Detection and Identification of the First Viruses in Chia (Salvia hispanica)
Abstract
:1. Introduction
2. Materials and Methods

3. Results and Discussion
| Virus | Acronym | Origin | GenBank Accession Number | % of Identity |
|---|---|---|---|---|
| Tomato yellow spot virus | ToYSV | Argentina | FJ538207 | 95.3% |
| Tomato yellow spot virus | ToYSV | Brazil | JX513952 | 93.9% |
| Leonurus mosaic virus | LeMV | Brazil | JX863082 | 93.6% |
| Leonurus mosaic virus | LeMV | Brazil | JX863081 | 93.5% |
| Leonurus mosaic virus | LeMV | Brazil | JQ429791 | 93.4% |
| Leonurus mosaic virus | LeMV | Paraguay | KC683374 | 91.6% |
| Tomato yellow spot virus | ToYSV | Brazil | KC706628 | 89.9% |
| Tomato yellow spot virus | ToYSV | Brazil | DQ336350/NC_007726 | 89.8% |
| Sida micrantha mosaic virus | SimMV | Brazil | AJ557450 | 89.2% |
| Sida mottle virus | SiMoV | Brazil | AY090555/NC_004637 | 88.9% |
| Sida micrantha mosaic virus | SimMV | Brazil | FN557522 | 88.6% |
| Sida mottle virus | SiMoV | Brazil | JX871378 | 88.1% |
| Sida mottle virus | SiMoV | Brazil | JX871377 | 88.0% |
| Okra mottle virus | OMoV | Brazil | EU914817 | 87.1% |
| Okra mottle virus | OMoV | Brazil | EU914819 | 87.1% |
| Okra mottle virus | OMoV | Brazil | FJ686695 | 87.0% |
| Sida yellow net virus | SiYNV | Brazil | JX871376 | 86.6% |
| Sida yellow mosaic virus | SiYMV | Brazil | JX871369 | 83.1% |
| Sida yellow mosaic virus | SiYMV | Brazil | AY090558/NC_004639 | 83.1% |
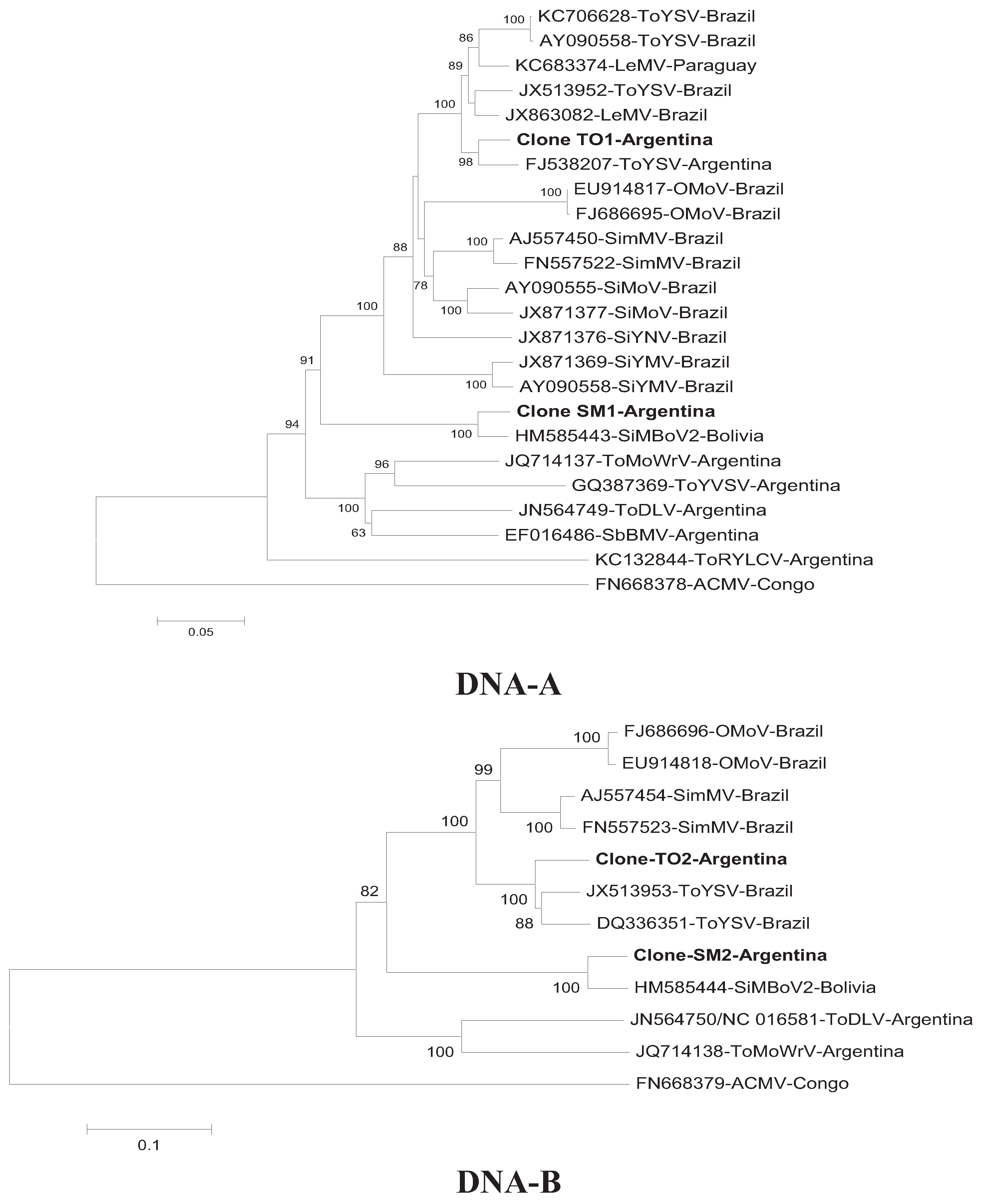
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ayerza, R.; Wayne, C. Chía, redescubriendo un olvidado alimento de los Aztecas, 1st ed.; Del Nuevo Extremo: Buenos Aires, Argentina, 2006; p. 232. [Google Scholar]
- Lobo Zavalía, R.; Alcocer, M.G.; Fuentes, F.J.; Rodriguez, W.A.; Morandini, M.; Devani, M.R. Desarrollo del cultivo de chía en Tucumán. República Argentina. Av. Agroind. 2011, 32, 27–30. [Google Scholar]
- La Gaceta. Economía—En el NOA se perdió entre el 70% y el 80% de la producción de chía, debido a la sequía. 2013. Available online: http://www.lagaceta.com.ar (accessed on 16 August 2013).
- Ayerza, R. Seed composition of two chía (Salvia hispanica L.) genotypes which differ in seed color. Emir. J. Food Agric. 2013, 25, 495–500. [Google Scholar]
- Ayerza, R.; Coates, W. Dietary levels of chia: Influence on yolk cholesterol, lipid content and fatty acid composition, for two strains of hens. Poult. Sci. 2000, 78, 724–739. [Google Scholar] [CrossRef]
- Ayerza, R.; Coates, W. The omega-3 enriched eggs: The influence of dietary linolenic fatty acid source combination on egg production and composition. Can. J. Anim. Sci. 2001, 81, 355–362. [Google Scholar] [CrossRef]
- Ayerza, R.; Coates, W.; Lauria, M. Chia seed (Salvia hispanica L.) as an ω-3 fatty acid source for broilers: Influence on fatty acid composition, cholesterol and fat content of white and dark meat, growth performance and sensory characteristics. Poult. Sci. 2002, 81, 826–837. [Google Scholar]
- Holcomb, G.E.; Valverde, R.A. Natural Infection of Salvia uliginosa with Cucumber mosaic cucumovirus. HortScience 1998, 33, 1215–1216. [Google Scholar]
- Ali, S.; Khan, A.A.; Afreen, B.; Sharma, R.; Jahan, T.; Naqvi, Q.A. Natural occurrence of Cucumber mosaic virus associated with mottling and mosaic disease on Salvia splendens, a new record from India. Australas. Plant Dis. Notes 2008, 3, 118–120. [Google Scholar] [CrossRef]
- Mumford, R.A.; Jarvis, B.; Harju, V.; Boonham, N.; Skelton, A. The first report of Broad bean wilt virus 2 in the UK: findings in foxglove and salvia. Plant Pathol. 2006, 55, 819. [Google Scholar]
- Ara, M.R.; Masud, M.M.H.; Akanda, A.M. Detection of plant viruses in some ornamental plants that act as alternate hosts. Agriculturists 2012, 10, 46–54. [Google Scholar]
- Valverde, R.A.; Singh, R.; Sabanadzovic, S. Detection and identification of Clerodendron golden mosaic China virus in Salvia splendens. Eur. J. Plant Pathol. 2012, 133, 499–503. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef]
- Rojas, M.R.; Gilbertson, R.L.; Russell, D.R.; Maxwell, D.P. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 1993, 77, 340–347. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A simple method for cloning the complete begomovirus genome using the bacteriophage phi 29 DNA polymerase. J. Virol. Method 2004, 116, 209–211. [Google Scholar] [CrossRef]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York, NY, USA, 1982; p. 1626. [Google Scholar]
- GenBank, National Center for Biotechnology Information – NCBI. Available online: http://www.ncbi.nlm.nih.gov (accessed on 15 June 2014).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Brown, J.K.; Fauquet, C.M.; Briddon, R.W.; Zerbini, M.; Moriones, E.; Navas Castillo, J. Geminiviridae. In Virus taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: London, UK, 2012; pp. 351–373. [Google Scholar]
- Wyant, P.S.; Gotthardt, D.; Schäfer, B.; Krenz, B.; Jeske, H. The genomes of four novel begomoviruses and a new Sida micrantha mosaic virus strain from Bolivian weeds. Arch. Virol. 2011, 156, 347–352. [Google Scholar] [CrossRef]
- Rodríguez Pardina, P.E.; Hanada, K.; Laguna, I.G.; Zerbini, F.M.; Ducasse, D.A. Molecular characterization and relative incidence of bean- and soybean-infecting begomoviruses in northwestern Argentina. Ann. Appl. Biol. 2011, 158, 69–78. [Google Scholar] [CrossRef]
- Barbosa, J.C.; Eckstein, B.; Bergamin Filho, A.; Rezende, J.A.M.; Dallagnol, L.J. First Report of Tomato yellow spot virus Infecting Leonurus sibiricus in Brazil. Plant Dis. 2013, 97, 289. [Google Scholar] [CrossRef]
- Calegario, R.F.; Ferreira, S.S.; Andrade, E.C.; Zerbini, F.M. Characterization of Tomato yellow spot virus, a novel tomato-infecting begomovirus in Brazil. Pesquisa Agropecuária Brasileira 2007, 42, 1335–1343. [Google Scholar]
- Fernandes, N.A.N.; Boiteux, L.S.; Fonseca, M.E.N.; Segnana, L.G.; Kitajima, E.W. Report of Tomato yellow spot virus infecting Leonurus sibiricus in Paraguay and within tomato fields in Brazil. Plant Dis. 2013, 97, 289. [Google Scholar]
- Alemandri, V.; Rodriguez Pardina, P.; Izaurralde, J.; García Medina, S.; Argüello Caro, E.; Mattio, M.F.; Dumón, A.; Rodriguez, S.M.; Truol, G. Incidence of begomoviruses and climatic characterisation of Bemisia tabaci-geminivirus complex in soybean and bean in Argentina. Agriscientia 2012, 29, 31–39. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Celli, M.G.; Perotto, M.C.; Martino, J.A.; Flores, C.R.; Conci, V.C.; Pardina, P.R. Detection and Identification of the First Viruses in Chia (Salvia hispanica). Viruses 2014, 6, 3450-3457. https://doi.org/10.3390/v6093450
Celli MG, Perotto MC, Martino JA, Flores CR, Conci VC, Pardina PR. Detection and Identification of the First Viruses in Chia (Salvia hispanica). Viruses. 2014; 6(9):3450-3457. https://doi.org/10.3390/v6093450
Chicago/Turabian StyleCelli, Marcos G., Maria C. Perotto, Julia A. Martino, Ceferino R. Flores, Vilma C. Conci, and Patricia Rodriguez Pardina. 2014. "Detection and Identification of the First Viruses in Chia (Salvia hispanica)" Viruses 6, no. 9: 3450-3457. https://doi.org/10.3390/v6093450
APA StyleCelli, M. G., Perotto, M. C., Martino, J. A., Flores, C. R., Conci, V. C., & Pardina, P. R. (2014). Detection and Identification of the First Viruses in Chia (Salvia hispanica). Viruses, 6(9), 3450-3457. https://doi.org/10.3390/v6093450




