Synthetic RNAs Mimicking Structural Domains in the Foot-and-Mouth Disease Virus Genome Elicit a Broad Innate Immune Response in Porcine Cells Triggered by RIG-I and TLR Activation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Plasmids
2.2. RNA Synthesis
2.3. Transfection of HEK 293 Cells and Luciferase Assay
2.4. Transfection of SK6 Cells and RT-PCR
2.5. PBMCs Transfection, RT-PCR and Cytokine Detection
2.6. Ethical Statement
2.7. Immunoblot Analysis
2.8. Statistical Analysis
3. Results
3.1. The FMDV ncRNAs Activate the Human IFN-β Promoter in a RIG-I-Dependent Manner
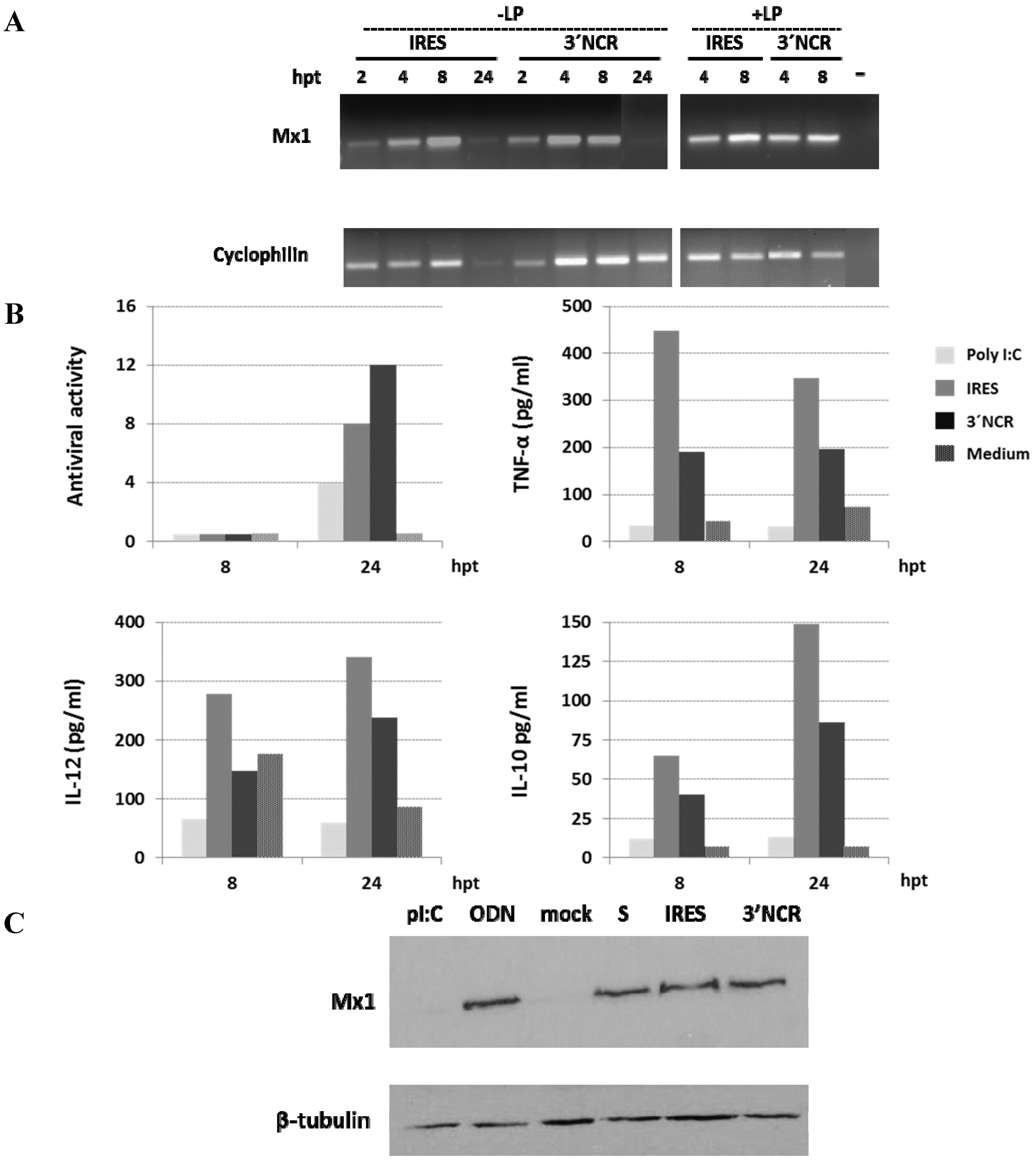
3.2. Induction of Mx1 in Porcine Epithelial Cultured Cells Transfected with the 3′NCR RNA

3.4. Effect of ncRNA Dose and Enrichment in pDCs on Cytokine Production by Swine PBMCs
3.5. The Innate Immune Response Induced by the FMDV ncRNAs in Porcine PBMCs is Significantly Decreased by Inhibition of Endosomal Acidification

4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barbalat, R.; Ewald, S.E.; Mouchess, M.L.; Barton, G.M. Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 2011, 29, 185–214. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Nan, G.; Zhang, Y.J. Interferon induction by rna viruses and antagonism by viral pathogens. Viruses 2014, 6, 4999–5027. [Google Scholar] [CrossRef] [PubMed]
- Lester, S.N.; Li, K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef] [PubMed]
- Dixit, E.; Kagan, J.C. Intracellular pathogen detection by RIG-I-like receptors. Adv. Immunol. 2013, 117, 99–125. [Google Scholar] [PubMed]
- Schlee, M. Master sensors of pathogenic RNA—RIG-I like receptors. Immunobiology 2013, 218, 1322–1335. [Google Scholar] [CrossRef] [PubMed]
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Berke, I.C.; Li, Y.; Modis, Y. Structural basis of innate immune recognition of viral RNA. Cell. Microbiol. 2013, 15, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Gay, N.J.; Symmons, M.F.; Gangloff, M.; Bryant, C.E. Assembly and localization of toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014, 14, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded rna viruses by toll-like receptor 7. Proc. Natl. Acad. Sci. USA 2004, 101, 5598–5603. [Google Scholar] [CrossRef] [PubMed]
- Olive, C. Pattern recognition receptors: Sentinels in innate immunity and targets of new vaccine adjuvants. Expert Rev. Vaccines 2012, 11, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Rothenfusser, S.; Hopfner, K.P. Sensing of viral nucleic acids by RIG-I: From translocation to translation. Eur. J. Cell Biol. 2012, 91, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pulido, M.; Borrego, B.; Sobrino, F.; Saiz, M. RNA structural domains in non-coding regions of foot-and-mouth disease virus genome trigger innate immunity in porcine cells and mice. J. Virol. 2011, 85, 6492–6501. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pulido, M.; Sobrino, F.; Borrego, B.; Saiz, M. Inoculation of newborn mice with non-coding regions of foot-and-mouth disease virus RNA can induce a rapid, solid and wide-range protection against viral infection. Antivir. Res. 2011, 92, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pulido, M.; Martin-Acebes, M.A.; Escribano-Romero, E.; Blazquez, A.B.; Sobrino, F.; Borrego, B.; Saiz, M.; Saiz, J.C. Protection against west nile virus infection in mice after inoculation with type I interferon-inducing rna transcripts. PLoS ONE 2012, 7, e49494. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, G.; Rodriguez-Pulido, M.; Lopez-Gil, E.; Sobrino, F.; Borrego, B.; Saiz, M.; Brun, A. Protection against rift valley fever virus infection in mice upon administration of interferon-inducing rna transcripts from the FMDV genome. Antivir. Res. 2014, 109, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Serrano, P.; Pulido, M.R.; Saiz, M.; Martinez-Salas, E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 2006, 87, 3013–3022. [Google Scholar] [CrossRef] [PubMed]
- Witwer, C.; Rauscher, S.; Hofacker, I.L.; Stadler, P.F. Conserved rna secondary structures in picornaviridae genomes. Nucleic Acids Res. 2001, 29, 5079–5089. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Lozano, G.; Diaz-Toledano, R. Picornavirus ires elements: Rna structure and host protein interactions. Virus Res. 2015, 206, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Escarmis, C.; Toja, M.; Medina, M.; Domingo, E. Modifications of the 5′ untranslated region of foot-and-mouth disease virus after prolonged persistence in cell culture. Virus Res. 1992, 26, 113–125. [Google Scholar] [CrossRef]
- Borrego, B.; Rodriguez-Pulido, M.; Mateos, F.; de la Losa, N.; Sobrino, F.; Saiz, M. Delivery of synthetic RNA can enhance the immunogenicity of vaccines against foot-and-mouth disease virus (FMDV) in mice. Vaccine 2013, 31, 4375–4381. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.; Mendoza, C.; Sanchez-Vizcaino, J.M.; Alonso, F. Inhibitory effect of african swine fever virus on lectin-dependent swine lymphocyte proliferation. Vet. Immunol. Immunopathol. 1990, 26, 71–80. [Google Scholar] [CrossRef]
- Nistal-Villan, E.; Gack, M.U.; Martinez-Delgado, G.; Maharaj, N.P.; Inn, K.S.; Yang, H.; Wang, R.; Aggarwal, A.K.; Jung, J.U.; Garcia-Sastre, A. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J. Biol. Chem. 2010, 285, 20252–20261. [Google Scholar] [CrossRef] [PubMed]
- Mibayashi, M.; Martinez-Sobrido, L.; Loo, Y.M.; Cardenas, W.B.; Gale, M., Jr.; Garcia-Sastre, A. Inhibition of retinoic acid-inducible gene i-mediated induction of β interferon by the NS1 protein of influenza a virus. J. Virol. 2007, 81, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Sachidanandam, R.; Garcia-Sastre, A. Preference of RIG-I for short viral rna molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 16303–16308. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.; Martinez-Salas, E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES). RNA 1999, 5, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Briones, M.M.; Blanco, E.; Chiva, C.; Andreu, D.; Ley, V.; Sobrino, F. Immunogenicity and T cell recognition in swine of foot-and-mouth disease virus polymerase 3D. Virology 2004, 322, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Gil, S.; Spagnuolo-Weaver, M.; Canals, A.; Sepulveda, N.; Oliveira, J.; Aleixo, A.; Allan, G.; Leitao, A.; Martins, C.L. Expression at mRNA level of cytokines and A238l gene in porcine blood-derived macrophages infected in vitro with african swine fever virus (ASFV) isolates of different virulence. Arch. Virol. 2003, 148, 2077–2097. [Google Scholar] [CrossRef] [PubMed]
- Guzylack-Piriou, L.; Balmelli, C.; McCullough, K.C.; Summerfield, A. Type-A CpG oligonucleotides activate exclusively porcine natural interferon-producing cells to secrete interferon-α, tumour necrosis factor-α and interleukin-12. Immunology 2004, 112, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Armas-Portela, R.; Parrales, M.A.; Albar, J.P.; Martinez, A.C.; Avila, J. Distribution and characteristics of betaii tubulin-enriched microtubules in interphase cells. Exp. Cell Res. 1999, 248, 372–380. [Google Scholar] [CrossRef] [PubMed]
- MacMicking, J.D. Ifn-inducible gtpases and immunity to intracellular pathogens. Trends Immunol. 2004, 25, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Reizis, B.; Bunin, A.; Ghosh, H.S.; Lewis, K.L.; Sisirak, V. Plasmacytoid dendritic cells: Recent progress and open questions. Annu. Rev. Immunol. 2011, 29, 163–183. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Dolganiuc, A. The role of plasmacytoid dendritic cell-derived ifn alpha in antiviral immunity. Crit. Rev. Immunol. 2008, 28, 61–94. [Google Scholar] [CrossRef] [PubMed]
- Summerfield, A.; McCullough, K.C. The porcine dendritic cell family. Dev. Comp. Immunol. 2009, 33, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kaiser, V.; Beier, E.; Bechheim, M.; Guenthner-Biller, M.; Ablasser, A.; Berger, M.; Endres, S.; Hartmann, G.; Hornung, V. Self-priming determines high type I IFN production by plasmacytoid dendritic cells. Eur. J. Immunol. 2014, 44, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Guiducci, C.; Ott, G.; Chan, J.H.; Damon, E.; Calacsan, C.; Matray, T.; Lee, K.D.; Coffman, R.L.; Barrat, F.J. Properties regulating the nature of the plasmacytoid dendritic cell response to toll-like receptor 9 activation. J. Exp. Med. 2006, 203, 1999–2008. [Google Scholar] [CrossRef] [PubMed]
- Braciale, T.J.; Hahn, Y.S. Immunity to viruses. Immunol. Rev. 2013, 255, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.P.; Farrar, J.D. Regulation of effector and memory T-cell functions by type I interferon. Immunology 2011, 132, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, K.; Oropallo, M.A.; Cancro, M.P.; Marshak-Rothstein, A. Role of type i interferons in the activation of autoreactive b cells. Immunol. Cell Biol. 2012, 90, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Tough, D.F. Modulation of T-cell function by type i interferon. Immunol. Cell Biol. 2012, 90, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.B.; Salazar-Mather, T.P.; Dalod, M.Y.; van Deusen, J.B.; Wei, X.Q.; Liew, F.Y.; Caligiuri, M.A.; Durbin, J.E.; Biron, C.A. Coordinated and distinct roles for IFN-α β, IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 2002, 169, 4279–4287. [Google Scholar] [CrossRef] [PubMed]
- Querec, T.; Bennouna, S.; Alkan, S.; Laouar, Y.; Gorden, K.; Flavell, R.; Akira, S.; Ahmed, R.; Pulendran, B. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J. Exp. Med. 2006, 203, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Doel, T.R. Natural and vaccine induced immunity to fmd. Curr. Top. Microbiol. Immunol. 2005, 288, 103–131. [Google Scholar] [PubMed]
- Shi, H.; Fu, Q.; Ren, Y.; Wang, D.; Qiao, J.; Wang, P.; Zhang, H.; Chen, C. Both foot-and-mouth disease virus and bovine viral diarrhea virus replication are inhibited by MX1 protein originated from porcine. Anim. Biotechnol. 2015, 26, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Yang, H.; Yang, D.; Zhao, B.; Ouyang, Z.; Liu, Z.; Fan, N.; Ouyang, H.; Gu, W.; Lai, L. Production of transgenic pigs over-expressing the antiviral gene MX1. Cell Regen Lond. 2014, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Diaz-San Segundo, F.; Moraes, M.P.; de Los Santos, T.; Dias, C.C.; Grubman, M.J. Interferon-induced protection against foot-and-mouth disease virus infection correlates with enhanced tissue-specific innate immune cell infiltration and interferon-stimulated gene expression. J. Virol. 2010, 84, 2063–2077. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borrego, B.; Rodríguez-Pulido, M.; Revilla, C.; Álvarez, B.; Sobrino, F.; Domínguez, J.; Sáiz, M. Synthetic RNAs Mimicking Structural Domains in the Foot-and-Mouth Disease Virus Genome Elicit a Broad Innate Immune Response in Porcine Cells Triggered by RIG-I and TLR Activation. Viruses 2015, 7, 3954-3973. https://doi.org/10.3390/v7072807
Borrego B, Rodríguez-Pulido M, Revilla C, Álvarez B, Sobrino F, Domínguez J, Sáiz M. Synthetic RNAs Mimicking Structural Domains in the Foot-and-Mouth Disease Virus Genome Elicit a Broad Innate Immune Response in Porcine Cells Triggered by RIG-I and TLR Activation. Viruses. 2015; 7(7):3954-3973. https://doi.org/10.3390/v7072807
Chicago/Turabian StyleBorrego, Belén, Miguel Rodríguez-Pulido, Concepción Revilla, Belén Álvarez, Francisco Sobrino, Javier Domínguez, and Margarita Sáiz. 2015. "Synthetic RNAs Mimicking Structural Domains in the Foot-and-Mouth Disease Virus Genome Elicit a Broad Innate Immune Response in Porcine Cells Triggered by RIG-I and TLR Activation" Viruses 7, no. 7: 3954-3973. https://doi.org/10.3390/v7072807
APA StyleBorrego, B., Rodríguez-Pulido, M., Revilla, C., Álvarez, B., Sobrino, F., Domínguez, J., & Sáiz, M. (2015). Synthetic RNAs Mimicking Structural Domains in the Foot-and-Mouth Disease Virus Genome Elicit a Broad Innate Immune Response in Porcine Cells Triggered by RIG-I and TLR Activation. Viruses, 7(7), 3954-3973. https://doi.org/10.3390/v7072807