Abstract
One of the main characteristics of the human immunodeficiency virus is its genetic variability and rapid adaptation to changing environmental conditions. This variability, resulting from the lack of proofreading activity of the viral reverse transcriptase, generates mutations that could be fixed either by random genetic drift or by positive selection. Among the forces driving positive selection are antiretroviral therapy and CD8+ T-cells, the most important immune mechanism involved in viral control. Here, we describe mutations induced by these selective forces acting on the pol gene of HIV in a group of infected individuals. We used Maximum Likelihood analyses of the ratio of non-synonymous to synonymous mutations per site (dN/dS) to study the extent of positive selection in the protease and the reverse transcriptase, using 614 viral sequences from Colombian patients. We also performed computational approaches, docking and algorithmic analyses, to assess whether the positively selected mutations affected binding to the HLA molecules. We found 19 positively-selected codons in drug resistance-associated sites and 22 located within CD8+ T-cell epitopes. A high percentage of mutations in these epitopes has not been previously reported. According to the docking analyses only one of those mutations affected HLA binding. However, algorithmic methods predicted a decrease in the affinity for the HLA molecule in seven mutated peptides. The bioinformatics strategies described here are useful to identify putative positively selected mutations associated with immune escape but should be complemented with an experimental approach to define the impact of these mutations on the functional profile of the CD8+ T-cells.
1. Introduction
Human immunodeficiency virus 1 (HIV-1) is characterized by high levels of diversity and rapid evolution. For HIV-1, four genetic groups have been described: group M causes most of the infections worldwide [] and is divided into nine subtypes: A–D, F–H, J and K []; groups O, N and P which are restricted to the West and Central Africa and have a lower epidemiological impact [,,]. Intersubtype recombinants originated in individuals co-infected with two or more subtypes are also common []. Two studies carried out in the cities of Medellín and Bogotá (Colombia) with HIV sequences of 2001 and 2002 classified the circulating strains within subtype B [,].
The genetic diversity of HIV is partially due to the high rates of replication and mutation []. The lack of proofreading activity of the viral reverse transcriptase (RT) is the major mechanism of mutation [,]. The mutation rate is defined as the number of substitutions per nucleotide site per replicative cycle (s/s/c); in HIV, this rate varies between 2.4 × 10−5 and 3.4 × 10−5 (s/s/c) [,,]. From this, it follows that between one third and one fourth of the new viral particles carry at least one mutation.
These mutations contribute to HIV diversification and have been associated with evasion of the immune response [], defective virus particles [,], changes of cellular tropism and resistance to antiretroviral drugs []. Some mutations are positively selected by the antiretroviral therapy or the immune response, driving HIV evolution [,]. Specifically, CD8+ T-cell responses to HIV-1, restricted through human leukocyte antigen (HLA) molecules, are responsible for the positive selection of escape mutations observed in HIV [,]. These mutations are often located in sites of inter- and intra-subtype variability []. The rapid generation of escape mutants by HIV-1 strains is an enormous challenge to be overcome by the immune system, and also in the design of a globally effective HIV vaccine [].
Moore and collaborators determined the association between specific HLA alleles in HIV-1-infected individuals and the presence or absence of some amino acid replacements in the virus sequences at particular positions []; this study confirmed selection of escape mutations restricted by HLA class I molecules, and underscores the high proportion of viral codons whose evolution is driven by immune escape. In fact, between 10% and 40% of the variation in codons of HIV-1 subtype B strains is driven by immune pressure associated with class I HLA alleles []. The clinical implication of these mutations associated with the immune escape is the increase in the rate of progression of the infection; this phenomenon has most frequently been observed in individuals expressing HLA-B*27 and HLA-B*51 in which amino acid substitutions in immunodominant epitopes led to high levels of HIV replication and rapid progression to AIDS [,,].
The aim of our study was to explore the presence of mutations presumably associated with immune escape in the protease (PRO) and RT proteins of HIV-1 of chronically infected individuals in Colombia. For this purpose, we designed an in silico strategy consisting in detecting positively selected mutations in T CD8+ epitopes, followed by the prediction of changes in the affinity between the mutated peptide and the corresponding HLA molecule. We identified some selected peptides not previously reported as escape mutants within epitopes restricted to class I HLA molecules with high prevalence in the Colombian population and predicted their consequences in the process of antigenic presentation.
2. Materials and Methods
2.1. Data Sources and Sequence Alignments
We analyzed 614 sequences from the pol gene (genome positions 2262–2549 and 2661–3290) coding for the PRO and part of the RT. These sequences were provided by the Centro de Análisis Molecular, a specialized laboratory located in Bogotá, the capital, which performed antiviral sensitivity tests in samples from different regions of Colombia. These samples were collected between 2000 and 2007 from individuals receiving antiretroviral therapy. Sequencing was performed by the TRUGENE® kit (Siemens Healthcare Diagnostics, Tarrytown, NY, USA) []. A code was assigned to each sequence in order to be used anonymously; clinical data and result of additional laboratory studies were not available. Sequences were aligned by the Clustal W method implemented in the BioEdit package (Ibis Bioscience, Carlsbad, CA, USA), using the reference sequence HXB2 of HIV-1 (GenBank accession number K03455.1). Analysis of antiviral resistance data, subtyping and phylodynamics of this database have already been published [,]. Most (82%) sequences exhibit resistance to at least one antiviral drug []. Almost all of these pol gene sequences were subtype B; the exception was one subtype F. No evidence of inter-subtype recombination was found [].
2.2. Tests of Positive Selection
We first used the Z-test, as implemented in the MEGA 5.2.2 software [], to explore evidence of natural selection in the dataset. The statistics (dN–dS), where dS and dN are the numbers of synonymous substitutions per synonymous site and nonsynonymous substitutions per nonsynonymous site, respectively, were estimated using the Nei-Gojobori method []. The variance of the difference was computed using the bootstrap method (10,000 replicates).
We also searched for natural selection by HyPhy, a Maximum Likelihood (ML) based method []. For each codon, estimates of dS and dN were produced using the joint ML reconstructions of ancestral states under the Muse-Gaut model [] of codon substitution and the General Time Reversible model [] of nucleotide substitution. Positive selection was inferred for codons with dN/dS values significantly greater than 1 and the probability of rejecting the null hypothesis of neutral evolution (p-value) was calculated [,]; p values < 0.05 were considered significant.
In both the Z-test and HyPhy analyses, all positions with less than 90% site coverage were eliminated. That is, fewer than 10% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 304 codons in the final dataset.
To confirm the results obtained with the MEGA implementation of HyPhy, the Single Likelihood Ancestor Counting (SLAC) method implemented in the Datamonkey webserver [] was used []. Since Datamonkey does not process datasets with more than 500 sequences, for this analysis the sequences were arbitrarily divided in two groups. The nucleotide and codons substitution models were the same used in the analysis performed in MEGA 5.2.2 software.
2.3. Identification of Peptides
We identified HLA-class I restricted epitopes using the HIV Molecular Immunology Database of Los Alamos []. Each epitope was located in the reference sequence HXB2 of HIV-1 (GenBank accession number: K03455.1) and compared to the homologous sequences in our dataset. For each epitope sequence, we identified the mutations, measured their frequency and classified them as susceptible forms (SF), immune escape (IE) or drug resistance associated (DRAS) mutations. Susceptible forms, defined as epitopes with mutations that are still able to induce a CD8+ T-cell response, and immune escape, defined as mutated epitopes with documented or inferred escape capabilities, were derived from the Los Alamos Database. Substitutions were considered as drug resistance mutations if they were included in the Stanford HIV drug resistance database [,] or in the Update of the Drug Resistance Mutations in HIV-1 of the International AIDS Society []; other substitutions observed but not included in any of the previous definitions were dubbed as “non-classified” (NC) mutations.
2.4. Collection and Preparation of the Three-Dimensional Structure of HLA Molecules
From the Protein Data Bank (PDB) database [] we downloaded the three-dimensional structure of certain HLA molecules found with high frequency in Colombia. We only considered proteins with a resolution ≤ 2.0 Å. The proteins were edited using the AutoDock Tools program []; external ligands and crystallized water were eliminated. AutoDock tools were used to add hydrogen to polar atoms of the molecules, and Gasteiger changes to HLA proteins, facilitating its potential interactions with the peptides. The PDB codes were 1E27, 1DUZ, 3UTQ, 3MRK, 1EFX, 1M6O, 3BO8, 3RL1, 1XR9, 3C9N and 4HX1.
2.5. Structural Prediction of Peptides
Since we required the 3D structures of all peptides included in the study for the in silico docking simulation assay, we used the homology-modeling I-TASSER [,] and ESyPred3D [] servers. With this strategy, we predicted the 3D structure of the wild type (WT) and mutated peptides in order to assess how the mutations affect their folding and the affinity of their binding to the MHC molecules.
2.6. HLA-Peptide Binding Predictions
We predicted the affinities between the HLA protein and the WT or mutant peptides using the receptor-ligand docking simulation AutoDockVina software [] and three different algorithms of affinity estimation: NetMHCpan 2.8 [,], NetMHC [] and stabilized matrix method (SMM) [,]. For AutoDockVina, we evaluated the affinity constant based on the indirect score of the free energy of binding. The conformational search space in the AutoDockVina simulation was limited to the vicinity of the major histocompatibility complex (MHC) pocket. For each peptide, the docking simulations were run 20 times and the best scores of each run were selected and averaged; the interactions between residues from both, ligand and receptor were verified in detail. Statistical comparison of the free energy of binding of the WT and mutated epitopes were performed by the Kruskal-Wallis and Mann-Whitney U tests.
For the NetMHCpan, NetMHC and SMM algorithms, we considered as good, intermediate and poor bindings when the Inhibitory Concentration 50 (IC50) was < 50 nM, 50–500 nM or > 500 nM, respectively. Two-fold or larger differences between the affinities of the WT and mutated peptides with the corresponding HLA molecule were considered as significant []. Correlation analyses among the different methods of affinity assessment were performed by the Spearman test. All statistics were performed using GraphPrism 5.0 package; p-values lower than 0.05 were considered significant.
3. Results
3.1. Z-Test and Maximum Likelihood Analysis of Positive Selection
We analyzed 614 pol gene sequences corresponding to the PRO (codons 4–99) and the RT (codons 38–247) obtained from HIV-infected individuals living in 16 Colombian cities, mainly Bogotá (44.4%), Cali (12.6%) and Medellín (10.6%). There were 280 variable sites out of a total of 304 codons.
Initially, we searched for evidence of selection using the Z-test in the overall average of all sequences. The null hypothesis of strict neutrality (dN = dS) was rejected in favor of the alternative hypothesis (dN ≠ dS) with a p = 0.000. The value for the statistic dN–dS was −10.5, indicating a higher rate of synonymous than non-synonymous substitutions. When tested for positive selection the null hypothesis of neutrality in favor of the alternative (dN > dS) could not be rejected (p = 1.000). Conversely, when tested for purifying selection the null hypothesis of neutrality was rejected (p = 0.000) in favor of the alternative (dN < dS), indicating that negative selection predominates along the studied segments of pol.
However, it is well known that a purifying selection in most parts of the sequence could hide positive selection occurring at specific codons [,]. To obtain an assessment of codon-specific selective pressures, we performed an analysis of dN/dS by the Maximum Likelihood method using the HyPhy implementation in MEGA. The test of positive selection was significant (p < 0.05) in 35 codons (18 in PRO and 17 in RT) (Table 1). For both proteins, sites under positive selection were spread throughout the gene.

Table 1.
Mutations identified as positively selected by the Maximum Likelihood method in protease (PRO) and reverse transcriptase (RT) of 614 human immunodeficiency virus (HIV) infected individuals.
| Codons Position (HXB2) | Substitution | dN/dSa | p Value | Location b |
|---|---|---|---|---|
| Protease | ||||
| 12 | T → P/S | 4.4 | 0.0000 * | Epitope |
| 13 | I → V | 5.0 | 0.0002 | Epitope |
| 19 | L → I/Q/V | 1.7 | 0.0015 | Epitope |
| 35 | E → D | 1.9 | 0.0038 | Epitope |
| 37 | S→N/E/D | 5.7 | 0.0000 * | Epitope |
| 41 | R→K | 2.9 | 0.0000 * | Epitope |
| 54 | I → L/V/M/T/A/S | 3.0 | 0.0000 * | Epitope/DRAS |
| 62 | I → V | 2.9 | 0.0002 | DRAS |
| 64 | I → L/M/V | 6.8 | 0.0000 * | DRAS |
| 71 | A → V/I/T/L | 2.3 | 0.0000 * | DRAS |
| 72 | I → V/T/L/R | 2.1 | 0.0115 | Epitope |
| 73 | G → C/S/T/A | 2.1 | 0.0146 | DRAS |
| 74 | T → P | 15.6 | 0.0000 * | DRAS |
| 77 | V → I | 5.3 | 0.0000 * | Epitope/DRAS |
| 82 | V → A/F/S/T | 4.2 | 0.0000 * | Epitope/DRAS |
| 85 | I → V | 36 | 0.0460 | DRAS |
| 90 | L → M | 4.4 | 0.0000 * | DRAS |
| 93 | I → L/M | 8.2 | 0.0000 * | DRAS |
| Reverse transcriptase | ||||
| 39 | T → A/K/S/L | 4.4 | 0.0000 * | Epitope |
| 48 | S → T | 2.6 | 0.0203 | Epitope |
| 69 | T → S/N/D/A/G | 2.2 | 0.0008 | DRAS |
| 74 | L → V/I | 3.0 | 0.0000 * | Epitope/DRAS |
| 75 | V → I | 2.4 | 0.0024 | DRAS |
| 98 | A → S | 2.0 | 0.0106 | Epitope |
| 102 | K → R/Q/E/N/H | 20.1 | 0.0000 * | DRAS |
| 103 | K → N/S | 2.0 | 0.0003 | DRAS |
| 118 | V → I | 1.7 | 0.0179 | Epitope |
| 135 | I → T/V/L/R/M/K | 3.3 | 0.0000 * | Epitope |
| 162 | S → C/A/Y/D/N/H | 2.1 | 0.0003 | Epitope |
| 184 | M → V | 2.5 | 0.0000 * | Epitope/DRAS |
| 188 | Y → L | 4.8 | 0.0002 | DRAS |
| 200 | T → A/I/E | 17.4 | 0.0000 * | Epitope |
| 202 | I → V | 606.1 | 0.0083 | Epitope |
| 211 | R → K/Q/G/T | 1.5 | 0.0051 | Epitope |
| 215 | T → I | 3.4 | 0.0000 * | Epitope |
a Ratio of nonsynonymous (dN) to synonymous (dS) substitutions per site. b DRAS: Drug resistance associated sites according to the Stanford HIV drug resistance database [,] or the last Update of the Drug Resistance Mutations in HIV-1 of the International AIDS Society []. Epitope: peptide recognized by CD8+ T-cells according to the HIV Molecular Immunology Database of Los Alamos []. * p-values below the Bonferroni corrected (0.00016) significance level.
Eleven out of 18 selected codons in the PRO and 7 out of 17 in the RT have been previously associated with drug resistance; one of them (position 102 in the RT) was associated with drug resistance in a previous report, although it is not included in the last update of the Drug Resistance Mutations in HIV-1 or in the Stanford HIV drug resistance database [,,]. On the other hand, 10 of the selected codons in the PRO and 12 in the RT were located in recognized CD8+ T-cell epitopes. Three codons in the PRO and two in the RT were located in drug resistance-associated sites that are also located within CD8+ T-cell epitopes. Similar results were obtained when the selection analysis was performed in the divided dataset using the SLAC method in Datamonkey; this method identified as positively selected the same positions previously identified by the HyPhy method implemented in MEGA 5.2.2, except for codon 202 of the RT.
3.2. Identification of CD8+ T-cell Epitopes with Amino Acid Substitutions
Based on the previous analysis, we selected the mutations located within CD8+ T-cell epitopes with significant (p < 0.05) evidence of positive selection and with a frequency greater than 5% among the 614 sequences. There were 20 such mutations, 11 in the PRO and 9 in the RT (Table 2).

Table 2.
Identification of epitopes affected by the positively selected mutations in in PRO and RT.
| Mutations | Frequency (%) | Epitope Affected (HLA Alleles) a | Association b |
|---|---|---|---|
| Protease | |||
| I13V | 20.4 | QRPLVTIKI (A*01:01) | NC |
| QRPLVTIKIG (B51) | NC | ||
| VTIKIGGQLK (A*11:01) | SF | ||
| TIKIGGQLK (A3 supertype) | NC | ||
| L19I | 9.0 | VTIKIGGQLK (A*11:01, A*03:01) | SF |
| TIKIGGQLK (A3 supertype) | NC | ||
| E35D | 28.7 | DTVLEEMSL (A*68:02) | NC |
| EEMSLPGRW (B*44:02, B*44:03, B18, B40) | IE | ||
| S37N | 56.0 | DTVLEEMSL (A*68:02) | SF |
| EEMSLPGRW (B*44:02, B*44:03, B18, B40) | SF | ||
| S37D | 14.1 | DTVLEEMSL (A*68:02) | NC |
| EEMSLPGRW (B*44:02, B*44:03, B18, B40) | SF | ||
| R41K | 42.8 | EEMSLPGRW (B*44:02, B*44:03, B18, B40) | NC |
| LPGRWKPKMI (Cw3) | NC | ||
| I54Vc | 19.3 | KMIGGIGGFI (B62) | IE |
| I72V | 9.3 | IEICGHKAIG (B18, B40, B44) | NC |
| GHKAIGTVL (B15) | NC | ||
| I72T | 5.5 | IEICGHKAIG (B18, B40, B44) | NC |
| GHKAIGTVL (B15) | NC | ||
| V77Ic | 27.8 | LVGPTPVNI (A2) | NC |
| V82A c | 13.8 | LVGPTPVNI (A2) | IE |
| Reverse Transcriptase | |||
| T39A | 7.1 | ALVEICTEM (A*02, A*02:01, A2) | NC |
| A98S | 8.7 | GIPHPAGLK (A*03:01) | NC |
| V118I | 19.1 | VLDVGDAYFSV (A*02:01) | NC |
| DAYFSVPL (A24, B*51:01) | NC | ||
| I135T | 30.7 | KYTAFTIPSI (A2) | NC |
| TAFTIPSI (B*51) | IE | ||
| I35V | 7.7 | KYTAFTIPSI (A2) | NC |
| TAFTIPSI (B*51) | IE | ||
| S162C | 10.1 | SPAIFQSSM (B7, B35) | SF |
| AIFQSSMTK (A*03:01) | SF | ||
| T200A | 19.0 | DLEIGQHRTK (A3) | NC |
| I202V | 6.8 | KIEELRQHL (A2) | NC |
| KIEELRQHLL (B58) | NC | ||
| IEELRQHLL (B*40:01, B60, B61) | IE | ||
| R211K | 49.3 | EELRQHLLRW (B44) | NC |
a The residues in the epitope affected by the substitutions are in bold. b IE: Immune escape, i.e., mutations with documented or inferred escape; SF: susceptibility forms, defined as epitope sequences with mutations able to induce a CTL response; NC: none classified, i.e., mutations not included in the previous definitions. c Mutations associated with both, immune escape and drug resistance.
Some of these mutations were located in more than one overlapping epitope; for instance, the I13V mutation was found in four peptides, although only VTIKIGGQLK had been previously recognized as an epitope binding to different HLA molecules and influencing the immune response []. As a consequence 31 epitopes were affected by mutations at a high frequency. In addition, there was more than one mutation in the some epitopes, for example in DTVLEDMNL and EDMNLPGRW.
Only five mutant epitopes were previously recognized as immune escape (IE) and seven as susceptible forms (SF). Most (19) of the mutated epitopes have not been classified as IE or SF. All epitopes identified in this study as positively selected are presented by HLA alleles that have been reported in our population, except the epitope LPGRWKPKMI which is presented by HLA-Cw3; frequencies of alleles at the HLA-C locus have not been reported for Colombia.
3.3. Docking Simulation and Algorithmic Estimation of the Affinity of Peptides Binding to HLA Molecules
We implemented an in silico screening system to assess the affinity between the HLA proteins and the corresponding WT or mutated peptides, using a combination of different computational tools. This protocol was applied to peptides selected in Table 2. First, each peptide with the respective HLA molecule was subjected to docking analysis in AutoDockVina and the average of the data was plotted (Figure 1). All the evaluated peptides, either WT or mutated, were docked to the respective HLA molecule (Figure 1). Average free energy of binding ranged from −4.0 to −7.7 Kcal/mol for the PRO and RT peptides. No significant differences in values of free energy between the WT and the mutated forms of the epitopes where found, except for the DTVLEEMNL variant, which has a decrease of 0.44 Kcal/mol (p < 0.05) from the wild type DTVLEEMSL form (Figure 1A,B).
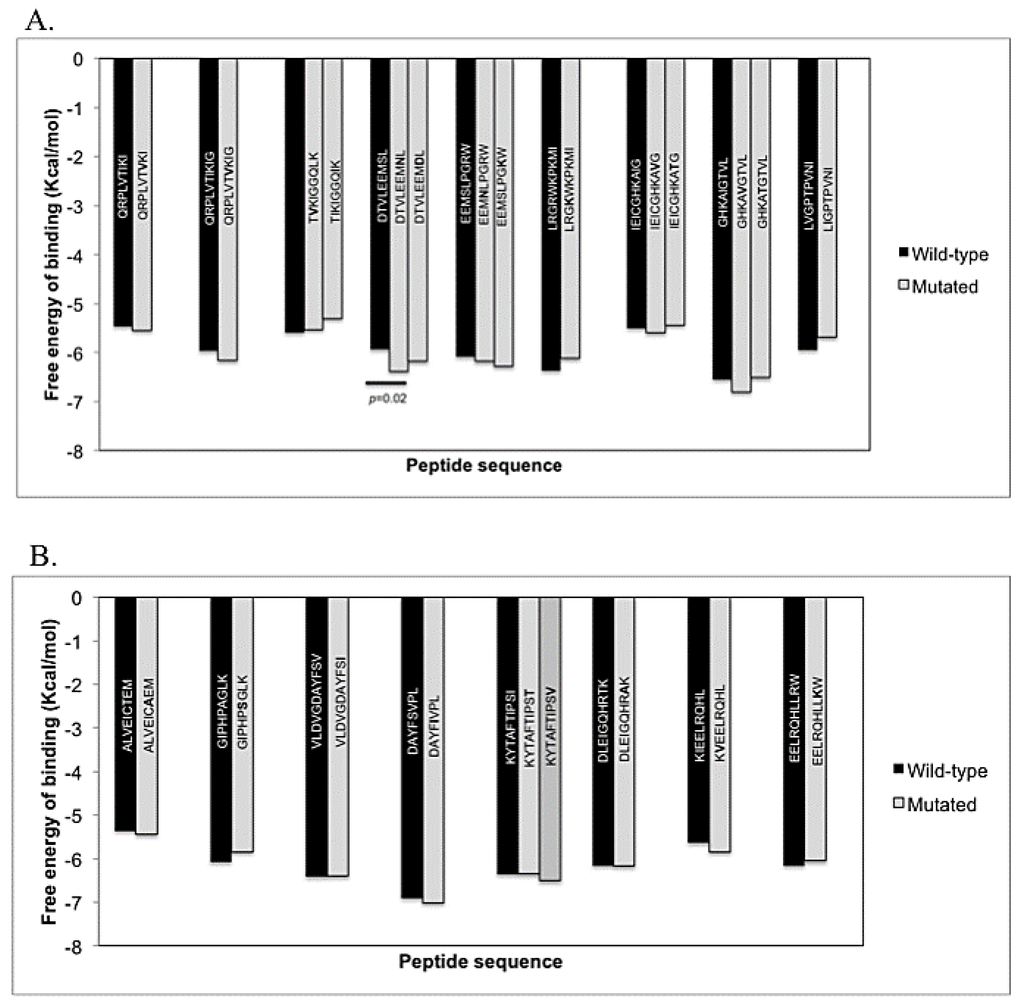
Figure 1.
Free energy of binding to HLA molecules of peptides using the in silico assay. The mean of the lowest free energy of binding in 20 runs was plotted for each epitope derived from the PRO (A); and RT (B).
Next, we estimated the binding affinity using three algorithmic tools: SMM, NetMHC and NetMHCpan. Many epitopes had an IC50 > 500 nM, indicating a low binding affinity; only one epitope in the PRO and six in the RT had good predicted binding affinities (IC50 < 50 nM) by at least one of the algorithms (Table 3). Four mutated epitopes in the PRO and three in the RT exhibited a significant decrease in the predicted affinity with respect to the WT form, as assessed by a two-fold or greater increase in the IC50 in at least two algorithms. Four of these epitopes, two in the PRO (TIKIGGQIK, DTVLEEMDL) and two in the RT (VLDVGDAYFSI, KYTAFTIPST) had not been reported previously as IE mutations (Table 3).
The IC50 estimated by the three algorithmic methods were highly correlated, with r ≥ 0.6991 and p < 0.0001. Conversely, the correlations of these values with the docking’s ΔGs were all negatively but not significantly correlated (Supplementary Table S1).

Table 3.
Epitope-HLA binding affinities by three computational algorithms (SMM, NetMHC and NetMHCpan).
| Amino Acid Sequence | Alleles | SMM | NetMHC | NetMHCpan | ||||
|---|---|---|---|---|---|---|---|---|
| Affinity (nM) | Affinity (nM) | Affinity (nM) | ||||||
| LPPVVAKEI a | B*51 | 172 | 102 | 797 | ||||
| NLVPMVATV a | A*02 | 66 | 29 | 21 | ||||
| Protease | ||||||||
| QRPLVTIKI | A*01:01 | 194334 | 21837 | 36562 | ||||
| QRPLVTVKI | 231499 | 21639 | 37194 | |||||
| QRPLVTIKIG | B51 | 166360 | 30751 | 43536 | ||||
| QRPLVTVKIG | 168286 | 30708 | 43446 | |||||
| TIKIGGQLK | A3 | 432 | 582 | 537 | ||||
| TVKIGGQLK | 456 | 720 | 856 | |||||
| TIKIGGQIK | 1041 b | 2126 b | 1654 b | |||||
| DTVLEEMSL | A*68:02 | 874 | 2686 | 1709 | ||||
| DTVLEEMNL | 1012 | 3795 | 2078 | |||||
| DTVLEEMDL | 2278 b | 12370 b | 6676 b | |||||
| EEMSLPGRW | B*44:02 | 30 | 25 | 14 | ||||
| EEMNLPGRW | 32 | 28 | 21 | |||||
| EEMSLPGKW | 30 | 22 | 14 | |||||
| EDMSLPGRW | 431 b | 565 b | 578 b | |||||
| IEICGHKAIG | B44 | 2093 | 6969 | 6674 | ||||
| IEICGHKAVG | 2122 | 5792 | 5601 | |||||
| IEICGHKATG | 2103 | 5060 | 5162 | |||||
| GHKAIGTVL | B15 | 13091 | 14972 | 15778 | ||||
| GHKAVGTVL | 9840 | 13657 | 14015 | |||||
| GHKATGTVL | 8222 | 12947 | 14673 | |||||
| LVGPTPVNI | A2 | 3027 | 3829 | 4005 | ||||
| LIGPTPVNI | 1945 | 2195 | 1596 | |||||
| LVGPTPANI | 3555 | 3014 | 3912 | |||||
| KMIGGIGGFI | B62 | 514 | 415 | 769 | ||||
| KMIGGIGGFV | 1493 b | 937 b | 1744 b | |||||
| Reverse transcriptase | ||||||||
| ALVEICTEM | A2 | 116 | 70 | 41 | ||||
| ALVEICAEM | 101 | 50 | 32 | |||||
| GIPHPAGLK | A*03:01 | 290 | 108 | 316 | ||||
| GIPHPSGLK | 266 | 99 | 334 | |||||
| VLDVGDAYFSV | A*02:01 | 4314 | 284 | 9 | ||||
| VLDVGDAYFSI | 8788 b | 534 | 33 b | |||||
| DAYFSVPL | B*51:01 | 7628 | 6527 | 4202 | ||||
| DAYFSIPL | 15150 | 7805 | 4148 | |||||
| KYTAFTIPSI | A2 | 1470 | 5199 | 9216 | ||||
| KYTAFTIPST | 4925 b | 15627 b | 27884 b | |||||
| KYTAFTIPSV | 381 | 1509 | 5743 | |||||
| Reverse transcriptase | ||||||||
| TAFTIPSI | B51 | 996 | 2399 | 1153 | ||||
| TAFTIPST | 1128 | 16898 b | 17009 b | |||||
| TAFTIPSV | 1372 | 4052 | 2887 b | |||||
| DLEIGQHRTK | A3 | 798 | 8188 | 15564 | ||||
| DLEIGQHRAK | 839 | 9011 | 15773 | |||||
| KIEELRQHL | A2 | 5689 | 10072 | 8180 | ||||
| KVEELRQHL | 8853 | 13644 | 14234 | |||||
| KIEELRQHL | A2 | 5689 | 10072 | 8180 | ||||
| KVEELRQHL | 8853 | 13644 | 14234 | |||||
| EELRQHLLRW | B44 | 78 | 104 | 29 | ||||
| EELRQHLLKW | 78 | 84 | 29 | |||||
a Known epitopes of HIV-1 GAG and cytomegalovirus pp65, respectively, included as positive controls of the analyses performance. b Two-fold or larger differences between the affinities of the WT and the mutated peptide.
4. Discussion
In the present study, we explored immune escape mutations in the PRO and the RT, using HIV-1 sequences from 614 Colombian patients who were genotyped for antiviral sensitivity purposes. We found evidence of positive selection in many codons, associated or not with drug resistance (Table 1). All 17 selected substitutions not previously associated to drug resistance were located in CD8+ T-cell epitopes. Of these, 14 have not been previously identified as immune escape in the HIV Molecular Immunology Database of Los Alamos [], opening the possibility of being considered as newly identified escape mutations, and prompting us to consider alternative explanations.
One possibility is that some of these mutations are indeed associated to drug resistance, but they have not been included in the drug resistance databases [] due to the lack of strong evidence of this association. For instance, the I13V mutation in the PRO, that had been reported as a minor mutation associated with resistance to tipranavir and ritonavir, was removed from the 2010 update []. A related explanation is that these mutations could have compensatory effects that had not been previously detected. Since most HIV drug resistance mutations are naturally deleterious, such compensatory mutations are frequently selected. Similarly, compensatory mutations that restore fitness losses have previously been reported in CD8+ T-cell escape mutants [,].
An alternative explanation is that the selection test could be detecting significant elevations in the dN/dS ratio just for stochastic reasons; in a 304 codon-long sequence with a significance threshold of 0.05 one could expect 15 codons with a dN/dS < 0.05 just by chance. However, even with the strict Bonferroni correction for multiple comparisons, most (11/18 in the PRO and 7/17 in the RT) codons remain with a dN/dS ratio with a p-value below the corrected (0.00016) significance level in the test for positive selection, as shown in Table 1.
A more arguable explanation for this finding is the potential “hitch-hiking” effect of selective sweeps induced by the selective pressure exerted by antiviral drugs. In the presence of antiviral therapy, a drug resistance mutation will increase its frequency in the viral population up to fixation; other mutations arising in the same molecule, could hitch-hike along with it to fixation even if they are slightly deleterious []. Previous reports indicate that such hitch-hiking mutations persist for about 18 months in HIV-infected patients receiving antiviral therapy []. This effect is attenuated by the high rate of recombination, which is even higher than the mutation rate in terms of events per genome per HIV-1 cycle []; this could limit the effect of selective sweeps to closely linked sites. However, the extent of this effect is difficult to assess and we cannot rule out the possibility that this phenomenon has played a role in the mutations observed in our dataset.
Even so, the consistent location of non-synonymous substitutions in CD8+ T-cell epitopes, and not in other sites that could also be affected by hitch-hiking, still makes the immune escape hypothesis a plausible explanation for the high frequency of such mutations. Although not all mutations were located at the second position or at the C-terminal amino acid of the peptide chain that are the critical positions for the stability of the HLA-epitope interaction [], the immune escape could also be associated with lack of T-cell receptor (TCR) recognition. Previous reports indicate that some mutations in the epitope can alter TCR recognition [,], resulting in a reduced functional capacity of CD8+ T-cells [,]. Functional studies are required to explore whether or not these mutations alter the strength of the interaction between the TCR and the HLA-peptide complex.
CD8+ T-cells play a crucial role in the control of HIV replication [], although complete viral suppression is not achieved for a number of different reasons, including the appearance of mutations in immunodominant epitopes []. Several studies have demonstrated that HIV-specific CD8+ T-cell response selects escape mutants during the acute and chronic phases of HIV and simian immunodeficiency virus (SIV) infections [,,], inducing loss of immune control and disease progression []. The selection of escape mutations depends on the HLA molecules. Indeed, the escape mutations could be predicted based on the host MHC alleles []. Most escape mutations revert when the virus is transmitted to another host with a different HLA make up. However, for HLA alleles that are common in a population, the corresponding escape mutations may become predominant in the strains circulating in that particular region, contributing to viral evolution and inducing an imprinting effect [,].
In this study, most CD8+ T-cell epitopes affected by positively selected mutations are restricted to the HLA alleles found in high frequency in the Colombian population (Supplementary Table S2). For instance, several mutated peptides are presented by the HLA-A*02 and -A*03 molecules, which in Colombia have frequencies of 22.2% and 7.9%, respectively []. In the case of HLA-B alleles, we observed mutations in epitopes restricted to the molecules -B*18 (7.1%), -B*35 (17.8%), -B*44 (9.1%) and -B*51 (6.4%) that are found in high frequencies (shown in parenthesis) []. However, the correlation between the frequency of the HLA alleles and the frequency of the mutation is not perfect. Only one HLA-A*24-restricted and one HLA-A*68-restricted epitopes showed substitutions in our study; these alleles had frequencies of 19.1% and 5.6%, respectively in Colombia; this could be due to a higher functional constraint against mutations in these peptides. Moreover, some epitopes restricted to rare HLA alleles in our population, such as HLA-B*15 (0.6%), were mutated with a significant (5.5%) frequency; these mutants could have appeared in a viral lineage before its introduction to Colombia and its frequency could have persisted or even increased by random genetic drift.
To predict the possible functional consequences of the observed amino acid substitutions in CD8+ T-cell epitopes, and further substantiate their immune escape potential, we performed an in silico docking simulation assay between the HLA molecules and the WT and mutated peptides. In addition, we predicted changes in the affinities of binding using three computational algorithms. We observed discrepancies between the inferred affinities obtained by docking simulation and by the computational algorithms; no correlation was observed in the values obtained by these two strategies. While the docking showed almost no changes in the free energy of binding in the mutant peptides, the algorithms predicted a significant decrease in affinities in seven peptides, four of which (TIKIGGQIK, DTVLEEMDL, VLDVGDAYFSI and KYTAFTIPST) had not been previously reported as immune escape mutants. Moreover, the results of the three algorithmic methods were strongly correlated (Supplementary Table S1).
The discrepancies between strategies could be explained by their different approaches. In general, the algorithms generate a prediction from a set of experimental data [] derived from previous reports in-house experiments or specialized databases containing epitope-related information; estimates are expressed as a function of the concentration of a binding inhibitor []. The docking strategy is based on computational structural analysis; it predicts the affinity based on the spatial disposition of the atoms and chemical bonding, treating the protein as a rigid structure and estimating the free energy of binding; the accuracy of this kind of prediction could be affected by the size of the ligand, the dimension of the motion space or the rotational degrees of freedom of the ligand [].
In this study, the algorithmic methods detected some (three out of five) of the previously reported IE mutations and allowed to infer four other mutations not previously reported as such. The docking strategy did not identify any of these IE mutations; instead, it detected a modest change of 0.44 kcal/mol between the WT and mutated forms in an epitope catalogued as SF. This suggests that algorithmic methods are more sensitive as tools for predicting potential IE mutants; however, we observed that some epitopes that experimentally had high binding to HLA molecules and high immunogenic capacity, exhibited low predicted affinities by the computational algorithms. For example, LVGPTPVNI was previously reported with a high HLA-A2 binding affinity and cytotoxic response of CD8+ T cells []; however, all the predicted affinities had IC50s > 500 nM (SMM: 3027.68 nM, NetMHC: 3829.00 nM and NetMHCpan: 4005.25 nM). Taking this into account, the four mutations found here with both, strong evidence of positive selection and significant decrease in the affinity of the epitopes, should be regarded only as candidate IE mutations; in vitro assays are required to determine whether they lead to destabilization of the HLA-peptide-TCR complex or to changes in the functional profiles in CD8+ T-cells.
The main limitations of this study were the absence of HLA typing, CD4+ T-cell counts and clinical data of the patients from whom the HIV-1 sequences were obtained, which would have allowed to make associations between mutations and disease progression. Nevertheless we can assume that all patients had viral loads ≥ 1000 RNA copies/µL, the minimal viral load that permits sequencing with the TRUGENE® kit. On the other hand, the strength of the bioinformatic approach to predict immune escape mutants that we introduce here is the combination of several analytical tools of different nature, and its application to a large dataset.
In summary, we introduced a strategy aimed at screening immune escape mutations that combine methods for positive selection, protein-protein docking and computational algorithms for affinity prediction. This allowed us to postulate four possible new mutations associated with immune escape in HIV-1-infected patients. Functional studies should be performed to confirm the effect of these mutations on the response of CD8+ T-cells; prospective clinical studies could test the biological relevance of these findings.
Supplementary Files
Supplementary File 1Acknowledgments
This work was financed by Colciencias (grant 1115-569-33380) and the program “Estrategia de Sostenibilidad del Grupo Inmunovirología 2014–2015 de la Universidad de Antioquia”. L.A.-S. is recipient of a doctoral scholarship from Colciencias. We are indebted to two anonymous reviewers for many useful suggestions and to Anne-Lise Haenni for her grammar corrections.
Author Contributions
F.J.D., R.O., M.T.R., P.O.-G., P.A.V.-H. and L.A.-S. conceived and designed the experiment; F.J.D., R.O and L.A.-S. performed the analyses; F.J.D., R.O. and L.A.-S. analyzed the date; F.J.D. and L.A.-S. wrote the manuscript; F.J.D., M.T.R., R.O., P.A.V.-H. and L.A.-S. proof-read the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References and Notes
- Hemelaar, J.; Gouws, E.; Ghys, P.D.; Osmanov, S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. Aids 2006, 20, W13–W23. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Anderson, J.P.; Bradac, J.A.; Carr, J.K.; Foley, B.; Funkhouser, R.K.; Gao, F.; Hahn, B.H.; Kalish, M.L.; Kuiken, C.; et al. Hiv-1 nomenclature proposal. Science 2000, 288, 55–56. [Google Scholar]
- Ayouba, A.; Mauclere, P.; Martin, P.M.; Cunin, P.; Mfoupouendoun, J.; Njinku, B.; Souquieres, S.; Simon, F. HIV-1 group o infection in cameroon, 1986 to 1998. Emerg. Infect. Dis. 2001, 7, 466–467. [Google Scholar] [CrossRef] [PubMed]
- Ayouba, A.; Souquieres, S.; Njinku, B.; Martin, P.M.; Muller-Trutwin, M.C.; Roques, P.; Barre-Sinoussi, F.; Mauclere, P.; Simon, F.; Nerrienet, E. HIV-1 group n among HIV-1-seropositive individuals in cameroon. Aids 2000, 14, 2623–2625. [Google Scholar] [CrossRef] [PubMed]
- Plantier, J.C.; Leoz, M.; Dickerson, J.E.; De Oliveira, F.; Cordonnier, F.; Lemee, V.; Damond, F.; Robertson, D.L.; Simon, F. A new human immunodeficiency virus derived from gorillas. Nat. Med. 2009, 15, 871–872. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Sharp, P.M.; McCutchan, F.E.; Hahn, B.H. Recombination in hiv-1. Nature 1995, 374, 124–126. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, G.I.; Bautista, C.T.; Eyzaguirre, L.; Carrion, G.; Arias, S.; Sateren, W.B.; Negrete, M.; Montano, S.M.; Sanchez, J.L.; Carr, J.K. Molecular epidemiology of human immunodeficiency virus-infected individuals in Medellin, Colombia. Am. J. Trop. Med. Hyg. 2006, 74, 674–677. [Google Scholar] [PubMed]
- Montano, S.M.; Sanchez, J.L.; Laguna-Torres, A.; Cuchi, P.; Avila, M.M.; Weissenbacher, M.; Serra, M.; Vinoles, J.; Russi, J.C.; Aguayo, N.; et al. Prevalences, genotypes, and risk factors for HIV transmission in south america. J. Acquir. Immune Defic. Syndr. 2005, 40, 57–64. [Google Scholar] [CrossRef]
- Ho, D.D.; Neumann, A.U.; Perelson, A.S.; Chen, W.; Leonard, J.M.; Markowitz, M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 1995, 373, 123–126. [Google Scholar] [CrossRef] [PubMed]
- O'Neil, P.K.; Sun, G.; Yu, H.; Ron, Y.; Dougherty, J.P.; Preston, B.D. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J. Biol. Chem. 2002, 277, 38053–38061. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.P.; Loeb, L.A. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 1992, 31, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Abram, M.E.; Ferris, A.L.; Shao, W.; Alvord, W.G.; Hughes, S.H. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 2010, 84, 9864–9878. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Temin, H.M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 1995, 69, 5087–5094. [Google Scholar] [PubMed]
- Simon, V.; Ho, D.D. HIV-1 dynamics in vivo: Implications for therapy. Nat. Rev. Microbiol. 2003, 1, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Leslie, A.J.; Pfafferott, K.J.; Chetty, P.; Draenert, R.; Addo, M.M.; Feeney, M.; Tang, Y.; Holmes, E.C.; Allen, T.; Prado, J.G.; et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 2004, 10, 282–289. [Google Scholar]
- Marozsan, A.J.; Fraundorf, E.; Abraha, A.; Baird, H.; Moore, D.; Troyer, R.; Nankja, I.; Arts, E.J. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. J. Virol. 2004, 78, 11130–11141. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, D.S.; Willey, R.L.; Sato, H.; Chang, L.J.; Blumenthal, R.; Martin, M.A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 1993, 67, 2182–2190. [Google Scholar] [PubMed]
- Hemelaar, J. Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 2013, 66, 391–400. [Google Scholar] [CrossRef] [PubMed]
- De, S.L.E.; Holmes, E.C.; Zanotto, P.M. Distinct patterns of natural selection in the reverse transcriptase gene of HIV-1 in the presence and absence of antiretroviral therapy. Virology 2004, 325, 181–191. [Google Scholar] [CrossRef] [PubMed]
- McMichael, A.J.; Borrow, P.; Tomaras, G.D.; Goonetilleke, N.; Haynes, B.F. The immune response during acute HIV-1 infection: Clues for vaccine development. Nat. Rev. Immunol. 2010, 10, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, Y.; Pfafferott, K.; Frater, J.; Matthews, P.; Payne, R.; Addo, M.; Gatanaga, H.; Fujiwara, M.; Hachiya, A.; Koizumi, H.; et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 2009, 458, 641–645. [Google Scholar]
- Moore, C.B.; John, M.; James, I.R.; Christiansen, F.T.; Witt, C.S.; Mallal, S.A. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2002, 296, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Matthews, P.C.; Leslie, A.J.; Katzourakis, A.; Crawford, H.; Payne, R.; Prendergast, A.; Power, K.; Kelleher, A.D.; Klenerman, P.; Carlson, J.; et al. HLA footprints on human immunodeficiency virus type 1 are associated with interclade polymorphisms and intraclade phylogenetic clustering. J. Virol. 2009, 83, 4605–4615. [Google Scholar]
- Hemelaar, J. The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 2012, 18, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Brumme, Z.L.; Brumme, C.J.; Heckerman, D.; Korber, B.T.; Daniels, M.; Carlson, J.; Kadie, C.; Bhattacharya, T.; Chui, C.; Szinger, J.; et al. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007, 3, e94. [Google Scholar]
- Kelleher, A.D.; Long, C.; Holmes, E.C.; Allen, R.L.; Wilson, J.; Conlon, C.; Workman, C.; Shaunak, S.; Olson, K.; Goulder, P.; et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 2001, 193, 375–386. [Google Scholar]
- Feeney, M.E.; Tang, Y.; Roosevelt, K.A.; Leslie, A.J.; McIntosh, K.; Karthas, N.; Walker, B.D.; Goulder, P.J. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 2004, 78, 8927–8930. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, Y.; Kuse, N.; Gatanaga, H.; Naruto, T.; Fujiwara, M.; Dohki, S.; Akahoshi, T.; Maenaka, K.; Goulder, P.; Oka, S.; et al. Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. J. Virol. 2010, 84, 7151–7160. [Google Scholar]
- Grant, R.M.; Kuritzkes, D.R.; Johnson, V.A.; Mellors, J.W.; Sullivan, J.L.; Swanstrom, R.; D’Aquila, R.T.; van Gorder, M.; Holodniy, M.; Lloyd, R.M., Jr.; et al. Accuracy of the trugene HIV-1 genotyping kit. J. Clin. Microbiol. 2003, 41, 1586–1593. [Google Scholar]
- Gómez, S.; Olaya-García, P.; Díaz, F. Resistencia a los medicamentos antirretrovirales en pacientes que reciben tratamiento para vih-sida en colombia. Infectio 2010, 14, 248–256. [Google Scholar] [CrossRef]
- Pineda-Peña, A.; Rodrigues Faria, N.; Diaz, F.; Olaya-García, P.; Moller Frederiksen, C.; Guangdi, L.; Gomez-Lopez, A.; Lemey, P.; Vandamme, A. The colombian pidemic is dominated by HIV-1subtype b over time: A molecular epidemiolog and phylodynamics study. In Proceedings of the 20th International HIV Dynamics and Evolution, Utrecht, The Netherlands, 8–11 May 2013.
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Pond, S.L.; Frost, S.D.; Muse, S.V. Hyphy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Muse, S.V.; Gaut, B.S. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 1994, 11, 715–724. [Google Scholar] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press, Inc: New York, NY, USA, 2000; pp. 147–163. [Google Scholar]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef]
- Suzuki, Y.; Gojobori, T. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 1999, 16, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Delport, W.; Poon, A.F.; Frost, S.D.; Kosakovsky Pond, S.L. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- HIV molecular immunology database. Available online: http://www.hiv.lanl.gov/ (accessed on 12 February 2014).
- Liu, T.F.; Shafer, R.W. Web Resources for HIV type 1 genotypic-resistance Test Interpretation. Clin. Infect. Dis. 2006, 42, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- HIV drug resistance database – Stanford University. Available online: http://hivdb.stanford.edu/ (accessed on 16 May 2014).
- Johnson, V.A.; Calvez, V.; Gunthard, H.F.; Paredes, R.; Pillay, D.; Shafer, R.W.; Wensing, A.M.; Richman, D.D. Update of the drug resistance mutations in HIV-1: March 2013. Top. Antivir. Med. 2013, 21, 6–14. [Google Scholar] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic. Acids. Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Green, L.G.; Radic, Z.; Taylor, P.; Sharpless, K.B.; Olson, A.J.; Grynszpan, F. Automated docking with protein flexibility in the design of femtomolar “click chemistry” inhibitors of acetylcholinesterase. J. Chem. Inf. Model. 2013, 53, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Leonard, N.; de Bolle, X.; Depiereux, E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics 2002, 18, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. Netmhcpan, a method for mhc class I binding prediction beyond humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lundegaard, C.; Blicher, T.; Lamberth, K.; Harndahl, M.; Justesen, S.; Roder, G.; Peters, B.; Sette, A.; Lund, O.; et al. Netmhcpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2007, 2, e796. [Google Scholar]
- Buus, S.; Lauemoller, S.L.; Worning, P.; Kesmir, C.; Frimurer, T.; Corbet, S.; Fomsgaard, A.; Hilden, J.; Holm, A.; Brunak, S. Sensitive quantitative predictions of peptide-MHC binding by a “query by committee” artificial neural network approach. Tissue Antigens 2003, 62, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.; Sette, A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform. 2005, 6, 132. [Google Scholar] [CrossRef]
- Immune epitope database and analysis resource. Available online: http://www.iedb.org/ (accessed on 12 January 2015).
- Sette, A.; Vitiello, A.; Reherman, B.; Fowler, P.; Nayersina, R.; Kast, W.M.; Melief, C.J.; Oseroff, C.; Yuan, L.; Ruppert, J.; et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994, 153, 5586–5592. [Google Scholar]
- Hachiya, A.; Gatanaga, H.; Kodama, E.; Ikeuchi, M.; Matsuoka, M.; Harada, S.; Mitsuya, H.; Kimura, S.; Oka, S. Novel patterns of nevirapine resistance-associated mutations of human immunodeficiency virus type 1 in treatment-naive patients. Virology 2004, 327, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.; Santoro, M.M.; Abbate, I.; Rozera, G.; Bruselles, A.; Bartolini, B.; Gori, C.; Forbici, F.; Orchi, N.; Tozzi, V.; et al. “Sentinel” mutations in standard population sequencing can predict the presence of HIV-1 reverse transcriptase major mutations detectable only by ultra-deep pyrosequencing. J. Antimicrob. Chemother. 2011, 66, 2615–2623. [Google Scholar]
- Sidney, J.; Southwood, S.; Sette, A. HLA Binding Peptides and Their Uses. U.S. Patent 20,060,079,453, 13 April 2006. [Google Scholar]
- Manosuthi, W.; Thongyen, S.; Nilkamhang, S.; Manosuthi, S.; Sungkanuparph, S. HIV-1 drug resistance-associated mutations among antiretroviral-naive thai patients with chronic HIV-1 infection. J. Med. Virol. 2013, 85, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Brockman, M.A.; Schneidewind, A.; Lahaie, M.; Schmidt, A.; Miura, T.; Desouza, I.; Ryvkin, F.; Derdeyn, C.A.; Allen, S.; Hunter, E.; et al. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 gag alter capsid interactions with cyclophilin a. J. Virol. 2007, 81, 12608–12618. [Google Scholar]
- Smith, J.M.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 2007, 89, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Pennings, P.S.; Kryazhimskiy, S.; Wakeley, J. Loss and recovery of genetic diversity in adapting populations of HIV. PLoS Genet. 2014, 10, e1004000. [Google Scholar] [CrossRef] [PubMed]
- Neher, R.A.; Leitner, T. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput. Biol. 2010, 6, e1000660. [Google Scholar] [CrossRef] [PubMed]
- Madden, D.R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 1995, 13, 587–622. [Google Scholar] [CrossRef] [PubMed]
- Ladell, K.; Hashimoto, M.; Iglesias, M.C.; Wilmann, P.G.; McLaren, J.E.; Gras, S.; Chikata, T.; Kuse, N.; Fastenackels, S.; Gostick, E.; et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 2013, 38, 425–436. [Google Scholar]
- Reid, S.W.; McAdam, S.; Smith, K.J.; Klenerman, P.; O'Callaghan, C.A.; Harlos, K.; Jakobsen, B.K.; McMichael, A.J.; Bell, J.I.; Stuart, D.I.; et al. Antagonist HIV-1 Gag peptides induce structural changes in HLA B8. J. Exp. Med. 1996, 184, 2279–2286. [Google Scholar]
- Davenport, M.P. Antagonists or altruists: Do viral mutants modulate T-cell responses? Immunol. Today 1995, 16, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.M.; O’Connor, D.H.; Jing, P.; Dzuris, J.L.; Mothe, B.R.; Vogel, T.U.; Dunphy, E.; Liebl, M.E.; Emerson, C.; Wilson, N.; et al. Tat-specific cytotoxic T lymphocytes select for siv escape variants during resolution of primary viraemia. Nature 2000, 407, 386–390. [Google Scholar]
- Koup, R.A.; Safrit, J.T.; Cao, Y.; Andrews, C.A.; McLeod, G.; Borkowsky, W.; Farthing, C.; Ho, D.D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994, 68, 4650–4655. [Google Scholar] [PubMed]
- Borrow, P.; Lewicki, H.; Wei, X.; Horwitz, M.S.; Peffer, N.; Meyers, H.; Nelson, J.A.; Gairin, J.E.; Hahn, B.H.; Oldstone, M.B.; et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 1997, 3, 205–211. [Google Scholar]
- Brumme, Z.L.; John, M.; Carlson, J.M.; Brumme, C.J.; Chan, D.; Brockman, M.A.; Swenson, L.C.; Tao, I.; Szeto, S.; Rosato, P.; et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 2009, 4, e6687. [Google Scholar]
- Prado, J.G.; Honeyborne, I.; Brierley, I.; Puertas, M.C.; Martinez-Picado, J.; Goulder, P.J. Functional consequences of human immunodeficiency virus escape from an HLA-B*13-restricted CD8+ T-cell epitope in p1 Gag protein. J. Virol. 2009, 83, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.D.; Korber, B.T. Immune control of HIV: The obstacles of HLA and viral diversity. Nat. Immunol. 2001, 2, 473–475. [Google Scholar] [CrossRef]
- Carlson, J.M.; Brumme, Z.L. HIV evolution in response to HLA-restricted CTL selection pressures: A population-based perspective. Microbes Infect./Inst. Pasteur 2008, 10, 455–461. [Google Scholar] [CrossRef]
- Goulder, P.J.; Brander, C.; Tang, Y.; Tremblay, C.; Colbert, R.A.; Addo, M.M.; Rosenberg, E.S.; Nguyen, T.; Allen, R.; Trocha, A.; et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 2001, 412, 334–338. [Google Scholar]
- Rodriguez, L.M.; Giraldo, M.C.; Garcia, N.; Velasquez, L.; Paris, S.C.; Alvarez, C.M.; Garcia, L.F. Human leucocyte antigen gene (HLA-A, HLA-B, HLA-DRB1) frequencies in deceased organ donors. Biomedica 2007, 27, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Ponomarenko, J.; Zhu, Z.; Tamang, D.; Wang, P.; Greenbaum, J.; Lundegaard, C.; Sette, A.; Lund, O.; Bourne, P.E.; et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012, 40, W525–W530. [Google Scholar]
- Peters, B.; Bui, H.H.; Frankild, S.; Nielson, M.; Lundegaard, C.; Kostem, E.; Basch, D.; Lamberth, K.; Harndahl, M.; Fleri, W.; et al. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput. Biol. 2006, 2, e65. [Google Scholar]
- Dhanik, A.; McMurray, J.S.; Kavraki, L.E. Dinc: A new autodock-based protocol for docking large ligands. BMC Struct. Biol. 2013, 13, S11. [Google Scholar] [CrossRef] [PubMed]
- Konya, J.; Stuber, G.; Bjorndal, A.; Fenyo, E.M.; Dillner, J. Primary induction of human cytotoxic lymphocytes against a synthetic peptide of the human immunodeficiency virus type 1 protease. J. Gen. Virol. 1997, 78, 2217–2224. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).