identification of novel, essential host genes affecting bromovirus RNA replication. PLoS ONE 2011, 6, e23988. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Serviene, E.; Gal, J.; Panavas, T.; Nagy, P.D. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 2006, 80, 7394–7404. [Google Scholar] [CrossRef] [PubMed]
- Shah Nawaz-Ul-Rehman, M.; Reddisiva Prasanth, K.; Baker, J.; Nagy, P.D. Yeast screens for host factors in positive-strand RNA virus replication based on a library of temperature-sensitive mutants. Methods 2013, 59, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Shah Nawaz-Ul-Rehman, M.; Martinez-Ochoa, N.; Pascal, H.; Sasvari, Z.; Herbst, C.; Xu, K.; Baker, J.; Sharma, M.; Herbst, A.; Nagy, P.D. Proteome-wide overexpression of host proteins for identification of factors affecting tombusvirus RNA replication: An inhibitory role of protein kinase C. J. Virol. 2012, 86, 9384–9935. [Google Scholar] [CrossRef] [PubMed]
- Serviene, E.; Jiang, Y.; Cheng, C.P.; Baker, J.; Nagy, P.D. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 2006, 80, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sasvari, Z.; Nagy, P.D. Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology 2011, 415, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sasvari, Z.; Nagy, P.D. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J. Virol. 2010, 84, 2270–2281. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pogany, J.; Panavas, T.; Xu, K.; Esposito, A.M.; Kinzy, T.G.; Nagy, P.D. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 2009, 385, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Barajas, D.; Panavas, T.; Herbst, D.A.; Nagy, P.D. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 2008, 82, 6911–6926. [Google Scholar] [CrossRef] [PubMed]
- Mendu, V.; Chiu, M.; Barajas, D.; Li, Z.; Nagy, P.D. Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology 2010, 406, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D. The roles of host factors in tombusvirus RNA recombination. Adv. Virus Res. 2011, 81, 63–84. [Google Scholar] [PubMed]
- Nagy, P.D.; Pogany, J. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv. Virus Res. 2010, 76, 123–177. [Google Scholar] [PubMed]
- Xu, K.; Nagy, P.D. RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc. Natl. Acad. Sci. USA 2015, 112, E1782–E1791. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Xu, K.; Sharma, M.; Wu, C.Y.; Nagy, P.D. Tombusviruses upregulate phospholipid biosynthesis via interaction between p33 replication protein and yeast lipid sensor proteins during virus replication in yeast. Virology 2014, 471, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Carman, G.M.; Han, G.S. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 2011, 80, 859–883. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.T.; McCartney, A.W.; Gidda, S.K.; Mullen, R.T. Localization of the Carnation Italian ringspot virus replication protein p36 to the mitochondrial outer membrane is mediated by an internal targeting signal and the TOM complex. BMC Cell Biol. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Xu, K.; de Castro Martin, I.F.; Sasvari, Z.; Brandizzi, F.; Risco, C.; Nagy, P.D. Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog. 2014, 10, e1004388. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.; Toulmay, A.; Prinz, W.A. Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 2015, 33, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Stork, J.; Li, Z.; Nagy, P.D. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. PNAS 2008, 105, 19956–19961. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Nagy, P.D. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 2008, 82, 5967–5980. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Nagy, P.D. p33-Independent activation of a truncated p92 RNA-dependent RNA polymerase of Tomato bushy stunt virus in yeast cell-free extract. J. Virol. 2012, 86, 12025–12038. [Google Scholar] [CrossRef] [PubMed]
- Pogany, J.; Nagy, P.D. Activation of Tomato bushy stunt virus RNA-dependent RNA polymerase by cellular heat shock protein 70 is enhanced by phospholipids in vitro. J. Virol. 2015, 89, 5714–5723. [Google Scholar] [CrossRef] [PubMed]
- Pathak, K.B.; Pogany, J.; Xu, K.; White, K.A.; Nagy, P.D. Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro. J. Virol. 2012, 86, 156–171. [Google Scholar] [CrossRef] [PubMed]
- Belov, G.A. Less grease, please. Phosphatidylethanolamine is the only lipid required for replication of a (+)RNA virus. Viruses 2015, 7, 3500–3505. [Google Scholar] [CrossRef] [PubMed]
- Hyodo, K.; Taniguchi, T.; Manabe, Y.; Kaido, M.; Mise, K.; Sugawara, T.; Taniguchi, H.; Okuno, T. Phosphatidic acid produced by phospholipase D promotes RNA replication of a plant RNA virus. PLoS Pathog. 2015, 11, e1004909. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M.; Ahlquist, P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 2003, 77, 12819–12828. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Diaz, A.; Mao, L.; Ahlquist, P.; Wang, X. Host acyl coenzyme A binding protein regulates replication complex assembly and activity of a positive-strand RNA virus. J. Virol. 2012, 86, 5110–5121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Z.; Chukkapalli, V.; Nchoutmboube, J.A.; Li, J.; Randall, G.; Belov, G.A.; Wang, X. Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc. Natl. Acad. Sci. USA 2016, 113, E1064–E1073. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Nagy, P.D. Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr. Opin. Virol. 2014, 9, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef] [PubMed]
| Gene | Cellular Function | Interaction |
|---|---|---|
| CHO2 | Phosphatidylethanolamine methyltransferase (PEMT) | |
| ERG4 | delta24 (24-1) sterol reductase | |
| ERG25 | ergosterol biosynthesis | |
| ERG10 | Acetyl-CoA C-acetyltransferase, mevalonate and sterol biosynthesis | |
| FAS2 | Alpha subunit of fatty acid synthetase | |
| FEN1 | Fatty acid elongase, involved in sphingolipid biosynthesis | |
| FOX2 | peroxisomal fatty acid beta-oxidation pathway | p33/vRNA |
| GPT2 | Glycerol-3-phosphate acyltransferase, involved in lipid biosynthesis | |
| INO2 | Transcription factor; required for derepression of phospholipid biosynthetic genes | |
| INO4 | Transcription factor; required for derepression of phospholipid biosynthetic genes | |
| MCT1 | S-malonyltransferase/fatty acid metabolism | |
| OLE1 | Fatty acid desaturase, required for monounsaturated fatty acid synthesis | |
| OPI1 | Transcriptional regulator, function in negative regulation of phospholipid biosynthetic genes | p33 |
| OSH3 | Member of an oxysterol-binding protein family, function in sterol metabolism | p33 |
| OSH5 | Member of an oxysterol-binding protein family, function in sterol metabolism | p33 |
| OSH6 | Member of an oxysterol-binding protein family, function in sterol metabolism | p33 |
| OSH7 | Member of an oxysterol-binding protein family, function in sterol metabolism | p33 |
| PAH1 | phosphatidate (PA) phosphatase; dephosphorylates PA to yield diacylglycerol | |
| POX1 | Fatty-acyl coenzyme A oxidase, fatty acid beta-oxidation pathway in the peroxisomes | |
| SCS2 | VAP homolog, ER-PM contact site, regulates phospholipid biosynthesis | p33 |
| SCS22 | VAP homolog, regulates phospholipid biosynthesis | |
| TGL2 | triacylglycerol lipase/lipid metabolism |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagy, P.D.; Pogany, J.; Xu, K. Cell-Free and Cell-Based Approaches to Explore the Roles of Host Membranes and Lipids in the Formation of Viral Replication Compartment Induced by Tombusviruses. Viruses 2016, 8, 68. https://doi.org/10.3390/v8030068
Nagy PD, Pogany J, Xu K. Cell-Free and Cell-Based Approaches to Explore the Roles of Host Membranes and Lipids in the Formation of Viral Replication Compartment Induced by Tombusviruses. Viruses. 2016; 8(3):68. https://doi.org/10.3390/v8030068
Chicago/Turabian StyleNagy, Peter D., Judit Pogany, and Kai Xu. 2016. "Cell-Free and Cell-Based Approaches to Explore the Roles of Host Membranes and Lipids in the Formation of Viral Replication Compartment Induced by Tombusviruses" Viruses 8, no. 3: 68. https://doi.org/10.3390/v8030068
APA StyleNagy, P. D., Pogany, J., & Xu, K. (2016). Cell-Free and Cell-Based Approaches to Explore the Roles of Host Membranes and Lipids in the Formation of Viral Replication Compartment Induced by Tombusviruses. Viruses, 8(3), 68. https://doi.org/10.3390/v8030068