HPV16 E6 Controls the Gap Junction Protein Cx43 in Cervical Tumour Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Antibodies
2.2. Proximity Ligation Assay (PLA)
2.3. Co-Immunoprecipitation
2.4. Western Blotting
2.5. Plasmid Construction and Cell Transfection
2.6. W12T Cell Transfected with siRNA
2.7. Immunofluorescence Microscopy
3. Results
3.1. hDlg and Cx43 Interact In Vivo
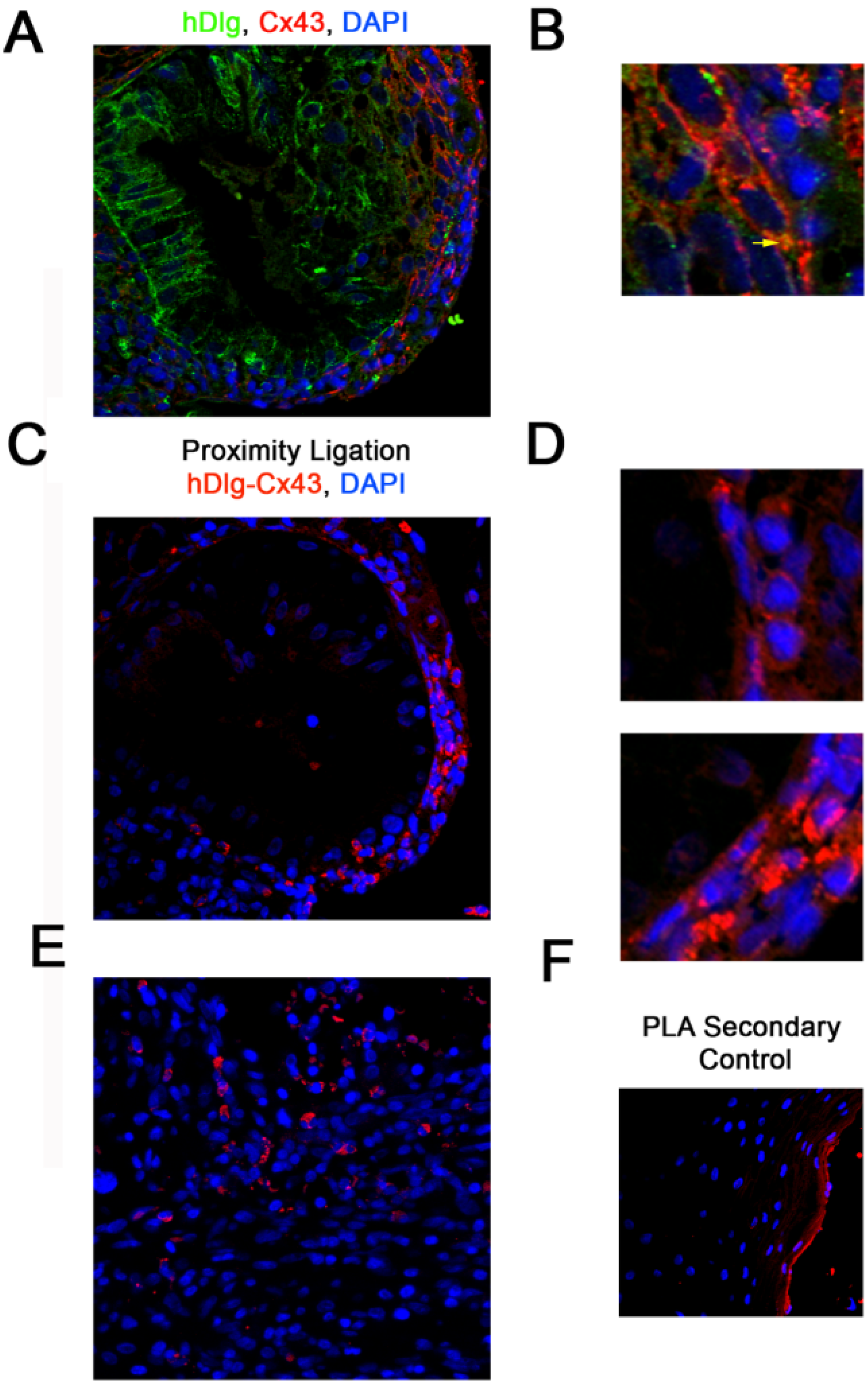
3.2. HPV16 E6 Is Present in a Complex Containing hDlg and Cx43 Cervical Tumour Cells
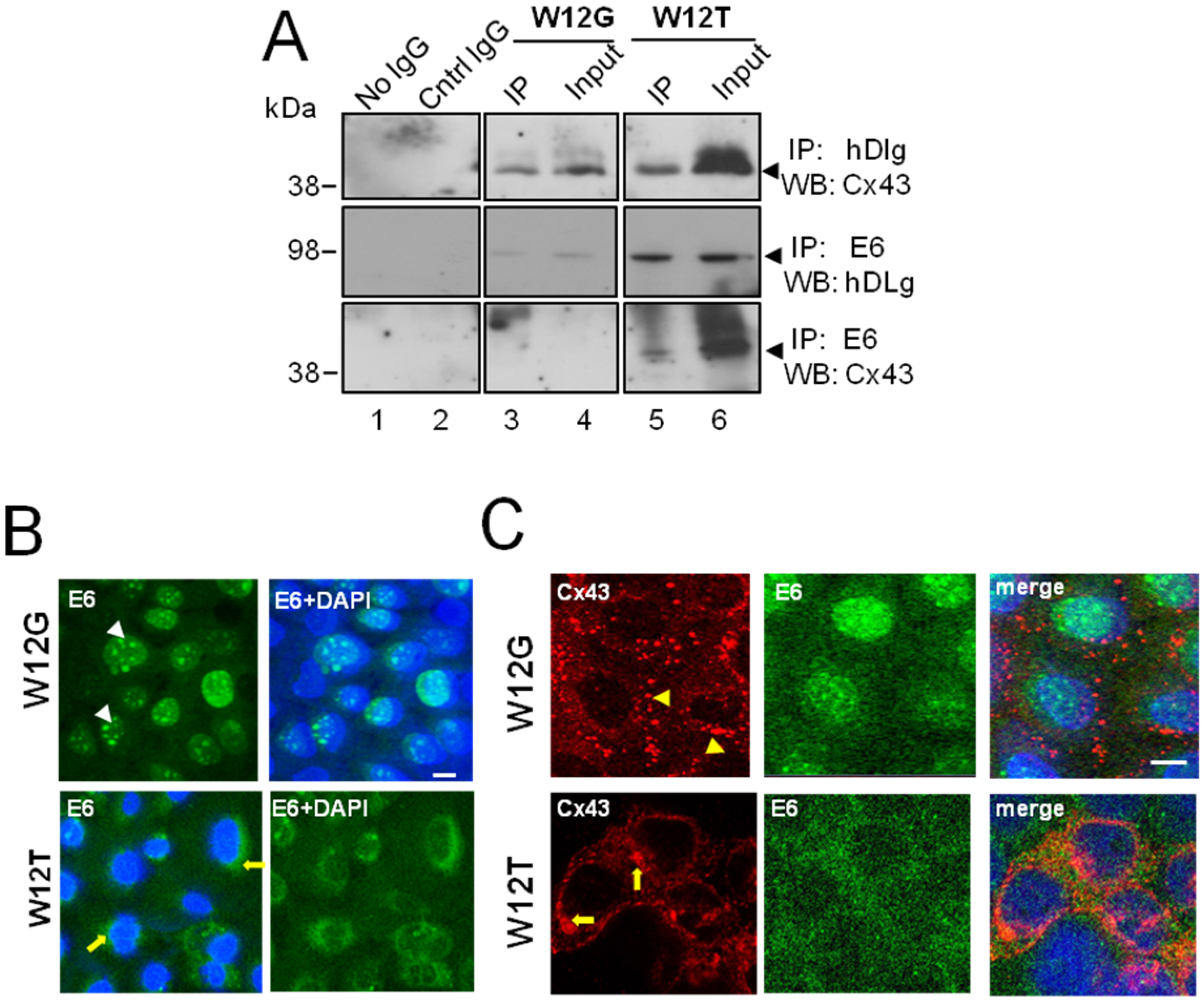
3.3. HPV16 E6 Controls Cx43 Trafficking in Cervical Tumour Cells

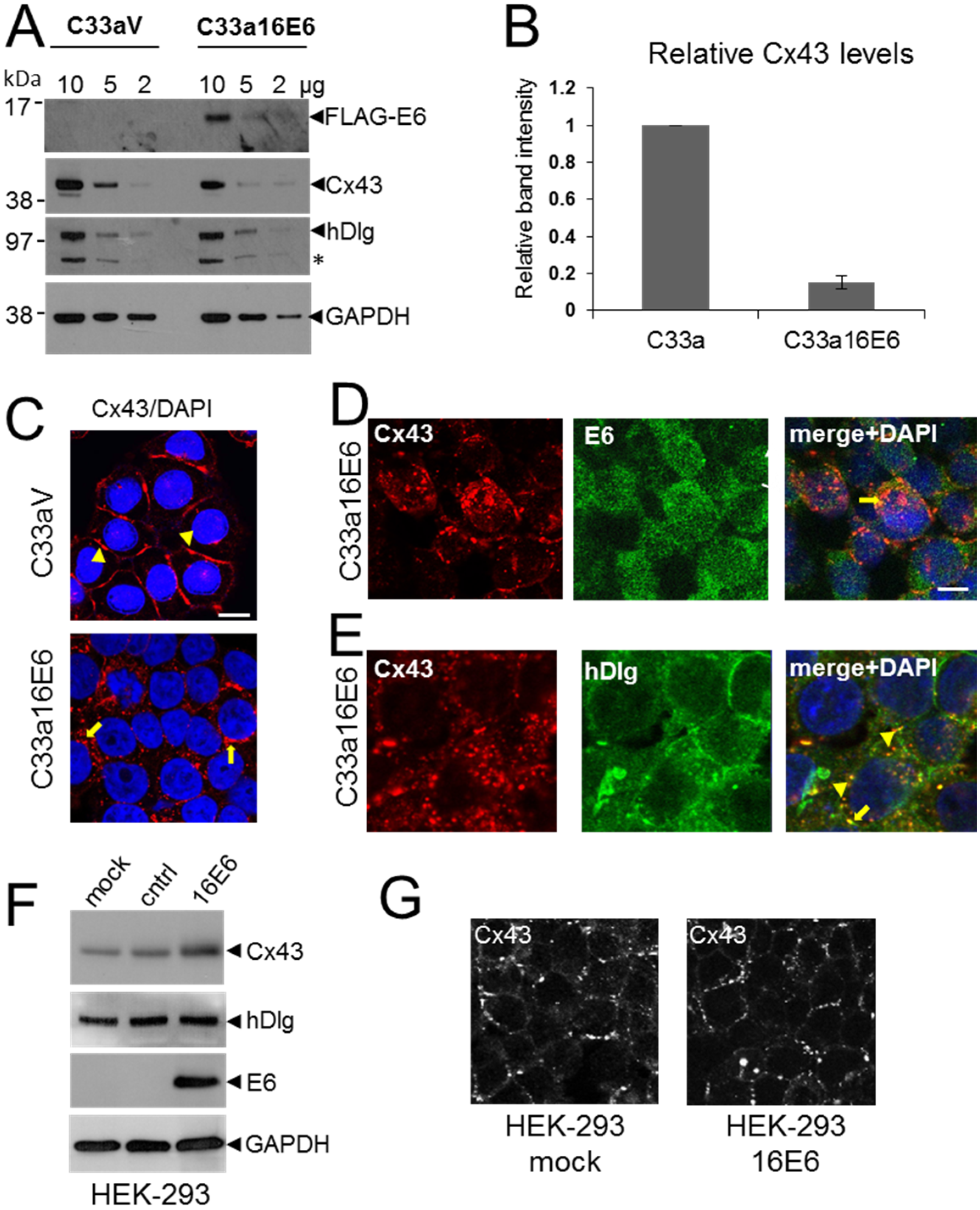
3.4. HPV E6 Causes Reduced Levels of Cx43 in C33a Cells
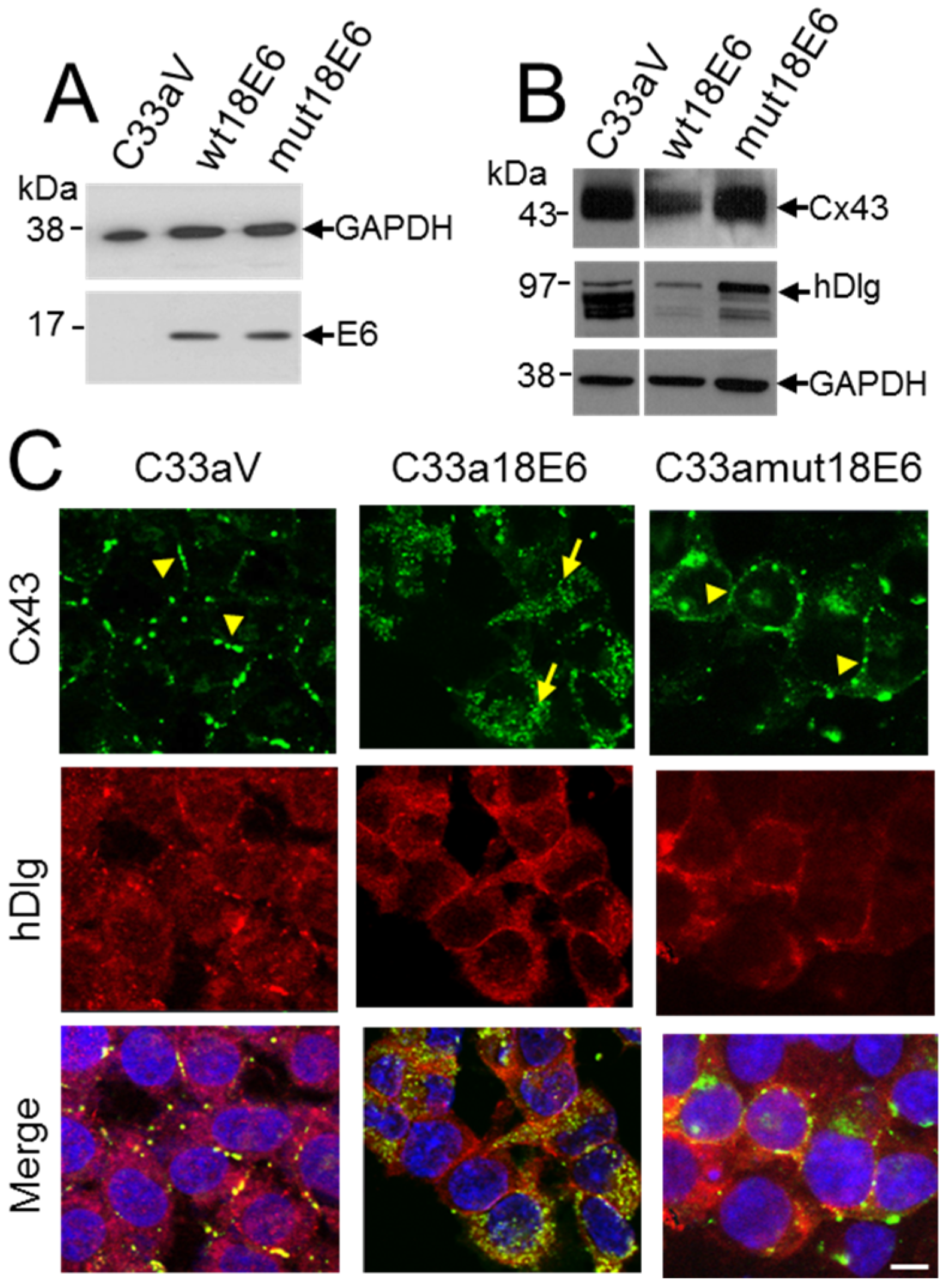
3.5. E6 PDZ Binding Motif Is Required for Relocation of Cx43 from the Membrane to the Cytoplasm
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Evans, W.H.; Martin, P.E. Gap junctions: Structure and function. Mol. Membr. Biol. 2002, 19, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Saez, J.C.; Berthoud, V.M.; Brañes, M.C.; Martínez, A.D.; Beyer, E.C. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol. Rev. 2003, 83, 1359–1400. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W. Life cycle of connexins in health and disease. Biochem. J. 2006, 394, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Kjenseth, A.; Fykerud, T.; Rivedal, E.; Leithe, E. Regulation of gap junction intercellular communication by the ubiquitin system. Cell. Signal. 2010, 22, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Evans, W.H.; de Vuyst, E.; Leybaert, L. The gap junction cellular internet: Connexin hemichannels enter the signalling limelight. Biochem. J. 2006, 397, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, S.R.; Billaud, M.; Lohman, A.W.; Taddeo, E.P.; Isakson, B.E. Posttranslational modifications in connexins and pannexins. J. Membr. Biol. 2012, 245, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T. Connexins: Junctional and non-junctional modulators of proliferation. Cell Tissue Res. 2015, 360, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Graham, S.V.; Edward, M.; Hodgins, M.B. Reduced expression of multiple gap junction proteins is a feature of cervical dysplasia. Mol. Cancer 2005, 4, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, S.; Going, J.J.; D’Arcy, R.; George, W.D. Expression of gap junction proteins connexin 26 and connexin 43 in normal human breast and in breast tumours. J. Pathol. 1998, 184, 37–43. [Google Scholar] [CrossRef]
- Bodily, J.; Laimins, L.A. Persistence of human papillomavirus infection: Keys to malignant progression. Trends Microbiol. 2011, 19, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Zur Hausen, H. Papillomaviruses in the causation of human cancers—A brief historical account. Virology 2009, 384, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Goon, P.K.; Stanley, M.A.; Ebmeyer, J.; Steinstrasser, L.; Upile, T.; Jerjes, W.; Bernal-Sprekelsen, M.; Gorner, M.; Sudhoff, H.H. HPV and head and neck cancer: A descriptive update. Head Neck Oncol. 2009, 1, 36. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Kiyono, T.; Hiraiwa, A.; Fujita, M.; Hayashi, Y.; Akiyama, T.; Ishibashi, M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 1997, 94, 11612–11616. [Google Scholar] [CrossRef]
- Lee, S.S.; Weiss, R.S.; Javier, R.T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 1997, 94, 6670–6675. [Google Scholar] [CrossRef] [PubMed]
- Subbaiah, V.K.; Kranjec, C.; Thomas, M.; Banks, L. PDZ domains: The building blocks regulating tumorigenesis. Biochem. J. 2011, 439, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Delury, C.; Marsh, E. The PDZ protein discs-large (DLG): The “Jekyll and Hyde” of the epithelial polarity proteins. FEBS J. 2012, 279, 3549–3558. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, L.; Davy, C.; Raj, K.; Kranjec, C.; Banks, L.; Doorbar, J. Stabilization of HPV16 E6 protein by PDZ proteins, and potential implications for genome maintenance. Virology 2011, 414, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Dimitratos, S.D.; Woods, D.F.; Stathakis, D.; Bryant, P.J. Signalling pathways are focused at specialised regions of the plasma membrane by scaffolding proteins of the MAGUK family. Bioessays 1999, 21, 912–921. [Google Scholar] [CrossRef]
- Kim, E.; Sheng, M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004, 5, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.F.; Bryant, P.J. The disc-large tumor supressor gene of drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell 1991, 66, 451–464. [Google Scholar] [CrossRef]
- Bilder, D.; Perrimon, N. Localisation of apical determinants by the basolateral PDZ protein Scribbled. Nature 2000, 403, 676–680. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, A.I.; Sun, P.; Hernandez-Lopez, H.; Aasen, T.; Hodgins, M.B.; Edward, M.; Roberts, S.; Massimi, P.; Thomas, M.; Banks, L.; et al. A functional interaction between the MAGUK protein hDlg and the gap junction protein connexin 43 in cervical tumour cells. Biochem. J. 2012, 446, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Hodgins, M.B.; Edward, M.; Graham, S.V. The relationship between connexins, gap junctions, tissue architecture and tumour invasion, as studied in a novel in vitro model of HPV-16-associated cervical cancer progression. Oncogene 2003, 22, 7969–7980. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Allen-Hoffman, B.L.; Lambert, P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995, 69, 2989–2997. [Google Scholar] [PubMed]
- Johnstone, S.R.; Kroncke, B.M.; Straub, A.C.; Best, A.K.; Dunn, C.A.; Mitchell, L.A.; Peskova, Y.; Nakamoto, R.K.; Koval, M.; Lo, C.W.; et al. MAPK phosphorylation of connexin 43 promotes binding of cyclin E and smooth muscle cell proliferation. Circ. Res. 2012, 111, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Gardiol, D.; Kühne, C.; Glausinger, B.; Lee, S.; Javier, R.; Banks, L. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 1999, 18, 5487–5496. [Google Scholar] [CrossRef] [PubMed]
- King, T.J.; Fukushima, L.H.; Donlon, T.A.; Hieber, A.D.; Shimabukuro, K.A.; Bertram, J.S. Correlation between growth control, neoplastic potential and endogenous connexin43 expression in HeLa cell lines: Implications for tumour progression. Carcinogenesis 2000, 21, 311–315. [Google Scholar] [CrossRef] [PubMed]
- King, T.J.; Fukushima, L.H.; Hieber, A.D.; Shimabukuro, K.A.; Sakr, W.A.; Bertram, J.S. Reduced levels of connexin 43 in cervical dysplasia: Inducible expression in a cervical carcinoma line decreases neoplastic potential with implications for tumour progression. Carcinogenesis 2000, 21, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.; Togtema, M.; Zehbe, I. Subcellular localization and quantitation of the human papillomavirus type 16 E6 oncoprotein through immunocytochemistry detection. Virology 2013, 435, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Lambert, P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: Implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 1654–1658. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, M.; MacDonald, A.I.; Stevenson, A.; Graham, S.V. Human Papillomavirus 16 Oncoprotein expression is controlled by the cellular splicing factor SRSF2 (SC35). J. Virol. 2015, 89, 5276–5287. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Glaunsinger, B.; Pim, D.; Javier, R.; Banks, L. HPV E6 and MAGUK protein interactions: Determination of the molecular basis for specific protein recognition and degradation. Oncogene 2001, 20, 5431–5439. [Google Scholar] [CrossRef] [PubMed]
- Giepmans, B.N.G.; Moolenar, W.H. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occulens-1 protein. Curr. Biol. 1998, 8, 931–934. [Google Scholar] [CrossRef]
- Toyofuku, T.; Yabuki, M.; Otsu, K.; Kuzuya, T.; Hori, M.; Tada, M.H. Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J. Biol. Chem. 1998, 273, 12725–12731. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.W.; Barker, R.; Zhu, C.; Gourdie, R. Zonal Occulens-1 alters connexin43 gap junction size and organisation by influencing channel accretion. Mol. Biol. Cell 2005, 16, 5686–5698. [Google Scholar] [CrossRef] [PubMed]
- Barker, R.J.; Price, R.L.; Gourdie, R.G. Increased association of ZO-1 with connexin43 during remodeling of cardiac gap junctions. Circ. Res. 2002, 90, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Segretain, D.; Decrouy, X.; Dompierre, J.; Escalier, D.; Rahman, N.; Fiorini, C.; Mograbi, B.; Siffroi, J.-P.; Huhtaniemi, I.; Fenichel, O.; et al. Sequestration of connexin43 in the early endosomes: An early event of Leydig cell tumor progression. Mol. Carcinogen. 2003, 38, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Laing, J.G.; Chou, B.C.; Steinberg, T.H. ZO-1 alters the plasma membrane localization and function of Cx43 in osteoblastic cells. J. Cell. Sci. 2005, 118, 2167–2176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thomas, M.; Massimi, P.; Navarro, C.; Borg, J.-P.; Banks, L. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 2005, 24, 6222–6230. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Paredes, A.; de la Cruz-Hernandez, E.; Martinez-Ramirez, I.; Duenas-Gonzalez, A.; Lizano, M. E6 variants of human papillomavirus 18 differentially modulate the protein kinase B/phosphatidylinositol 3-kinase (akt/PI3K) signaling pathway. Virology 2009, 383, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Spangle, J.M.; Munger, K. The Human Papillomavirus Type 16 E6 Oncoprotein activates mTORC1 signaling and increases protein synthesis. J. Virol. 2010, 84, 9398–9407. [Google Scholar] [CrossRef] [PubMed]
- Spangle, J.M.; Munger, K. The HPV16 E6 oncoprotein causes prolonged receptor protein tyrosine kinase signaling and enhances internalization of phosphorylated receptor species. PLoS Pathog. 2013, 9, e1003237. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W.; Castillo, M.; Kasprzak, L. Gap junction turnover, intracellular trafficking, and phosphorylation of connexin43 in brefeldin A-treated rat mammary tumor cells. J. Cell Biol. 1995, 131, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Solan, J.L.; Lampe, P.D. Connexin43 phosphorylation: Structural changes and biological effects. Biochem. J. 2009, 419, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Beardslee, M.A.; Laing, J.G.; Beyer, E.C.; Saffitz, J.E. Rapid turnover of connexin43 in the adult rat heart. Circ. Res. 1998, 83, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.Y.; Sheng, M. PDZ domains: Structural modules for protein complex assembly. J. Biol. Chem. 2002, 277, 5699–5702. [Google Scholar] [CrossRef] [PubMed]
- Steinhoff, I.; Leykauf, K.; Bleyl, U.; Dürst, M.; Alonso, A. Phosphorylation of the gap junction protein Connexin43 in CIN III lesions and cervical carcinomas. Cancer Lett. 2006, 235, 291–297. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, P.; Dong, L.; MacDonald, A.I.; Akbari, S.; Edward, M.; Hodgins, M.B.; Johnstone, S.R.; Graham, S.V. HPV16 E6 Controls the Gap Junction Protein Cx43 in Cervical Tumour Cells. Viruses 2015, 7, 5243-5256. https://doi.org/10.3390/v7102871
Sun P, Dong L, MacDonald AI, Akbari S, Edward M, Hodgins MB, Johnstone SR, Graham SV. HPV16 E6 Controls the Gap Junction Protein Cx43 in Cervical Tumour Cells. Viruses. 2015; 7(10):5243-5256. https://doi.org/10.3390/v7102871
Chicago/Turabian StyleSun, Peng, Li Dong, Alasdair I. MacDonald, Shahrzad Akbari, Michael Edward, Malcolm B. Hodgins, Scott R. Johnstone, and Sheila V. Graham. 2015. "HPV16 E6 Controls the Gap Junction Protein Cx43 in Cervical Tumour Cells" Viruses 7, no. 10: 5243-5256. https://doi.org/10.3390/v7102871
APA StyleSun, P., Dong, L., MacDonald, A. I., Akbari, S., Edward, M., Hodgins, M. B., Johnstone, S. R., & Graham, S. V. (2015). HPV16 E6 Controls the Gap Junction Protein Cx43 in Cervical Tumour Cells. Viruses, 7(10), 5243-5256. https://doi.org/10.3390/v7102871






