Genomic and Phylogenetic Characterization of Novel, Recombinant H5N2 Avian Influenza Virus Strains Isolated from Vaccinated Chickens with Clinical Symptoms in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chicken Population
2.2. Virus Detection and Isolation
2.3. RT-PCR and Sequencing
2.4. Genomic and Phylogenetic Analysis
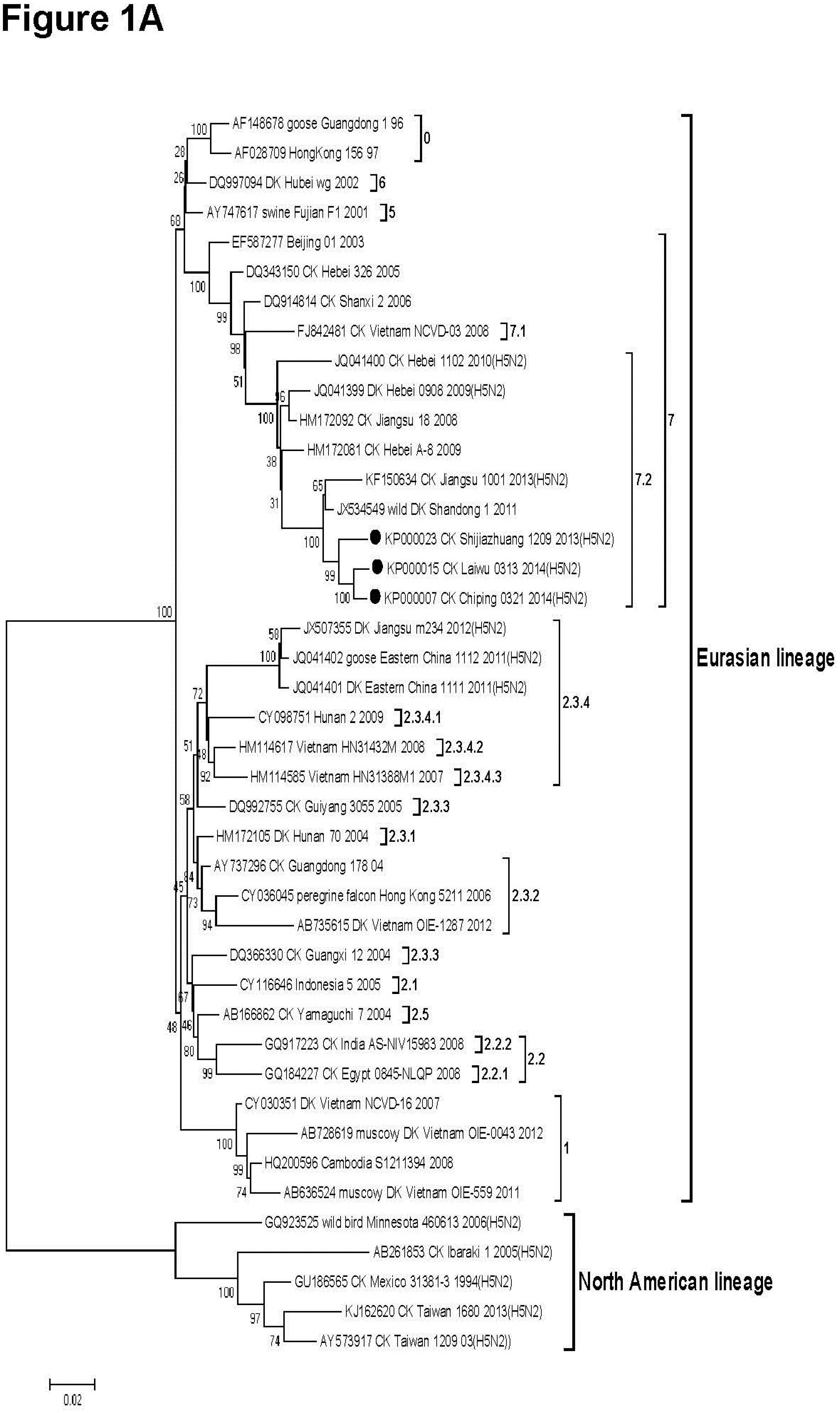
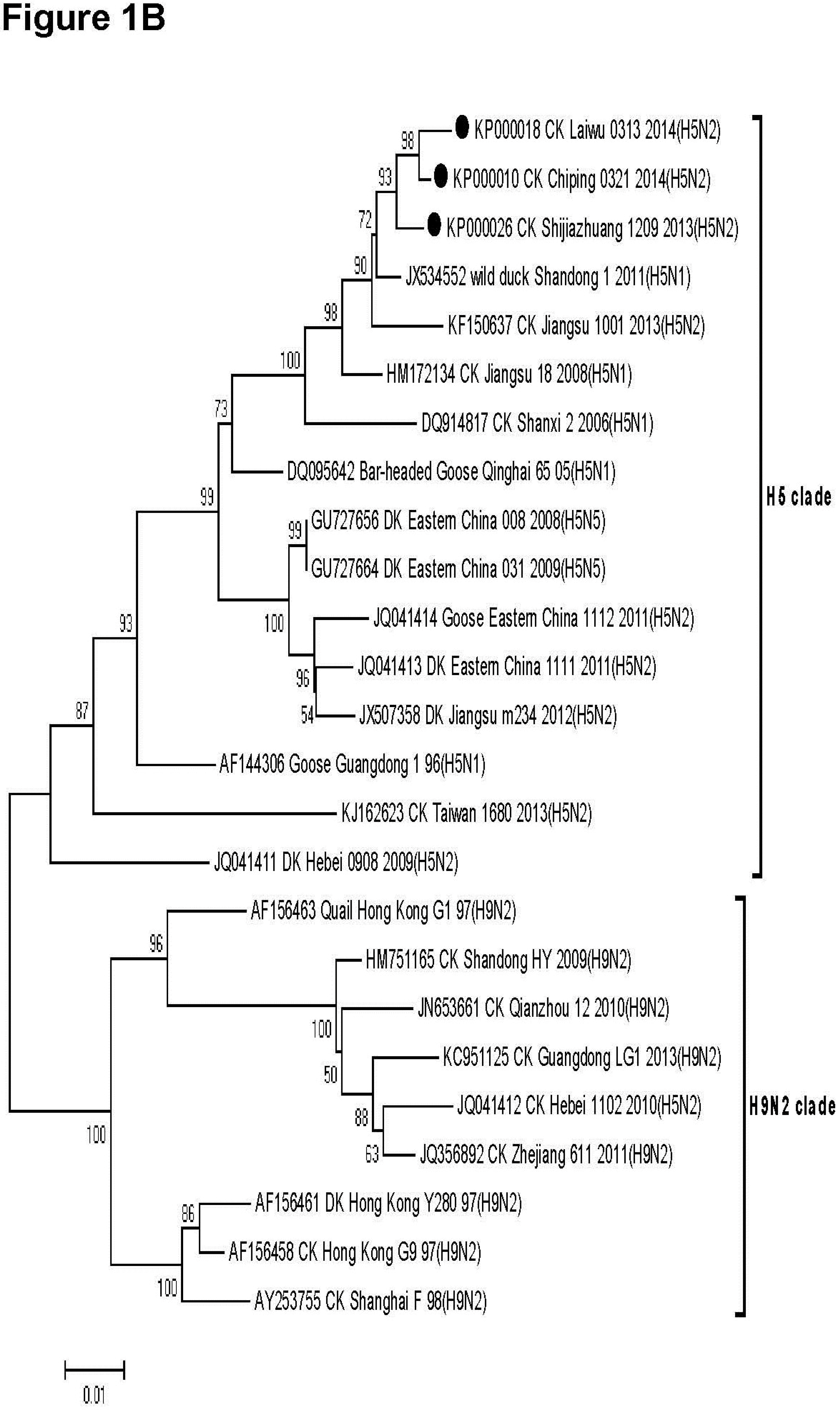
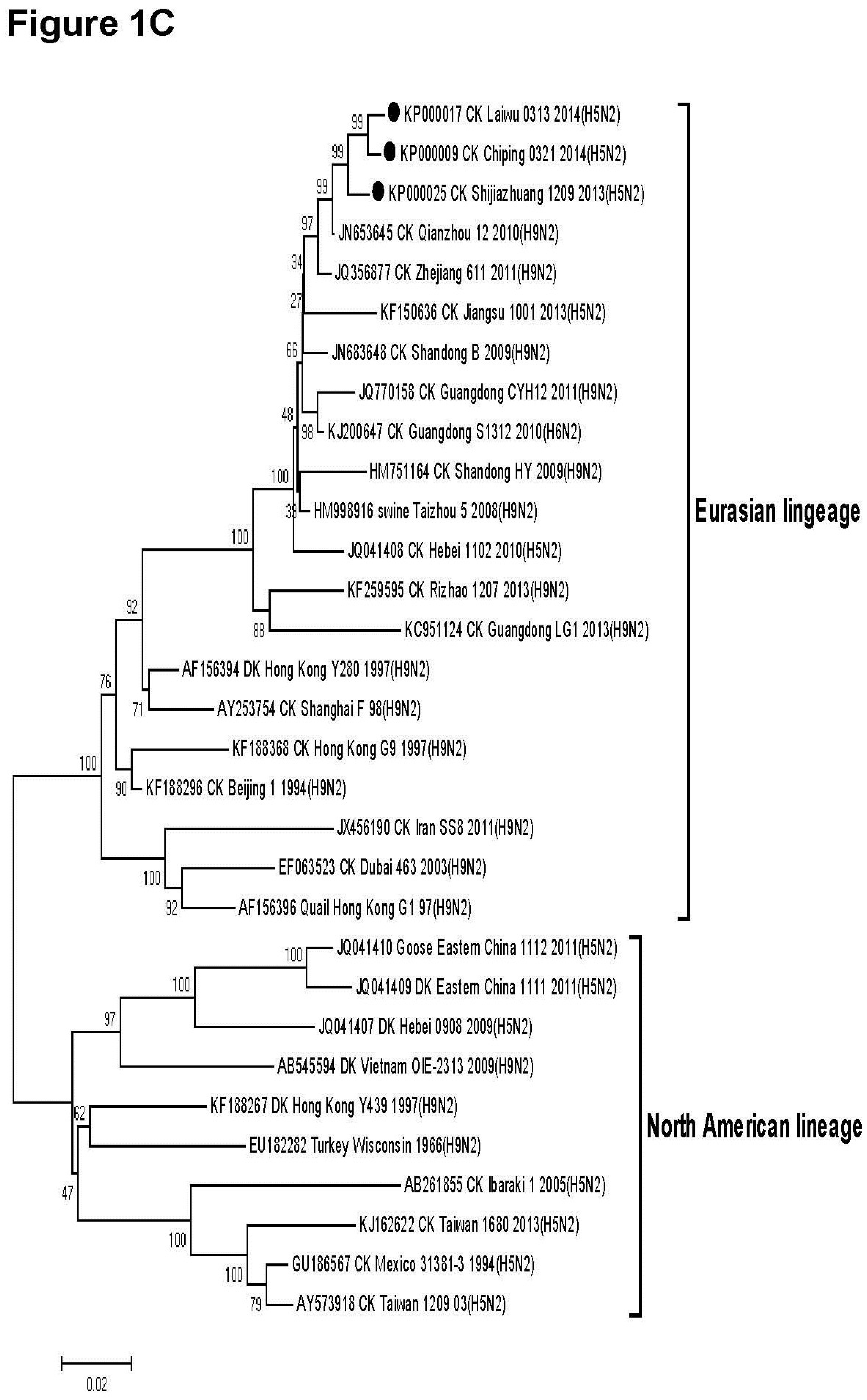
2.5. Ethical Approval
3. Results
3.1. Virus Detection and Isolation
3.2. Phylogenetic Analysis
| Gene * Segment | CK/CP/0321/2014 | CK/LW/0313/2014 | CK/SJZ/1209/2013 | |||
|---|---|---|---|---|---|---|
| Closest Virus | Nucleotide Identity, % | Closest Virus | Nucleotide Identity, % | Closest Virus | Nucleotide Identity, % | |
| PB2 | A/chicken/Qianzhou/12/2010(H9N2) | 98 | A/chicken/Qianzhou/12/2010(H9N2) | 98 | A/chicken/Qianzhou/12/2010(H9N2) | 98 |
| PB1 | A/chicken/Jiangsu/Q3/2010(H9N2) | 98 | A/chicken/Jiangsu/Q3/2010(H9N2) | 98 | A/chicken/Jiangsu/Q3/2010(H9N2) | 98 |
| PA | A/chicken/Shandong/397/2010(H9N2) | 99 | A/chicken/Shandong/397/2010(H9N2) | 99 | A/chicken/Shandong/397/2010(H9N2) | 99 |
| HA | A/wild duck/Shandong/1/2011(H5N1) | 98 | A/wild duck/Shandong/1/2011(H5N1) | 98 | A/wild duck/Shandong/1/2011(H5N1) | 98 |
| NP | A/chicken/Zhejiang/611/2011(H9N2) | 98 | A/chicken/Zhejiang/611/2011(H9N2) | 98 | A/chicken/Zhejiang/611/2011(H9N2) | 98 |
| NA | A/chicken/Qianzhou/12/2010(H9N2) | 99 | A/chicken/Qianzhou/12/2010(H9N2) | 99 | A/chicken/Qianzhou/12/2010(H9N2) | 99 |
| M | A/wild duck/Shandong/1/2011(H5N1) | 99 | A/wild duck/Shandong/1/2011(H5N1) | 99 | A/wild duck/Shandong/1/2011(H5N1) | 99 |
| NS | A/chicken/Qianzhou/12/2010(H9N2) | 98 | A/chicken/Qianzhou/12/2010(H9N2) | 98 | A/chicken/Qianzhou/12/2010(H9N2) | 98 |
3.3. Molecular Characteristics
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bean, W.J.; Kawaoka, Y.; Wood, J.M.; Pearson, J.E.; Webster, R.G. Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: Potential role of defective interfering RNAs in nature. J. Virol. 1985, 54, 151–160. [Google Scholar] [PubMed]
- Webster, R.G.; Kawaoka, Y.; Bean, W.J., Jr. Molecular changes in A/Chicken/Pennsylvania/83 (H5N2) influenza virus associated with acquisition of virulence. Virology 1986, 149, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Abolnik, C.; Olivier, A.J.; Grewar, J.; Gers, S.; Romito, M. Molecular analysis of the 2011 HPAI H5N2 outbreak in ostriches, South Africa. Avian Dis. 2012, 56, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.C.; Soda, K.; Lee, M.S.; Lee, S.H.; Sakoda, Y.; Kida, H.; Wang, C.H. Isolation and characterization of potentially pathogenic H5N2 influenza virus from a chicken in Taiwan in 2008. Avian Dis. 2010, 54, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Donatelli, I.; Campitelli, L.; di Trani, L.; Puzelli, S.; Selli, L.; Fioretti, A.; Alexander, D.J.; Tollis, M.; Krauss, S.; Webster, R.G. Characterization of H5N2 influenza viruses from Italian poultry. J. Gener. Virol. 2001, 82, 623–630. [Google Scholar]
- Gu, M.; Huang, J.; Chen, Y.; Chen, J.; Wang, X.; Liu, X.; Liu, X. Genome sequence of a natural reassortant H5N2 avian influenza virus from domestic mallard ducks in eastern China. J. Virol. 2012, 86, 12463–12464. [Google Scholar] [CrossRef] [PubMed]
- Horimoto, T.; Rivera, E.; Pearson, J.; Senne, D.; Krauss, S.; Kawaoka, Y.; Webster, R.G. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 1995, 213, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Wei, L.; Yuan, R.; Gong, L.; Cao, L.; Song, Y.; Luo, K.; Ren, T.; Liao, M. Complete genome sequence of an H5N2 avian influenza virus isolated from a parrot in southern China. J. Virol. 2012, 86, 8890–8891. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Park, C.K.; Oem, J.K.; Bae, Y.C.; Choi, J.G.; Lee, O.S.; Lee, Y.J. Characterization of H5N2 influenza viruses isolated in South Korea and their influence on the emergence of a novel H9N2 influenza virus. J. Gener. Virol. 2010, 91, 1978–1983. [Google Scholar] [CrossRef]
- Okamatsu, M.; Saito, T.; Yamamoto, Y.; Mase, M.; Tsuduku, S.; Nakamura, K.; Tsukamoto, K.; Yamaguchi, S. Low pathogenicity H5N2 avian influenza outbreak in Japan during the 2005–2006. Vet. Microbiol. 2007, 124, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Pasick, J.; Handel, K.; Robinson, J.; Bowes, V.; Li, Y.; Leighton, T.; Kehler, H.; Ridd, D.; Cottam-Birt, C. Relationship between H5N2 avian influenza viruses isolated from wild and domestic ducks in British Columbia, Canada. Avian Dis. 2007, 51, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Mi, Z.; Liu, W.; Fan, H.; An, X.; Pei, G.; Wang, W.; Xu, X.; Ma, M.; Zhang, Z.; Cao, W.; et al. Complete Genome Sequence of Avian Influenza Virus A/chicken/Jiangsu/1001/2013(H5N2), Demonstrating Continuous Reassortance of H5N2 in China. Genome Announc. 2013, 1. [Google Scholar] [CrossRef]
- Zhao, G.; Gu, X.; Lu, X.; Pan, J.; Duan, Z.; Zhao, K.; Gu, M.; Liu, Q.; He, L.; Chen, J.; et al. Novel reassortant highly pathogenic H5N2 avian influenza viruses in poultry in China. PLOS ONE 2012, 7, e46183. [Google Scholar] [CrossRef]
- Nishi, T.; Okamatsu, M.; Sakurai, K.; Chu, H.D.; Thanh, L.P.; van Nguyen, L.; van Hoang, N.; Thi, D.N.; Sakoda, Y.; Kida, H. Genetic analysis of an H5N2 highly pathogenic avian influenza virus isolated from a chicken in a live bird market in Northern Vietnam in 2012. J. Vet. Med. Sci. 2014, 76, 85–87. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Zhu, H.; Huang, P.Y.; Peng, L.; Chang, Y.C.; Yip, C.H.; Li, Y.T.; Cheung, C.L.; Compans, R.; Yang, C.; et al. Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan. J. Virol. 2014, 88, 5677–5686. [Google Scholar] [CrossRef]
- Wu, H.; Peng, X.; Xu, L.; Jin, C.; Cheng, L.; Lu, X.; Xie, T.; Yao, H.; Wu, N. Characterization of a novel highly pathogenic H5N2 avian influenza virus isolated from a duck in eastern China. Arch. Virol. 2014, 159, 3377–3383. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Chen, H.; Li, Q.; Huang, J.; Zhao, M.; Gu, X.; Jiang, K.; Wang, X.; Peng, D.; Liu, X. Enzootic genotype S of H9N2 avian influenza viruses donates internal genes to emerging zoonotic influenza viruses in China. Vet. Microbiol. 2014, 174, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, C.J.; Adeyanju, A.T.; de Landtsheer, S.; Ottosson, U.; Manu, S.; Hagemeijer, W.; Mundkur, T.; Muller, C.P. Reassortant low-pathogenic avian influenza H5N2 viruses in African wild birds. J. Gener. Virol. 2011, 92, 1172–1183. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Escorcia, M.; Vazquez, L.; Mendez, S.T.; Rodriguez-Ropon, A.; Lucio, E.; Nava, G.M. Avian influenza: Genetic evolution under vaccination pressure. Virol. J. 2008, 5, e15. [Google Scholar] [CrossRef]
- Zhao, S.; Suo, L.; Jin, M. Genetic characterization of a novel recombinant H5N2 avian influenza virus isolated from chickens in Tibet. J. Virol. 2012, 86, 13836–13837. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.X.; Jiang, W.M.; Liu, S.; Wang, S.C.; Zhuang, Q.Y.; Hou, G.Y.; Liu, X.M.; Sui, Z.H.; Chen, J.M. Subclinical Highly Pathogenic Avian Influenza Virus Infection among Vaccinated Chickens, China. Emerg. Infect. Dis. 2014, 20, 2152–2154. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Pascua, P.N.; Song, M.S.; Baek, Y.H.; Kim, C.J.; Choi, H.W.; Sung, M.H.; Webby, R.J.; Webster, R.G.; Poo, H.; et al. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J. Virol. 2009, 83, 4205–4215. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.S.; Yang, J.R.; Liu, M.T.; Yang, C.H.; Cheng, M.C.; Chang, F.Y. Influenza A(H5N2) virus antibodies in humans after contact with infected poultry, Taiwan, 2012. Emerg. Infect. Dis. 2014, 20, 857–860. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Meng, F.; Huang, D.; Sheng, X.; Wang, Y.; Zhang, W.; Chang, W.; Wang, L.; Qin, Z. Genomic and Phylogenetic Characterization of Novel, Recombinant H5N2 Avian Influenza Virus Strains Isolated from Vaccinated Chickens with Clinical Symptoms in China. Viruses 2015, 7, 887-898. https://doi.org/10.3390/v7030887
Xu H, Meng F, Huang D, Sheng X, Wang Y, Zhang W, Chang W, Wang L, Qin Z. Genomic and Phylogenetic Characterization of Novel, Recombinant H5N2 Avian Influenza Virus Strains Isolated from Vaccinated Chickens with Clinical Symptoms in China. Viruses. 2015; 7(3):887-898. https://doi.org/10.3390/v7030887
Chicago/Turabian StyleXu, Huaiying, Fang Meng, Dihai Huang, Xiaodan Sheng, Youling Wang, Wei Zhang, Weishan Chang, Leyi Wang, and Zhuoming Qin. 2015. "Genomic and Phylogenetic Characterization of Novel, Recombinant H5N2 Avian Influenza Virus Strains Isolated from Vaccinated Chickens with Clinical Symptoms in China" Viruses 7, no. 3: 887-898. https://doi.org/10.3390/v7030887
APA StyleXu, H., Meng, F., Huang, D., Sheng, X., Wang, Y., Zhang, W., Chang, W., Wang, L., & Qin, Z. (2015). Genomic and Phylogenetic Characterization of Novel, Recombinant H5N2 Avian Influenza Virus Strains Isolated from Vaccinated Chickens with Clinical Symptoms in China. Viruses, 7(3), 887-898. https://doi.org/10.3390/v7030887





