In Vitro and In Vivo Models for the Study of Human Polyomavirus Infection
Abstract
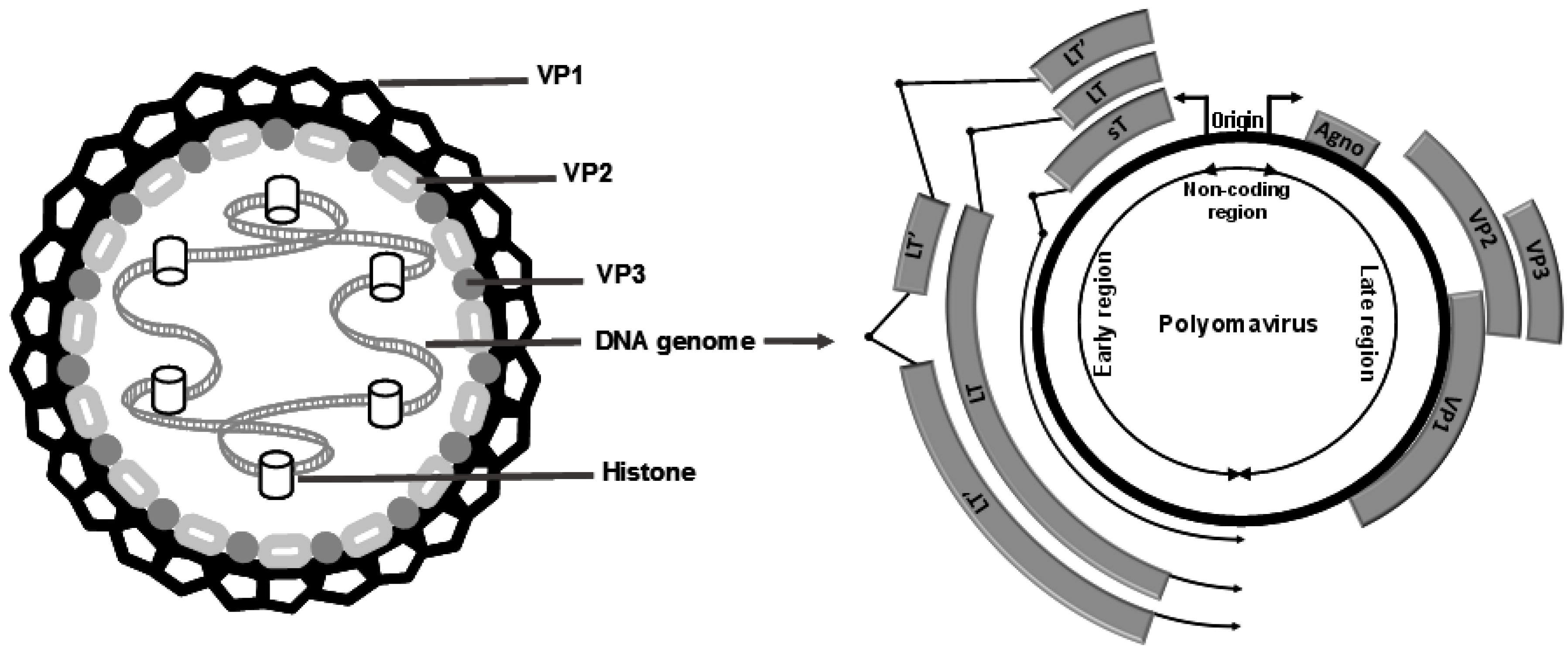
:1. The Growing Family of Human PyV
2. The PyV Life Cycle
3. PyV-Associated Pathologies
4. Experimental Model Systems to Study PyV Infection
4.1. VP1 Pentamers, PyV-Like Particles and PyV Pseudoparticles
4.2. Cell Culture-Derived Infectious PyV
4.3. Animal Models
4.3.1. Transgenic Mouse Model
4.3.2. Xenograft Mouse Model
4.3.3. Humanized Mouse Models
4.3.4. Mouse PyV Infection Model
4.3.5. Simian PyV SV40 Monkey Model
5. Conclusions and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- Gross, L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc Soc Exp Biol Med 1953, 83, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (BK) isolated from urine after renal transplantation. Lancet 1971, 297, 1253–1257. [Google Scholar] [CrossRef]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 297, 1257–1260. [Google Scholar] [CrossRef]
- Allander, T.; Andreasson, K.; Gupta, S.; Bjerkner, A.; Bogdanovic, G.; Persson, M.A.A.; Dalianis, T.; Ramqvist, T.; Andersson, B. Identification of a third human polyomavirus. J. Virol. 2007, 81, 4130–4136. [Google Scholar] [CrossRef] [PubMed]
- Gaynor, A.M.; Nissen, M.D.; Whiley, D.M.; Mackay, I.M.; Lambert, S.B.; Wu, G.; Brennan, D.C.; Storch, G.A.; Sloots, T.P.; Wang, D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007, 3, e64. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Schowalter, R.M.; Pastrana, D.V.; Pumphrey, K.A.; Moyer, A.L.; Buck, C.B. Merkel cell polyomavirus and two novel polyomaviruses are chronically shed from human skin. Cell Host Microbe 2010, 7, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Van der Meijden, E.; Janssens, R.W.A.; Lauber, C.; Bouwes Bavinck, J.N.; Gorbalenya, A.E.; Feltkamp, M.C.W. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010, 6, e1001024. [Google Scholar] [CrossRef] [PubMed]
- Scuda, N.; Hofmann, J.; Calvignac-Spencer, S.; Ruprecht, K.; Liman, P.; Kühn, J.; Hengel, H.; Ehlers, B. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 2011, 85, 4586–4590. [Google Scholar] [CrossRef] [PubMed]
- Siebrasse, E.A.; Reyes, A.; Lim, E.S.; Zhao, G.; Mkakosya, R.S.; Manary, M.J.; Gordon, J.I.; Wang, D. Identification of MW polyomavirus, a novel polyomavirus in human stool. J. Virol. 2012, 86, 10321–10326. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Phan, G.Q.; Raiji, M.T.; Murphy, P.M.; McDermott, D.H.; McBride, A.A. Complete genome sequence of a tenth human polyomavirus. J. Virol. 2012, 86, 10887. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Greninger, A.L.; Isa, P.; Phan, T.G.; Martínez, M.A.; de la Luz Sanchez, M.; Contreras, J.F.; Santos-Preciado, J.I.; Parsonnet, J.; Miller, S.; et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS ONE 2012, 7, e49449. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Reyes, A.; Antonio, M.; Saha, D.; Ikumapayi, U.N.; Adeyemi, M.; Stine, O.C.; Skelton, R.; Brennan, D.C.; Mkakosya, R.S.; et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 2013, 436, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Korup, S.; Rietscher, J.; Calvignac-Spencer, S.; Trusch, F.; Hofmann, J.; Moens, U.; Sauer, I.; Voigt, S.; Schmuck, R.; Ehlers, B. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS ONE 2013, 8, e58021. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Pereira, M.; Rhodes, R.H.; An, P.; Pipas, J.M.; Jain, K.; Kapoor, A.; Briese, T.; Faust, P.L.; Lipkin, W.I. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J. Infect. Dis. 2014, 210, 1595–1599. [Google Scholar] [CrossRef] [PubMed]
- DeCaprio, J.A.; Garcea, R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013, 11, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Nicol, J.T.J.; Leblond, V.; Arnold, F.; Guerra, G.; Mazzoni, E.; Tognon, M.; Coursaget, P.; Touzé, A. Seroprevalence of Human Malawi Polyomavirus. J. Clin. Microbiol. 2014, 52, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Meinerz, N.M.; Primi, B.; Wang, D.; Garcea, R.L. Common exposure to STL polyomavirus during childhood. Emerg. Infect. Dis. 2014, 20, 1559–1561. [Google Scholar] [CrossRef] [PubMed]
- Van der Meijden, E.; Bialasiewicz, S.; Rockett, R.J.; Tozer, S.J.; Sloots, T.P.; Feltkamp, M.C.W. Different serologic behavior of MCPyV, TSPyV, HPyV6, HPyV7 and HPyV9 polyomaviruses found on the skin. PLoS ONE 2013, 8, e81078. [Google Scholar] [CrossRef] [PubMed]
- Gossai, A.; Waterboer, T.; Nelson, H.H.; Michel, A.; Willhauck-Fleckenstein, M.; Farzan, S.F.; Hoen, A.G.; Christensen, B.C.; Kelsey, K.T.; Marsit, C.J.; et al. Seroepidemiology of human polyomaviruses in a US population. Am. J. Epidemiol. 2016, 183, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Berrios, C.; Jung, J.; Primi, B.; Wang, M.; Pedamallu, C.; Duke, F.; Marcelus, C.; Cheng, J.; Garcea, R.L.; Meyerson, M.; et al. Malawi polyomavirus is a prevalent human virus that interacts with known tumor suppressors. J. Virol. 2015, 89, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Cubitt, C. Molecular Genetics of the BK Virus. In Polyomaviruses and Human Diseases; Ahsan, N., Ed.; Springer: New York, NY, USA, 2006; Volume 577, pp. 85–95. [Google Scholar]
- Pavesi, A. African origin of polyomavirus JC and implications for prehistoric human migrations. J. Mol. Evol. 2003, 56, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Karalic, D.; Lazarevic, I.; Knezevic, A.; Cupic, M.; Jevtovic, D.; Jovanovic, T. Distribution of JC virus genotypes among serbian patients infected with HIV and in healthy donors. J. Med. Virol. 2014, 86, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Zanotta, N.; Delbue, S.; Rossi, T.; Pelos, G.; D’Agaro, P.; Monasta, L.; Ferrante, P.; Comar, M. Molecular epidemiology of JCV genotypes in patients and healthy subjects from Northern Italy. J. Med. Virol. 2013, 85, 1286–1292. [Google Scholar] [CrossRef] [PubMed]
- Chehadeh, W.; Kurien, S.S.; Nampoory, M.R. Molecular characterization of BK and JC viruses circulating among potential kidney donors in Kuwait. BioMed Res. Int. 2013, 2013, 683464. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, J.; Deckhut, A.; Joseph, B.; Eberwein, P.; Cubitt, C.; Ryschkewitsch, C.; Agostini, H.; Stoner, G. Chinese strains (Type 7) of JC Virus are afro-asiatic in origin but are phylogenetically distinct from the mongolian and indian strains (Type 2D) and the korean and japanese strains (Type 2A). J. Mol. Evol. 2004, 58, 568–583. [Google Scholar] [PubMed]
- Jobes, D.V.; Friedlaender, J.S.; Mgone, C.S.; Agostini, H.T.; Koki, G.; Yanagihara, R.; Ng, T.C.N.; Chima, S.C.; Ryschkewitsch, C.F.; Stoner, G.L. New JC virus (JCV) genotypes from Papua New Guinea and Micronesia (Type 8 and Type 2E) and evolutionary analysis of 32 complete JCV genomes. Arch. Virol. 2001, 146, 2097–2113. [Google Scholar] [CrossRef]
- Cubitt, C.; Cui, X.; Agostini, H.; Nerurkar, V.; Scheirich, I.; Yanagihara, R.; Ryschkewitsch, C.; Stoner, G. Predicted amino acid sequences for 100 JCV strains. J. Neurovirol. 2001, 7, 339–344. [Google Scholar] [PubMed]
- Schowalter, R.M.; Buck, C.B. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog. 2013, 9, e1003558. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.J.; Fink, L.H.L.; O’Hara, B.; Atwood, W.J.; Sullivan, C.S. Evolutionarily conserved function of a viral microRNA. J. Virol. 2008, 82, 9823–9828. [Google Scholar] [CrossRef] [PubMed]
- Theiss, J.M.; Gunther, T.; Alawi, M.; Neumann, F.; Tessmer, U.; Fischer, N.; Grundhoff, A. A comprehensive analysis of replicating merkel cell polyomavirus genomes delineates the viral transcription program and suggests a role for mcv-miR-M1 in episomal persistence. PLoS Pathog. 2015, 11, e1004974. [Google Scholar] [CrossRef] [PubMed]
- Rocca, A.; Martelli, F.; Delbue, S.; Ferrante, P.; Bartolozzi, D.; Azzi, A.; Giannecchini, S. The JCPYV DNA load inversely correlates with the viral microrna expression in blood and cerebrospinal fluid of patients at risk of PML. J. Clin. Virol. 2015, 70, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, T.; Nummi, M.; Auvinen, P.; Mannonen, L.; Auvinen, E. Expression of BKV and JCV encoded microRNA in human cerebrospinal fluid, plasma and urine. J. Clin. Virol. 2015, 65, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Low, J.A.; Magnuson, B.; Tsai, B.; Imperiale, M.J. Identification of gangliosides GD1b and GT1b as receptors for BK Virus. J. Virol. 2006, 80, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Allen, S.A.; Blaum, B.S.; Liu, Y.; Frank, M.; Palma, A.S.; Stroh, L.J.; Feizi, T.; Peters, T.; Atwood, W.J.; et al. A structure-guided mutation in the major capsid protein retargets BK polyomavirus. PLoS Pathog. 2013, 9, e1003688. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Maginnis, M.S.; Palma, A.S.; Ströh, L.J.; Nelson, C.D.S.; Feizi, T.; Atwood, W.J.; Stehle, T. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 2010, 8, 309–319. [Google Scholar] [CrossRef]
- Stroh, L.J.; Maginnis, M.S.; Blaum, B.S.; Nelson, C.D.; Neu, U.; Gee, G.V.; O’Hara, B.A.; Motamedi, N.; DiMaio, D.; Atwood, W.J.; et al. The greater affinity of JC polyomavirus capsid for alpha2,6-linked lactoseries tetrasaccharide c than for other sialylated glycans is a major determinant of infectivity. J. Virol. 2015, 89, 6364–6375. [Google Scholar] [CrossRef]
- Elphick, G.F.; Querbes, W.; Jordan, J.A.; Gee, G.V.; Eash, S.; Manley, K.; Dugan, A.; Stanifer, M.; Bhatnagar, A.; Kroeze, W.K.; et al. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 2004, 306, 1380–1383. [Google Scholar] [CrossRef] [PubMed]
- Assetta, B.; Maginnis, M.S.; Gracia Ahufinger, I.; Haley, S.A.; Gee, G.V.; Nelson, C.D.; O’Hara, B.A.; Allen Ramdial, S.A.; Atwood, W.J. 5-HT2 receptors facilitate JC polyomavirus entry. J. Virol. 2013, 87, 13490–13498. [Google Scholar] [CrossRef] [PubMed]
- Stroh, L.J.; Gee, G.V.; Blaum, B.S.; Dugan, A.S.; Feltkamp, M.C.; Atwood, W.J.; Stehle, T. Trichodysplasia spinulosa-associated polyomavirus uses a displaced binding site on VP1 to engage sialylated glycolipids. PLoS Pathog. 2015, 11, e1005112. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.M.; Liu, Y.; Neu, U.; Gilbert, M.; Ehlers, B.; Feizi, T.; Stehle, T. Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-Glycolyl neuraminic acid as a receptor candidate. J. Virol. 2014, 88, 6100–6111. [Google Scholar] [CrossRef] [PubMed]
- Erickson, K.D.; Garcea, R.L.; Tsai, B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J. Virol. 2009, 83, 10275–10279. [Google Scholar] [CrossRef] [PubMed]
- Schowalter, R.M.; Pastrana, D.V.; Buck, C.B. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011, 7, e1002161. [Google Scholar] [CrossRef] [PubMed]
- Ströh, L.J.; Neu, U.; Blaum, B.S.; Buch, M.H.C.; Garcea, R.L.; Stehle, T. Structure analysis of the major capsid proteins of human polyomaviruses 6 and 7 reveals an obstructed sialic acid binding site. J. Virol. 2014, 88, 10831–10839. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Wang, J.; Macejak, D.; Garcea, R.L.; Stehle, T. Structures of the major capsid proteins of the human Karolinska Institutet and Washington University polyomaviruses. J. Virol. 2011, 85, 7384–7392. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, T.; Marquez, J.P.; Wakatsuki, T.; Sorokin, A. Caveolar endocytosis is critical for BK virus infection of human renal proximal tubular epithelial cells. J. Virol. 2007, 81, 8552–8562. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Marciano, A.T.; Rivet, C.R.; Imperiale, M.J. Caveolin- and clathrin-independent entry of BKPyV into primary human proximal tubule epithelial cells. Virology 2016, 492, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Meneguzzi, G.; Pignatti, P.F.; Barbanti-Brodano, G.; Milanesi, G. Minichromosome from BK virus as a template for transcription in vitro. Proc. Natl. Acad. Sci. USA 1978, 75, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Wollebo, H.S.; Woldemichaele, B.; Khalili, K.; Safak, M.; White, M.K. Epigenetic regulation of polyomavirus JC. Virol. J. 2013, 10, 264. [Google Scholar] [CrossRef] [PubMed]
- Bialasiewicz, S.; Rockett, R.J.; Barraclough, K.A.; Leary, D.; Dudley, K.J.; Isbel, N.M.; Sloots, T.P. Detection of recently discovered human polyomaviruses in a longitudinal kidney transplant cohort. Am. J. Transplant. 2016, 16, 2734–2740. [Google Scholar] [CrossRef] [PubMed]
- Fratini, M.; Di Bonito, P.; La Rosa, G. Oncogenic papillomavirus and polyomavirus in water environments: Is there a potential for waterborne transmission? Food Environ. Virol. 2014, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Della Libera, S.; Petricca, S.; Iaconelli, M.; Briancesco, R.; Paradiso, R.; Semproni, M.; Di Bonito, P.; Bonadonna, L. First detection of papillomaviruses and polyomaviruses in swimming pool waters: Unrecognized recreational water-related pathogens? J. Appl. Microbiol. 2015, 119, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.C.; Major, E.O. Immune system involvement in the pathogenesis of JC Virus induced PML: What is learned from studies of patients with underlying diseases and therapies as risk factors. Front. Immunol. 2015, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.M.; Broekema, N.M.; Imperiale, M.J. BK polyomavirus: Emerging pathogen. Microbes Infect. 2012, 14, 672–683. [Google Scholar] [CrossRef] [PubMed]
- Babel, N.; Volk, H.D.; Reinke, P. BK polyomavirus infection and nephropathy: The virus-immune system interplay. Nat. Rev. Nephrol. 2011, 7, 399–406. [Google Scholar] [CrossRef] [PubMed]
- White, M.K.; Gordon, J.; Khalili, K. The rapidly expanding family of human polyomaviruses: Recent developments in understanding their life cycle and role in human pathology. PLoS Pathog. 2013, 9, e1003206. [Google Scholar] [CrossRef] [PubMed]
- Ferenczy, M.W.; Marshall, L.J.; Nelson, C.D.; Atwood, W.J.; Nath, A.; Khalili, K.; Major, E.O. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 2012, 25, 471–506. [Google Scholar] [CrossRef] [PubMed]
- Miskin, D.P.; Koralnik, I.J. Novel syndromes associated with JC virus infection of neurons and meningeal cells: No longer a gray area. Curr. Opin. Neurol. 2015, 28, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Kazem, S.; van der Meijden, E.; Feltkamp, M.C. The trichodysplasia spinulosa-associated polyomavirus: Virological background and clinical implications. Apmis 2013, 121, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Matthews, M.R.; Wang, R.C.; Reddick, R.L.; Saldivar, V.A.; Browning, J.C. Viral-associated trichodysplasia spinulosa: A case with electron microscopic and molecular detection of the trichodysplasia spinulosa-associated human polyomavirus. J. Cutan. Pathol. 2011, 38, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Moore, P.S. Merkel cell carcinoma: A virus-induced human cancer. Annu. Rev. Pathol. 2012, 7, 123–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yang, R.; Payne, A.S.; Schowalter, R.M.; Spurgeon, M.E.; Lambert, P.F.; Xu, X.; Buck, C.B.; You, J. Identifying the target cells and mechanisms of merkel cell polyomavirus infection. Cell Host Microbe 2016, 19, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Jedrych, J.J.; Feng, H.; Natalie, A.A.; Grandinetti, L.; Mirvish, E.; Crespo, M.M.; Yadav, D.; Fasanella, K.E.; Proksell, S.; et al. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J. Infect. Dis. 2015, 211, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Rennspiess, D.; Pujari, S.; Keijzers, M.; Abdul-Hamid, M.A.; Hochstenbag, M.; Dingemans, A.M.; Kurz, A.K.; Speel, E.J.; Haugg, A.; Pastrana, D.V.; et al. Detection of human polyomavirus 7 in human thymic epithelial tumors. J. Thorac. Oncol. 2015, 10, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.D.; Derdowski, A.; Maginnis, M.S.; O’Hara, B.A.; Atwood, W.J. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology 2012, 428, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, E.A.; de Raad, M.; Mastrobattista, E. Production and biomedical applications of virus-like particles derived from polyomaviruses. J. Control. Release 2013, 172, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-C.; Iwasaki, K.; Katano, H.; Kataoka, M.; Nagata, N.; Kobayashi, K.; Mizutani, T.; Takeda, N.; Wakita, T.; Suzuki, T. Characterization of self-assembled virus-like particles of merkel cell polyomavirus. PLoS ONE 2015, 10, e0115646. [Google Scholar] [CrossRef] [PubMed]
- Norkiene, M.; Stonyte, J.; Ziogiene, D.; Mazeike, E.; Sasnauskas, K.; Gedvilaite, A. Production of recombinant VP1-derived virus-like particles from novel human polyomaviruses in yeast. BMC Biotechnol. 2015, 15, 68. [Google Scholar] [CrossRef] [PubMed]
- Hale, A.D.; Bartkeviciute, D.; Dargeviciute, A.; Jin, L.; Knowles, W.; Staniulis, J.; Brown, D.W.; Sasnauskas, K. Expression and antigenic characterization of the major capsid proteins of human polyomaviruses BK and JC in Saccharomyces cerevisiae. J. Virol. Methods 2002, 104, 93–98. [Google Scholar] [CrossRef]
- Chang, D.; Fung, C.Y.; Ou, W.C.; Chao, P.C.; Li, S.Y.; Wang, M.; Huang, Y.L.; Tzeng, T.Y.; Tsai, R.T. Self-assembly of the JC virus major capsid protein, VP1, expressed in insect cells. J. Gen. Virol. 1997, 78, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Stolt, A.; Sasnauskas, K.; Koskela, P.; Lehtinen, M.; Dillner, J. Seroepidemiology of the human polyomaviruses. J. Gen. Virol. 2003, 84, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Lundstig, A.; Dillner, J. Serological diagnosis of human polyomavirus infection. Adv. Exp. Med. Biol. 2006, 577, 96–101. [Google Scholar] [PubMed]
- Viscidi, R.P.; Rollison, D.E.; Sondak, V.K.; Silver, B.; Messina, J.L.; Giuliano, A.R.; Fulp, W.; Ajidahun, A.; Rivanera, D. Age-specific seroprevalence of merkel cell polyomavirus, BK virus, and JC virus. Clin. Vaccine Immunol. 2011, 18, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Sroller, V.; Hamsikova, E.; Ludvikova, V.; Musil, J.; Nemeckova, S.; Salakova, M. Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV and KIPyV polyomaviruses among the healthy blood donors. J. Med. Virol. 2016, 88, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Plavina, T.; Castro, A.; Berman, M.; Jaiswal, D.; Rivas, S.; Schlain, B.; Subramanyam, M. A second-generation ELISA (STRATIFY JCV DxSelect) for detection of JC virus antibodies in human serum and plasma to support progressive multifocal leukoencephalopathy risk stratification. J. Clin. Virol. 2013, 57, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Plavina, T.; Subramanyam, M.; Bloomgren, G.; Richman, S.; Pace, A.; Lee, S.; Schlain, B.; Campagnolo, D.; Belachew, S.; Ticho, B. Anti-JC virus antibody levels in serum or plasma further define risk of natalizumab-associated progressive multifocal leukoencephalopathy. Ann. Neurol. 2014, 76, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Samimi, M.; Molet, L.; Fleury, M.; Laude, H.; Carlotti, A.; Gardair, C.; Baudin, M.; Gouguet, L.; Maubec, E.; Avenel-Audran, M.; et al. Prognostic value of antibodies to Merkel cell polyomavirus T antigens and VP1 protein in patients with Merkel cell carcinoma. Br. J. Dermatol. 2016, 174, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Gee, G.V.; O’Hara, B.A.; Derdowski, A.; Atwood, W.J. Pseudovirus mimics cell entry and trafficking of the human polyomavirus JCPyV. Virus Res. 2013, 178, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Schowalter, R.M.; Reinhold, W.C.; Buck, C.B. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS ONE 2012, 7, e42181. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, B.A.; Rupasinghe, C.; Yatawara, A.; Gaidos, G.; Mierke, D.F.; Atwood, W.J. Gallic acid-based small-molecule inhibitors of JC and BK polyomaviral infection. Virus Res. 2014, 189, 280–285. [Google Scholar] [CrossRef]
- Yatawara, A.; Gaidos, G.; Rupasinghe, C.N.; O’Hara, B.A.; Pellegrini, M.; Atwood, W.J.; Mierke, D.F. Small-molecule inhibitors of JC polyomavirus infection. J. Pept. Sci. 2015, 21, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Pastrana, D.V.; Brennan, D.C.; Cuburu, N.; Storch, G.A.; Viscidi, R.P.; Randhawa, P.S.; Buck, C.B. Neutralization serotyping of BK polyomavirus infection in kidney transplant recipients. PLoS Pathog. 2012, 8, e1002650. [Google Scholar] [CrossRef] [PubMed]
- Pastrana, D.V.; Ray, U.; Magaldi, T.G.; Schowalter, R.M.; Çuburu, N.; Buck, C.B. BK polyomavirus genotypes represent distinct serotypes with distinct entry tropism. J. Virol. 2013, 87, 10105–10113. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, P.; Pastrana, D.V.; Zeng, G.; Huang, Y.; Shapiro, R.; Sood, P.; Puttarajappa, C.; Berger, M.; Hariharan, S.; Buck, C.B. Commercially available immunoglobulins contain virus neutralizing antibodies against all major genotypes of polyomavirus BK. Am. J. Transplant. 2015, 15, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Ray, U.; Cinque, P.; Gerevini, S.; Longo, V.; Lazzarin, A.; Schippling, S.; Martin, R.; Buck, C.B.; Pastrana, D.V. JC polyomavirus mutants escape antibody-mediated neutralization. Sci. Transl. Med. 2015, 7, 306ra151. [Google Scholar] [CrossRef] [PubMed]
- Broekema, N.M.; Imperiale, M.J. Efficient propagation of archetype BK and JC polyomaviruses. Virology 2012, 422, 235–241. [Google Scholar] [CrossRef]
- Buck, C.B.; Pastrana, D.V.; Lowy, D.R.; Schiller, J.T. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 2004, 78, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Sharma, B.N.; Linder, S.; Gutteberg, T.J.; Hirsch, H.H.; Rinaldo, C.H. Characteristics of polyomavirus BK (BKPyV) infection in primary human urothelial cells. Virology 2013, 440, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Justice, J.L.; Verhalen, B.; Kumar, R.; Lefkowitz, E.J.; Imperiale, M.J.; Jiang, M. Quantitative proteomic analysis of enriched nuclear fractions from BK polyomavirus-infected primary renal proximal tubule epithelial cells. J. Proteome Res. 2015, 14, 4413–4424. [Google Scholar] [CrossRef] [PubMed]
- Verhalen, B.; Justice, J.L.; Imperiale, M.J.; Jiang, M. Viral DNA replication-dependent DNA damage response activation during BK polyomavirus infection. J. Virol. 2015, 89, 5032–5039. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhao, L.; Gamez, M.; Imperiale, M.J. Roles of ATM and ATR-mediated DNA damage responses during lytic BK polyomavirus infection. PLoS Pathog. 2012, 8, e1002898. [Google Scholar] [CrossRef] [PubMed]
- Jeffers-Francis, L.K.; Burger-Calderon, R.; Webster-Cyriaque, J. Effect of Leflunomide, Cidofovir and Ciprofloxacin on replication of BKPyV in a salivary gland in vitro culture system. Antiviral Res. 2015, 118, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Burger-Calderon, R.; Madden, V.; Hallett, R.A.; Gingerich, A.D.; Nickeleit, V.; Webster-Cyriaque, J. Replication of oral BK virus in human salivary gland cells. J. Virol. 2014, 88, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Alsanie, W.F.; Niclis, J.C.; Petratos, S. Human embryonic stem cell-derived oligodendrocytes: Protocols and perspectives. Stem Cells Dev. 2013, 22, 2459–2476. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, C.; O’Hara, B.A.; Lane, T.E.; Atwood, W.J. Human embryonic stem cell-derived oligodendrocyte progenitor cells express the serotonin receptor and are susceptible to JC virus infection. J. Virol. 2008, 82, 8896–8899. [Google Scholar] [CrossRef] [PubMed]
- Major, E.O.; Miller, A.E.; Mourrain, P.; Traub, R.G.; de Widt, E.; Sever, J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc. Natl. Acad. Sci. USA 1985, 82, 1257–1261. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, S.; Tylden, G.D.; Dumoulin, A.; Sharma, B.N.; Hirsch, H.H.; Rinaldo, C.H. The human fetal glial cell line SVG p12 contains infectious BK polyomavirus. J. Virol. 2014, 88, 7556–7568. [Google Scholar] [CrossRef] [PubMed]
- Brickelmaier, M.; Lugovskoy, A.; Kartikeyan, R.; Reviriego-Mendoza, M.M.; Allaire, N.; Simon, K.; Frisque, R.J.; Gorelik, L. Identification and characterization of mefloquine efficacy against JC virus in vitro. Antimicrob. Agents Chemother. 2009, 53, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Chalkias, S.; Koralnik, I.J. JC virus-iLOV fluorescent strains enable the detection of early and late viral protein expression. J. Virol. Methods 2015, 223, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Israel, M.A.; Chan, H.W.; Hourihan, S.L.; Rowe, W.P.; Martin, M.A. Biological activity of polyoma viral DNA in mice and hamsters. J. Virol. 1979, 29, 990–996. [Google Scholar] [PubMed]
- Small, J.A.; Scangos, G.A.; Cork, L.; Jay, G.; Khoury, G. The early region of human papovavirus JC induces dysmyelination in transgenic mice. Cell 1986, 46, 13–18. [Google Scholar] [CrossRef]
- Haas, S.; Haque, N.S.; Beggs, A.H.; Khalili, K.; Knobler, R.L.; Small, J. Expression of the myelin basic protein gene in transgenic mice expressing human neurotropic virus, JCV, early protein. Virology 1994, 202, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Trapp, B.D.; Small, J.A.; Pulley, M.; Khoury, G.; Scangos, G.A. Dysmyelination in transgenic mice containing JC virus early region. Ann. Neurol. 1988, 23, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Small, J.A.; Khoury, G.; Jay, G.; Howley, P.M.; Scangos, G.A. Early regions of JC virus and BK virus induce distinct and tissue-specific tumors in transgenic mice. Proc. Natl. Acad. Sci. USA 1986, 83, 8288–8292. [Google Scholar] [CrossRef] [PubMed]
- Shollar, D.; Del Valle, L.; Khalili, K.; Otte, J.; Gordon, J. JCV T-antigen interacts with the neurofibromatosis type 2 gene product in a transgenic mouse model of malignant peripheral nerve sheath tumors. Oncogene 2004, 23, 5459–5467. [Google Scholar] [CrossRef] [PubMed]
- Spurgeon, M.E.; Cheng, J.; Bronson, R.T.; Lambert, P.F.; DeCaprio, J.A. Tumorigenic activity of Merkel cell polyomavirus T antigens expressed in the stratified epithelium of mice. Cancer Res. 2015, 75, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Verhaegen, M.E.; Mangelberger, D.; Harms, P.W.; Vozheiko, T.D.; Weick, J.W.; Wilbert, D.M.; Saunders, T.L.; Ermilov, A.N.; Bichakjian, C.K.; Johnson, T.M.; et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Invest. Dermatol. 2015, 135, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Shuda, M.; Guastafierro, A.; Geng, X.; Shuda, Y.; Ostrowski, S.M.; Lukianov, S.; Jenkins, F.J.; Honda, K.; Maricich, S.M.; Moore, P.S.; et al. Merkel cell polyomavirus small T antigen induces cancer and embryonic Merkel cell proliferation in a transgenic mouse model. PLoS ONE 2015, 10, e0142329. [Google Scholar] [CrossRef]
- Herter-Sprie, G.S.; Kung, A.L.; Wong, K.K. New cast for a new era: Preclinical cancer drug development revisited. J. Clin. Investig. 2013, 123, 3639–3645. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Shuda, M.; Guastafierro, A.; Feng, H.; Toptan, T.; Tolstov, Y.; Normolle, D.; Vollmer, L.L.; Vogt, A.; Domling, A.; et al. Survivin is a therapeutic target in Merkel cell carcinoma. Sci. Transl. Med. 2012, 4, 133ra156. [Google Scholar] [CrossRef] [PubMed]
- Dresang, L.R.; Guastafierro, A.; Arora, R.; Normolle, D.; Chang, Y.; Moore, P.S. Response of Merkel cell polyomavirus-positive merkel cell carcinoma xenografts to a survivin inhibitor. PLoS ONE 2013, 8, e80543. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.S.; Broge, T.A., Jr.; Seung, E.; Vrbanac, V.; Viscidi, R.; Gordon, J.; Tager, A.M.; Koralnik, I.J. Detection of JC virus-specific immune responses in a novel humanized mouse model. PLoS ONE 2013, 8, e64313. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Windrem, M.S.; Zou, L.; Chandler-Militello, D.; Schanz, S.J.; Auvergne, R.M.; Betstadt, S.J.; Harrington, A.R.; Johnson, M.; Kazarov, A.; et al. Human glial chimeric mice reveal astrocytic dependence of JC virus infection. J. Clin. Investig. 2014, 124, 5323–5336. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.E.; Li, H.; Sur, G.; Carmillo, P.; Bushnell, S.; Tizard, R.; McAuliffe, M.; Tonkin, C.; Simon, K.; Goelz, S.; et al. Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients. J. Infect. Dis. 2011, 204, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, L.; Reid, C.; Testa, M.; Brickelmaier, M.; Bossolasco, S.; Pazzi, A.; Bestetti, A.; Carmillo, P.; Wilson, E.; McAuliffe, M.; et al. Progressive multifocal leukoencephalopathy (PML) development is associated with mutations in JC virus capsid protein VP1 that change its receptor specificity. J. Infect. Dis. 2011, 204, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Schaffhausen, B.S.; Roberts, T.M. Lessons from polyoma middle T antigen on signaling and transformation: A DNA tumor virus contribution to the war on cancer. Virology 2009, 384, 304–316. [Google Scholar] [CrossRef] [PubMed]
- van der Meijden, E.; Kazem, S.; Dargel, C.A.; van Vuren, N.; Hensbergen, P.J.; Feltkamp, M.C. Characterization of T antigens, including middle T and alternative T, expressed by the human polyomavirus associated with trichodysplasia spinulosa. J. Virol. 2015, 89, 9427–9439. [Google Scholar] [CrossRef]
- Kemball, C.C.; Lee, E.D.; Vezys, V.; Pearson, T.C.; Larsen, C.P.; Lukacher, A.E. Late priming and variability of epitope-specific CD8+ T cell responses during a persistent virus infection. J. Immunol. 2005, 174, 7950–7960. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Kemball, C.C.; Hadley, A.; Wilson, J.J.; Hofstetter, A.R.; Pack, C.D.; Lukacher, A.E. Heterogeneity among viral antigen-specific CD4+ T cells and their de novo recruitment during persistent polyomavirus infection. J. Immunol. 2010, 185, 1692–1700. [Google Scholar] [CrossRef] [PubMed]
- Frost, E.L.; Lukacher, A.E. The importance of mouse models to define immunovirologic determinants of progressive multifocal leukoencephalopathy. Front. Immunol. 2014, 5, 646. [Google Scholar] [CrossRef] [PubMed]
- Han Lee, E.D.; Kemball, C.C.; Wang, J.; Dong, Y.; Stapler, D.C.; Hamby, K.M.; Gangappa, S.; Newell, K.A.; Pearson, T.C.; Lukacher, A.E.; et al. A mouse model for polyomavirus-associated nephropathy of kidney transplants. Am. J. Transplant. 2006, 6, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, J.A.; Dong, Y.; Wang, J.; Breeden, C.; Farris, A.B., 3rd; Lukacher, A.E.; Newell, K.A. Adaptive immunity rather than viral cytopathology mediates polyomavirus-associated nephropathy in mice. Am. J. Transplant. 2012, 12, 1419–1428. [Google Scholar] [CrossRef]
- Horvath, C.J.; Simon, M.A.; Bergsagel, D.J.; Pauley, D.R.; King, N.W.; Garcea, R.L.; Ringler, D.J. Simian virus 40-induced disease in rhesus monkeys with simian acquired immunodeficiency syndrome. Am. J. Pathol. 1992, 140, 1431–1440. [Google Scholar] [PubMed]
- Dang, X.; Wuthrich, C.; Axthelm, M.K.; Koralnik, I.J. Productive simian virus 40 infection of neurons in immunosuppressed Rhesus monkeys. J. Neuropathol. Exp. Neurol. 2008, 67, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, C.; Koralnik, I.J. Frequent infection of cortical neurons by JC virus in patients with progressive multifocal leukoencephalopathy. J. Neuropathol. Exp. Neurol. 2012, 71, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Kaliyaperumal, S.; Dang, X.; Wuethrich, C.; Knight, H.L.; Pearson, C.; MacKey, J.; Mansfield, K.G.; Koralnik, I.J.; Westmoreland, S.V. Frequent infection of neurons by SV40 virus in SIV-infected macaque monkeys with progressive multifocal leukoencephalopathy and meningoencephalitis. Am. J. Pathol. 2013, 183, 1910–1917. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Wojno, K.; Imperiale, M.J. BK virus as a cofactor in the etiology of prostate cancer in its early stages. J. Virol. 2008, 82, 2705–2714. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, J.C.; Randhawa, P.; Rinaldo, C.H.; Drachenberg, C.B.; Alexiev, B.; Hirsch, H.H. BK polyomavirus infection and renourinary tumorigenesis. Am. J. Transplant. 2016, 16, 398–406. [Google Scholar] [CrossRef] [PubMed]
| Human Polyomavirus | Abbreviation | Year of Discovery | NCBI RefSeq or GenBank Accession | Source of Isolation | Seroprevalence (%) * | Associated Disease | Ref. |
|---|---|---|---|---|---|---|---|
| BK polyomavirus | BKPyV | 1971 | NC_001538 | Urine | 80–90 (a) | Nephropathy, hemorrhagic cystitis | [2] |
| JC polyomavirus | JCPyV | 1971 | NC_001699 | Brain | 40–55 (b) | Progressive multifocal leukoencephalopathy | [3] |
| Karolinska Institute polyomavirus | KIPyV | 2007 | NC_009238 | Respiratory tract | 55–90 | Not known | [4] |
| Washington University polyomavirus | WUPyV | 2007 | NC_009539 | Respiratory tract | 70–90 | Not known | [5] |
| Merkel cell polyomavirus | MCPyV | 2008 | NC_010277 | Skin tumor | 60–80 | Merkel cell carcinoma | [6] |
| Human polyomavirus 6 | HPyV6 | 2010 | NC_014406 | Normal skin | 70–75 | Not known | [7] |
| Human polyomavirus 7 | HPyV7 | 2010 | NC_014407 | Normal skin | 35–62 | Pruritic rash | [7] |
| Trichodysplasia spinulosa-associated polyomavirus | TSPyV | 2010 | NC_014361 | Skin lesion | 70–84 | Trichodysplasia spinulosa | [8] |
| Human polyomavirus 9 | HPyV9 | 2011 | NC_015150 | Blood and urine | 18–50 | Not known | [9] |
| Malawi polyomavirus | MWPyV | 2012 | NC_018102 | Stool | 42–75 | Not known | [10] |
| Human polyomavirus 10 | HPyV10 | 2012 | JX262162 | Condyloma | 99 | Not known | [11] |
| Mexico polyomavirus | MXPyV | 2012 | JX259273 | Stool | Not known | Not known | [12] |
| St Louis polyomavirus | STLPyV | 2012 | NC_020106 | Stool | 70 | Not known | [13] |
| Human polyomavirus 12 | HPyV12 | 2013 | NC_020890 | Liver | 23 | Not known | [14] |
| New Jersey polyomavirus | NJPyV | 2013 | NC_024118 | Muscle biopsy | Not known | Not known | [15] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barth, H.; Solis, M.; Kack-Kack, W.; Soulier, E.; Velay, A.; Fafi-Kremer, S. In Vitro and In Vivo Models for the Study of Human Polyomavirus Infection. Viruses 2016, 8, 292. https://doi.org/10.3390/v8100292
Barth H, Solis M, Kack-Kack W, Soulier E, Velay A, Fafi-Kremer S. In Vitro and In Vivo Models for the Study of Human Polyomavirus Infection. Viruses. 2016; 8(10):292. https://doi.org/10.3390/v8100292
Chicago/Turabian StyleBarth, Heidi, Morgane Solis, Wallys Kack-Kack, Eric Soulier, Aurélie Velay, and Samira Fafi-Kremer. 2016. "In Vitro and In Vivo Models for the Study of Human Polyomavirus Infection" Viruses 8, no. 10: 292. https://doi.org/10.3390/v8100292
APA StyleBarth, H., Solis, M., Kack-Kack, W., Soulier, E., Velay, A., & Fafi-Kremer, S. (2016). In Vitro and In Vivo Models for the Study of Human Polyomavirus Infection. Viruses, 8(10), 292. https://doi.org/10.3390/v8100292





