Role of Virus-Encoded microRNAs in Avian Viral Diseases
Abstract
:1. Introduction
2. MiRNA-Encoding Avian Viruses and Associated Diseases
3. Identification of miRNAs Encoded by Avian Viruses
| Virus | pre-miRs | mature miRs | miRNAs with proposed function | Target | Proposed function |
|---|---|---|---|---|---|
| MDV-1 | 14 | 26 | mdv1-mir-M3 | Smad2 | Anti-apoptotic [31]. |
| mdv1-mir-M4-5p | Pu.1, CEBPβ, HIVEP2, BCL2L13, PDCD6, GPM6B, RREB1, c-Myb, MAP3K7IP2 | Mimics cellular mir-155 [32,33,34]. | |||
| mdv1-mir-M4-3p | UL28 | Prevent lytic replication/promote latency [33] | |||
| UL32 | Prevent lytic replication/promote latency [33] | ||||
| mdv1-mir-M7-5p | ICP4 and ICP27 | Establish and/or maintain latency [ 35] | |||
| mdv1-mir-M2 | R-LORF8 | Lymphocyte growth [34] | |||
| MDV-2 | 18 | 36 | mdv2-mir-M29 | R-LORF2 | Lymphocyte growth [34] |
| HVT | 17 | 28 | |||
| ILTV | 7 | 10 | iltv-mir-I5 | ICP4 | Establish and/or maintain latency [36] |
| DEV | 24 | 33 | |||
| ALV-J | 1 | 2 | E(XSR)miR | Potential role in oncogenesis |
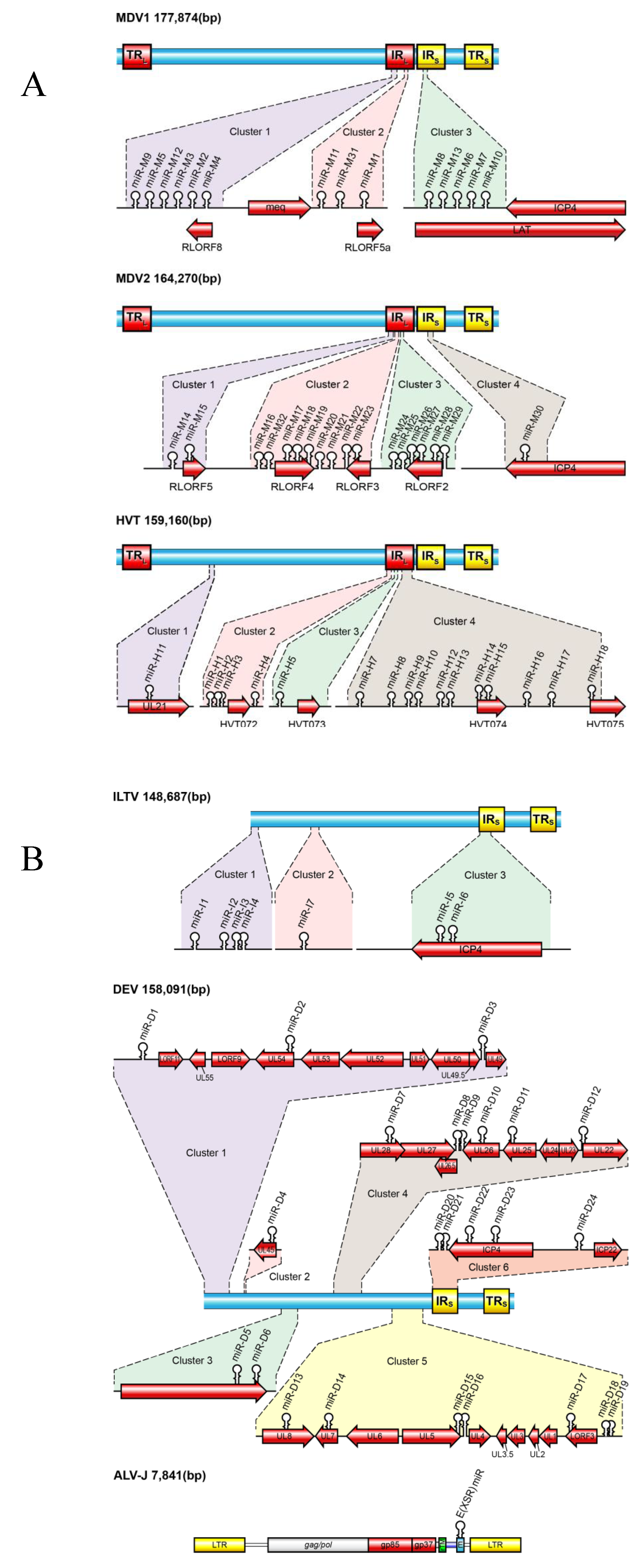
3.1. MDV-1 miRNAs
3.2. MDV-2 miRNAs
3.3. HVT miRNAs
3.4. ILTV-miRNAs
3.5. DEV miRNAs
3.6. ALV-J miRNAs
4. Viral Orthologs of Host miRNAs
5. Target Identification of Avian Herpesvirus miRNAs
6. Viral Targets of Viral miRNAs
7. Cellular Targets of Viral miRNAs
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef]
- Cullen, B.R. How do viruses avoid inhibition by endogenous cellular microRNAs. PLoS Pathog. 2013, 9, e1003694. [Google Scholar] [CrossRef]
- Libri, V.; Miesen, P.; van Rij, R.P.; Buck, A.H. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell. Mol. Life Sci. 2013, 70, 3525–3544. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Sullivan, C.S. Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef]
- Chen, C.J.; Cox, J.E.; Kincaid, R.P.; Martinez, A.; Sullivan, C.S. Divergent microrna targetomes of closely related circulating strains of a polyomavirus. J. Virol. 2013, 87, 11135–11147. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Burke, J.M.; Cox, J.C.; de Villiers, E.M.; Sullivan, C.S. A human torque teno virus encodes a microRNA that inhibits interferon signaling. PLoS Pathog. 2013, 9, e1003818. [Google Scholar] [CrossRef]
- Lin, J.; Cullen, B.R. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 2007, 81, 12218–12226. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Burke, J.M.; Sullivan, C.S. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. USA 2012, 109, 3077–3082. [Google Scholar] [CrossRef]
- Rosewick, N.; Momont, M.; Durkin, K.; Takeda, H.; Caiment, F.; Cleuter, Y.; Vernin, C.; Mortreux, F.; Wattel, E.; Burny, A.; et al. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 2306–2311. [Google Scholar] [CrossRef]
- Yao, Y.; Smith, L.P.; Nair, V.; Watson, M. An avian retrovirus uses canonical expression and processing mechanisms to generate viral microRNA. J. Virol. 2013. [Google Scholar] [CrossRef]
- Legione, A.R.; Coppo, M.J.; Lee, S.W.; Noormohammadi, A.H.; Hartley, C.A.; Browning, G.F.; Gilkerson, J.R.; O’Rourke, D.; Devlin, J.M. Safety and vaccine efficacy of a glycoprotein G deficient strain of infectious laryngotracheitis virus delivered in vovo. Vaccine 2012, 30, 7193–7198. [Google Scholar] [CrossRef]
- Morrow, C.; Fehler, F. Marek’s Disease: An Evolving Problem; Davison, F., Nair, V., Eds.; Elsevier Academic Press: Oxford, UK, 2004; pp. 49–61. [Google Scholar]
- Bublot, M.; Sharma, J. Vaccination against Marek’s disease. In Marek’s Disease: An Evolving Problem; Davison, F., Nair, V., Eds.; Elsevier Academic Press: Amsterdam, The Netherlands, 2004; pp. 168–185. [Google Scholar]
- Gimeno, I.M. Marek’s disease vaccines: A solution for today but a worry for tomorrow? Vaccine 2008, 26, C31–C41. [Google Scholar] [CrossRef]
- Nair, V. Evolution of Marek’s disease—A paradigm for incessant race between the pathogen and the host. Vet. J. 2005, 170, 175–183. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, A.; Wang, M.; Zhu, D.; Jia, R.; Chen, S.; Zhou, Y.; Chen, X. Comparative genomic analysis of duck enteritis virus strains. J. Virol. 2012, 86, 13841–13842. [Google Scholar] [CrossRef]
- Liu, J.; Chen, P.; Jiang, Y.; Deng, G.; Shi, J.; Wu, L.; Lin, Y.; Bu, Z.; Chen, H. Recombinant duck enteritis virus works as a single-dose vaccine in broilers providing rapid protection against H5N1 influenza infection. Antivir. Res. 2013, 97, 329–333. [Google Scholar] [CrossRef]
- Payne, L.N.; Howes, K.; Gillespie, A.M.; Smith, L.M. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: Support for a new avian retrovirus envelope subgroup, designated J. J. Gen. Virol. 1992, 73, 2995–2997. [Google Scholar] [CrossRef]
- Payne, L.N.; Brown, S.R.; Bumstead, N.; Howes, K.; Frazier, J.A.; Thouless, M.E. A novel subgroup of exogenous avian leukosis virus in chickens. J. Gen. Virol. 1991, 72, 801–807. [Google Scholar] [CrossRef]
- Gao, Y.; Yun, B.; Qin, L.; Pan, W.; Qu, Y.; Liu, Z.; Wang, Y.; Qi, X.; Gao, H.; Wang, X. Molecular epidemiology of avian leukosis virus subgroup J in layer flocks in China. J. Clin. Microbiol. 2012, 50, 953–960. [Google Scholar] [CrossRef]
- Burnside, J.; Bernberg, E.; Anderson, A.; Lu, C.; Meyers, B.C.; Green, P.J.; Jain, N.; Isaacs, G.; Morgan, R.W. Marek’s disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 2006, 80, 8778–8786. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Xu, H.; Smith, L.P.; Lawrie, C.H.; Watson, M.; Nair, V. MicroRNA profile of Marek’s disease virus-transformed T-cell line MSB-1: Predominance of virus-encoded microRNAs. J. Virol. 2008, 82, 4007–4015. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Xu, H.; Smith, L.P.; Lawrie, C.H.; Sewer, A.; Zavolan, M.; Nair, V. Marek’s disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 2007, 81, 7164–7170. [Google Scholar] [CrossRef]
- Waidner, L.A.; Morgan, R.W.; Anderson, A.S.; Bernberg, E.L.; Kamboj, S.; Garcia, M.; Riblet, S.M.; Ouyang, M.; Isaacs, G.K.; Markis, M.; et al. MicroRNAs of Gallid and Meleagrid herpesviruses show generally conserved genomic locations and are virus-specific. Virology 2009, 388, 128–136. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Watson, M.; Nair, V. Novel microRNAs (miRNAs) encoded by herpesvirus of Turkeys: Evidence of miRNA evolution by duplication. J. Virol. 2009, 83, 6969–6973. [Google Scholar] [CrossRef]
- Rachamadugu, R.; Lee, J.Y.; Wooming, A.; Kong, B.W. Identification and expression analysis of infectious laryngotracheitis virus encoding microRNAs. Virus Genes 2009, 39, 301–308. [Google Scholar] [CrossRef]
- Yao, Y.; Smith, L.P.; Petherbridge, L.; Watson, M.; Nair, V. Novel microRNAs encoded by duck enteritis virus. J. Gen. Virol. 2012, 93, 1530–1536. [Google Scholar] [CrossRef]
- Xu, S.; Xue, C.; Li, J.; Bi, Y.; Cao, Y. Marek’s disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 2011, 85, 276–285. [Google Scholar] [CrossRef]
- Zhao, Y.; Yao, Y.; Xu, H.; Lambeth, L.; Smith, L.P.; Kgosana, L.; Wang, X.; Nair, V. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J. Virol. 2009, 83, 489–492. [Google Scholar] [CrossRef]
- Muylkens, B.; Coupeau, D.; Dambrine, G.; Trapp, S.; Rasschaert, D. Marek’s disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch. Virol. 2010, 155, 1823–1837. [Google Scholar] [CrossRef]
- Parnas, O.; Corcoran, D.L.; Cullen, B.R. Analysis of the mRNA targetome of microRNAs expressed by Marek’s disease virus. Am. Soc. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Strassheim, S.; Stik, G.; Rasschaert, D.; Laurent, S. mdv1-miR-M7–5p, located in the newly identified first intron of the latency-associated transcript of Marek’s disease virus, targets the immediate-early genes ICP4 and ICP27. J. Gen. Virol. 2012, 93, 1731–1742. [Google Scholar] [CrossRef]
- Waidner, L.A.; Burnside, J.; Anderson, A.S.; Bernberg, E.L.; German, M.A.; Meyers, B.C.; Green, P.J.; Morgan, R.W. A microRNA of infectious laryngotracheitis virus can downregulate and direct cleavage of ICP4 mRNA. Virology 2011, 411, 25–31. [Google Scholar] [CrossRef]
- Morgan, R.; Anderson, A.; Bernberg, E.; Kamboj, S.; Huang, E.; Lagasse, G.; Isaacs, G.; Parcells, M.; Meyers, B.C.; Green, P.J.; et al. Sequence conservation and differential expression of Marek’s disease virus microRNAs. J. Virol. 2008, 82, 12213–12220. [Google Scholar]
- Burnside, J.; Morgan, R. Emerging roles of chicken and viral microRNAs in avian disease. BMC Proc. 2011, 5. [Google Scholar] [CrossRef]
- Coupeau, D.; Dambrine, G.; Rasschaert, D. Kinetic expression analysis of the cluster MDV1-mir-M9-M4, genes meq and vIL-8 differs between the lytic and latent phases of Marek’s disease virus infection. J. Gen. Virol. 2012, 93, 1519–1529. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, H.; Yao, Y.; Smith, L.P.; Kgosana, L.; Green, J.; Petherbridge, L.; Baigent, S.J.; Nair, V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 2011, 7, e1001305. [Google Scholar] [CrossRef]
- Yu, Z.H.; Teng, M.; Sun, A.J.; Yu, L.L.; Hu, B.; Qu, L.H.; Ding, K.; Cheng, X.C.; Liu, J.X.; Cui, Z.Z.; et al. Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek’s disease virus. Virology 2014, 448, 55–64. [Google Scholar] [CrossRef]
- Brown, A.C.; Nair, V.; Allday, M.J. Epigenetic regulation of the latency-associated region of Marek’s disease virus in tumor-derived T-cell lines and primary lymphoma. J. Virol. 2012, 86, 1683–1695. [Google Scholar]
- Stik, G.; Laurent, S.; Coupeau, D.; Coutaud, B.; Dambrine, G.; Rasschaert, D.; Muylkens, B. A p53-dependent promoter associated with polymorphic tandem repeats controls the expression of a viral transcript encoding clustered microRNAs. RNA 2010, 16, 2263–2276. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Lawrie, C.H.; Saunders, N.J.; Watson, M.; Nair, V. Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J. Gen. Virol. 2009, 90, 1551–1559. [Google Scholar] [CrossRef]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef]
- Xu, H.; Yao, Y.; Smith, L.P.; Nair, V. MicroRNA-26a-mediated regulation of interleukin-2 expression in transformed avian lymphocyte lines. Cancer Cell Int. 2010, 10, 15. [Google Scholar] [CrossRef]
- Hirai, K.; Yamada, M.; Arao, Y.; Kato, S.; Nii, S. Replicating Marek’s disease virus (MDV) serotype 2 DNA with inserted MDV serotype 1 DNA sequences in a Marek’s disease lymphoblastoid cell line MSB1-41C. Arch. Virol. 1990, 114, 153–165. [Google Scholar] [CrossRef]
- Xiang, J.; Cheng, A.; Wang, M.; Zhang, S.; Zhu, D.; Jia, R.; Chen, S.; Zhou, Y.; Wang, X.; Chen, X. Computational identification of microRNAs in anatid herpesvirus 1 genome. Virol. J. 2012, 9, e93. [Google Scholar] [CrossRef]
- Chesters, P.M.; Smith, L.P.; Nair, V. E (XSR) element contributes to the oncogenicity of Avian leukosis virus (subgroup J). J. Gen. Virol. 2006, 87, 2685–2692. [Google Scholar] [CrossRef]
- Zisoulis, D.G.; Lovci, M.T.; Wilbert, M.L.; Hutt, K.R.; Liang, T.Y.; Pasquinelli, A.E.; Yeo, G.W. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010, 17, 173–179. [Google Scholar] [CrossRef]
- Gottwein, E.; Mukherjee, N.; Sachse, C.; Frenzel, C.; Majoros, W.H.; Chi, J.T.; Braich, R.; Manoharan, M.; Soutschek, J.; Ohler, U.; et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007, 450, 1096–1099. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Samols, M.A.; Plaisance, K.B.; Boss, I.W.; Riva, A.; Lopez, M.C.; Baker, H.V.; Renne, R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007, 81, 12836–12845. [Google Scholar] [CrossRef]
- McClure, L.V.; Sullivan, C.S. Kaposi’s sarcoma herpes virus taps into a host microRNA regulatory network. Cell Host Microbe 2008, 3, 1–3. [Google Scholar] [CrossRef]
- Tili, E.; Croce, C.M.; Michaille, J.J. miR-155: On the crosstalk between inflammation and cancer. Int. Rev. Immunol. 2009, 28, 264–284. [Google Scholar] [CrossRef]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta 2009, 1792, 497–505. [Google Scholar]
- Rai, D.; Kim, S.W.; McKeller, M.R.; Dahia, P.L.; Aguiar, R.C. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 3111–3116. [Google Scholar]
- Valeri, N.; Gasparini, P.; Fabbri, M.; Braconi, C.; Veronese, A.; Lovat, F.; Adair, B.; Vannini, I.; Fanini, F.; Bottoni, A.; et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA 2010, 107, 6982–6987. [Google Scholar] [CrossRef]
- Lu, F.; Weidmer, A.; Liu, C.G.; Volinia, S.; Croce, C.M.; Lieberman, P.M. Epstein-Barr virus-induced miR-155 attenuates NF-κB signaling and stabilizes latent virus persistence. J. Virol. 2008, 82, 10436–10443. [Google Scholar] [CrossRef]
- Bolisetty, M.T.; Dy, G.; Tam, W.; Beemon, K.L. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J. Virol. 2009, 83, 12009–12017. [Google Scholar] [CrossRef]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef]
- Galardi, S.; Mercatelli, N.; Giorda, E.; Massalini, S.; Frajese, G.V.; Ciafre, S.A.; Farace, M.G. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007, 282, 23716–23724. [Google Scholar]
- Lambeth, L.S.; Yao, Y.; Smith, L.P.; Zhao, Y.; Nair, V. MicroRNAs 221 and 222 target p27Kip1 in Marek’s disease virus-transformed tumour cell line MSB-1. J. Gen. Virol. 2009, 90, 1164–1171. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget 2010, 1, 224–227. [Google Scholar]
- Carl, J.W., Jr.; Trgovcich, J.; Hannenhalli, S. Widespread evidence of viral miRNAs targeting host pathways. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar]
- Brown, K.M.; Chu, C.Y.; Rana, T.M. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005, 12, 469–470. [Google Scholar] [CrossRef]
- Grey, F.; Nelson, J. Identification and function of human cytomegalovirus microRNAs. J. Clin. Virol. 2008, 41, 186–191. [Google Scholar] [CrossRef]
- Boss, I.W.; Plaisance, K.B.; Renne, R. Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol. 2009, 17, 544–553. [Google Scholar] [CrossRef]
- Murphy, E.; Vanicek, J.; Robins, H.; Shenk, T.; Levine, A.J. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. USA 2008, 105, 5453–5458. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yao, Y.; Nair, V. Role of Virus-Encoded microRNAs in Avian Viral Diseases. Viruses 2014, 6, 1379-1394. https://doi.org/10.3390/v6031379
Yao Y, Nair V. Role of Virus-Encoded microRNAs in Avian Viral Diseases. Viruses. 2014; 6(3):1379-1394. https://doi.org/10.3390/v6031379
Chicago/Turabian StyleYao, Yongxiu, and Venugopal Nair. 2014. "Role of Virus-Encoded microRNAs in Avian Viral Diseases" Viruses 6, no. 3: 1379-1394. https://doi.org/10.3390/v6031379
APA StyleYao, Y., & Nair, V. (2014). Role of Virus-Encoded microRNAs in Avian Viral Diseases. Viruses, 6(3), 1379-1394. https://doi.org/10.3390/v6031379





