Emaravirus: A Novel Genus of Multipartite, Negative Strand RNA Plant Viruses
Abstract
:1. Introduction
| Virus Species | Particle Morphology | Vector (Putative) | Host Species |
|---|---|---|---|
| EMARaV | DMBs 80–120 nm | (Phytoptus pyri) | Sorbus aucuparia |
| FMV | DMBs 90–200 nm | Aceria ficus | Ficus carica |
| RRV | DMBs 120–150 nm | Phyllocoptes fructiphilus | Rosa multiflora cultivated hybrid roses |
| RLBV | indistinct filamentous bodies | Phyllocoptes gracilis | Rubus spp. |
| MRSV (HPV) | filamentous structures and enveloped particles 80–200 nm | Aceria tosichella | Zea mays, Triticum aestivum |
| PPSMV | filamentous structures and DMBs 100–150 nm | Aceria cajani | Cajanus cajan |
2. Viruses and Associated Diseases
2.1. EMARaV — Ringspot Disease of European Mountain Ash (Sorbus aucuparia L.)

2.2. FMV — Fig Mosaic Disease (Ficus carica L.)
2.3. RRV — Rose Rosette Disease (Rosa spp.)
2.4. RLBV — Raspberry Leaf Blotch Disorder (Rubus spp.)
2.5. MRSV (HPV) — High Plains Disease (Zea mays, Triticum aestivum)
2.6. PPSMV — Sterility Mosaic Disease of Pigeonpea (Cajanus cajan)
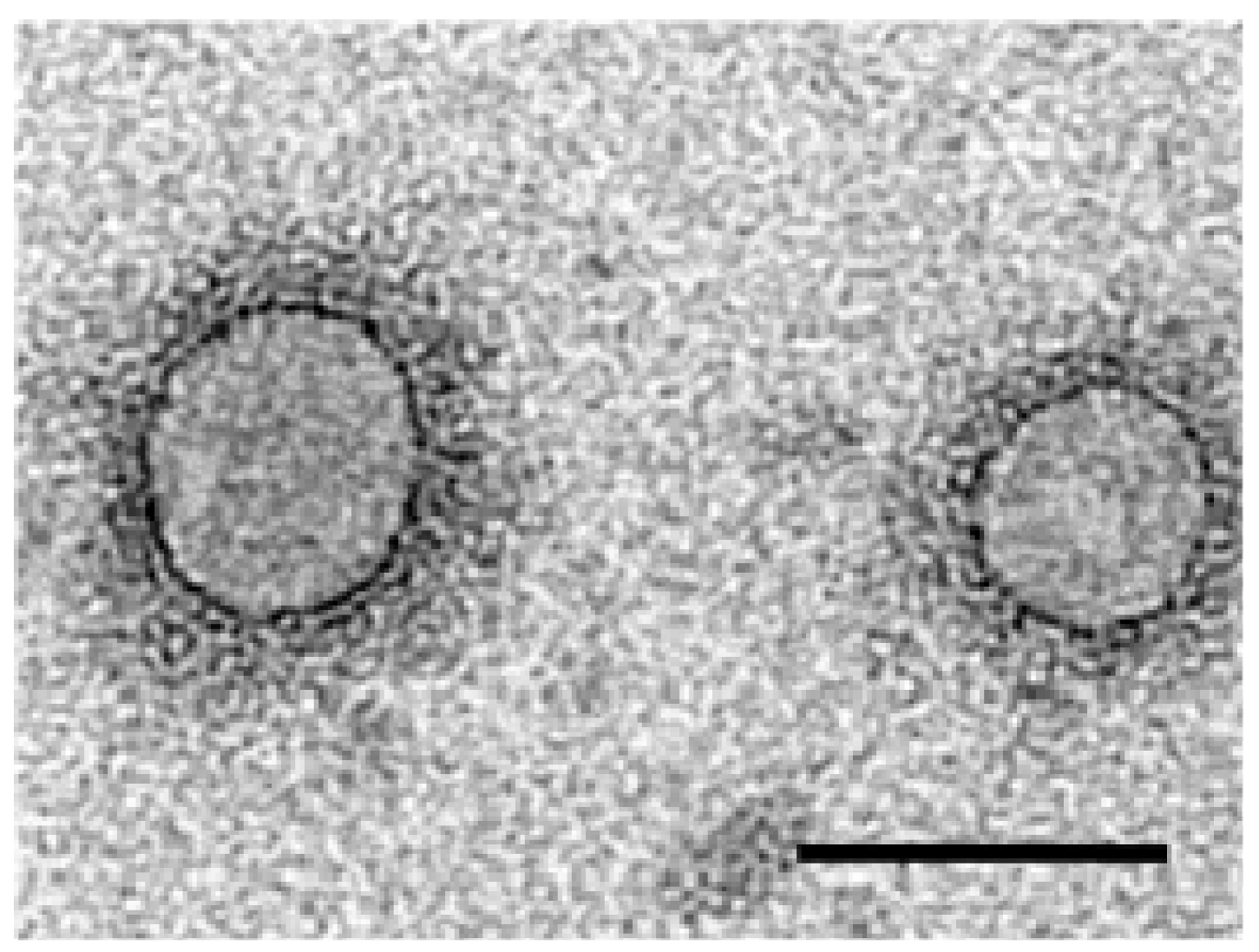
3. Virus Morphology
4. Genome Organization and Viral Proteins
| Virus Species | RNA 1 | RNA 2 | RNA 3 | RNA 4 | RNA 5 | RNA 6 |
|---|---|---|---|---|---|---|
| EMARaV | 7040 ntP1: 266 kDaRdRp | 2335 ntP2: 75 kDaGlycoproteinprecursor | 1559 ntP3: 35 kDaNucleocapsid | 1348 ntP4: 27 kDaunknown | - | - |
| FMV | 7093 ntP1: 264 kDaRdRp | 2252 ntP2: 73 kDaGlycoprotein precursor | 1490 ntP3: 35 kDaNucleocapsid | 1472 ntP4: 40.5 kDaunknown | 1752 ntP5: 59 kDaunknown | 1212 ntP6: 22 kDaunknown |
| RRV | 7026 nt P1: 265 kDaRdRp | 2220 ntP2: 74 kDaGlycoproteinprecursor | 1544 ntP3: 36 kDaNucleocapsid | 1541 ntP4: 41 kDaMP | - | - |
| RLBV | 7062 ntP1: 269 kDaRdRp | 2135 nt P2: 75 kDaGlycoprotein precursor | 1365 ntP3: 32 kDaNucleocapsid | 1675 ntP4: 42 kDaMP | 1718 ntP5: 56 kDaunknown | - |
| MRSV a | RNA-L 7–8 kbRdRp? | RNA-M (double band) 2.5/2 kb | RNA-S1.4 kb32 kDaNucleocapsid | ? | ? | ? |
| PPSMV b | 6.8 kb? | 2.7 kb? | 2.1 kb? | 1.6 kb? | 1.4 kbP5: 32 kDaNucleocapsid | 1.2 kb? |
4.1. EMARaV

4.2. FMV
4.3. RRV
4.4. RLBV
4.5. MRSV
4.6. PPSMV
4.7. Sequence Similarities among All Emaravirus Related Viruses
5. Serological Relationship and Diagnostic Procedures
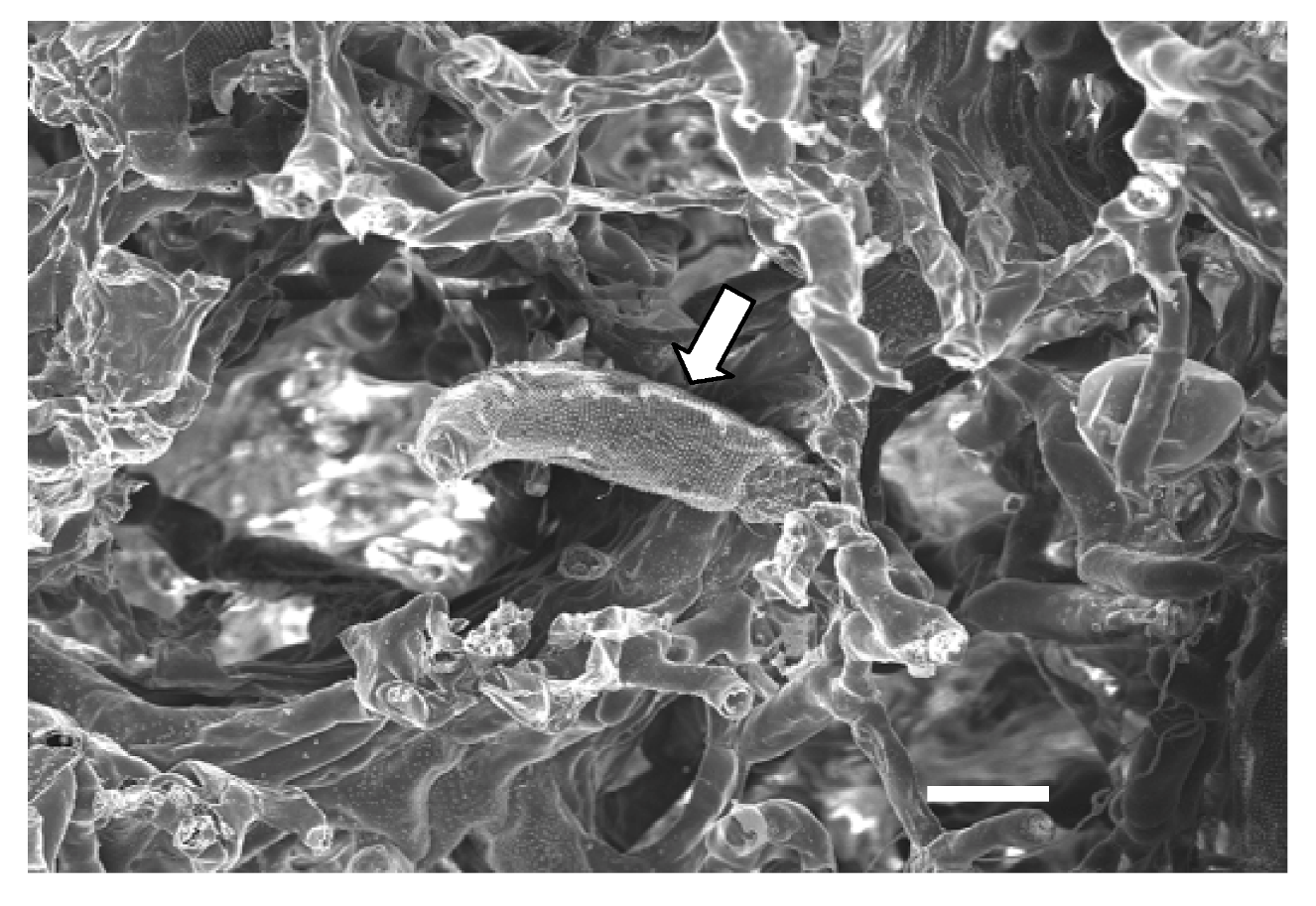
6. Virus Transmission
7. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Carstens, E.B. Introduction to virus taxonomy. In Virus Taxonomy. Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkovitz, E.J., Eds.; Elsevier: Oxford, UK, 2011; pp. 3–7. [Google Scholar]
- Mielke, N.; Muehlbach, H.-P. A novel, multipartite, negative-strand RNA virus is associated with the ringspot disease of European mountain ash (Sorbus aucuparia L.). J. Gen. Virol. 2007, 88, 1337–1346. [Google Scholar] [CrossRef]
- Mühlbach, H.-P.; Mielke-Ehret, N. Emaravirus. In Virus Taxonomy. Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkovitz, E.J., Eds.; Elsevier: Oxford, UK, 2011; pp. 767–770. [Google Scholar]
- Elbeaino, T.; Digiaro, M.; Martelli, G.P. Complete nucleotide sequence of four RNA segments of fig mosaic virus. Arch. Virol. 2009, 154, 1719–1727. [Google Scholar] [CrossRef]
- Walia, J.J.; Salem, N.M.; Falk, B.W. Partial sequence and survey analysis identify a multipartite, negative-sense RNA-virus associated with fig mosaic. Plant Dis. 2009, 93, 4–10. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Alabdullah, A.; de Stradis, A.; Minafra, A.; Mielke, N.; Castellano, M.A.; Martelli, G.P. A multipartite single-stranded negative-sense RNA virus is the putative agent of fig mosaic disease. J. Gen. Virol. 2009, 90, 1281–1288. [Google Scholar] [CrossRef]
- Adams, M.J.; Carstens, E.B. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2012). Arch. Virol. 2012, 157, 1411–1422. [Google Scholar] [CrossRef]
- Laney, A.G.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A discovery 70 years in the making: Characterization of the rose rosette virus. J. Gen. Virol. 2011, 92, 1727–1732. [Google Scholar] [CrossRef]
- McGavin, W.J.; Mitchell, C.; Cock, P.J.A.; Wright, K.M.; MacFarlane, S.A. Raspberry leaf blotch virus, a putative new member of the genus Emaravirus, encodes a novel genomic RNA. J. Gen. Virol. 2012, 93, 430–437. [Google Scholar] [CrossRef]
- Skare, J.M.; Wijkamp, I.; Denham, I.; Rezende, J.A.M.; Kitajima, E.W.; Park, J.W.; Desvoyes, B.; Rush, C.M.; Michels, G.; Scholthof, K.-B.G.; et al. A new eriophyid mite-borne membrane-enveloped virus-like complex isolated from plants. Virology 2006, 347, 343–353. [Google Scholar] [CrossRef]
- Kumar, P.L.; Jones, A.T.; Reddy, D.V.R. A novel mite-transmitted virus with a divided RNA genome closely associated with pigeonpea sterility mosaic disease. Phytopathology 2003, 93, 71–81. [Google Scholar] [CrossRef]
- Kegler, H. Das Ringfleckenmosaik der Eberesche (Sorbus aucuparia L.) (in German). Phytopathol. Z. 1960, 37, 214–216. [Google Scholar] [CrossRef]
- Benthack, W.; Mielke, N.; Büttner, C.; Mühlbach, H.-P. Double-stranded RNA pattern and partial sequence data indicate plant virus infection associated with ringspot disease of European mountain ash (Sorbus aucuparia L.). Arch. Virol. 2005, 150, 37–52. [Google Scholar] [CrossRef]
- Mielke, N.; Weber, M.; Khan, S.; Muehlbach, H.-P. Detection of European mountain ash ringspot associated virus (EMARAV) in Sorbus aucuparia L. by a specific antiserum and reverse transcription PCR. For. Pathol. 2008, 38, 371–380. [Google Scholar] [CrossRef]
- Schlatermund, N. Lokalisation und Quantifizierung der RNAs des European Mountain Ash Ringspot Associated Virus (EMARAV) in der Eberesche (Sorbus aucuparia L.) (in German).
- Polak, Z.; Prochazkova, Z.; Braniaova, H. Recent findings of viruses of forest trees on the territory of the Czech Republic. Arch. Phytopathol. Pflanzenschutz 1990, 26, 389–393. [Google Scholar] [CrossRef]
- Polak, Z.; Zieglerova, J. Towards ringspots and variegation in mountain ash leaves. Z. Pflanzenkr. Pflanzenschutz 1996, 103, 432–435. [Google Scholar]
- Kallinen, A.K.; Lindberg, I.L.; Tugume, A.K.; Valkonen, J.P.T. Detection, distribution and genetic variability of European mountain ash ringspot-associated virus. Phytopathology 2009, 99, 344–352. [Google Scholar] [CrossRef]
- Raspe, O.; Findlay, C.; Jaquemart, A.-L. Sorbus aucuparia L. J. Ecol. 2000, 88, 910–930. [Google Scholar] [CrossRef]
- Condit, I.J.; Horne, W.T. A mosaic of the fig in California. Phytopathology 1933, 23, 887–896. [Google Scholar]
- Serrano, L.; Ramon, J.; Segarra, J.; Medina, V.; Achón, M.A.; López, M. New approach in the identification of the causal agent of fig mosaic disease. Acta Hort. 2004, 657, 559–566. [Google Scholar]
- Martelli, G.P.; Castellano, M.A.; Lafortezza, R. An ultrastructural study of fig mosaic. Phytopathol. Mediterr. 1993, 32, 33–43. [Google Scholar]
- Castellano, M.A.; Gattoni, G.; Minafra, A.; Conti, M.; Martelli, G.P. Fig mosaic in Mexico and South Africa. J. Plant Pathol. 2007, 89, 441–444. [Google Scholar]
- Acikgöz, S.; Döken, T. The determination of sampling time for dsRNA isolation of the agent of fig mosaic disease prevalent in Aegean region: Turkey. Acta Hort. 2003, 605, 307–310. [Google Scholar]
- Elbeaino, T.; Digiaro, M.; de Stradis, A.; Martelli, G.P. Partial characterization of a closterovirus associated with a chlorotic mottling of fig. J. Plant Pathol. 2006, 88, 187–192. [Google Scholar]
- Elbeaino, T.; Digiaro, M.; de Stradis, A.; Martelli, G.P. Identification of a second member of the family closteroviridae in mosaic-diseased figs. J. Plant Pathol. 2007, 89, 119–124. [Google Scholar]
- Martelli, G.P. Fig mosaic disease and associated pathogens. In Virus and Virus-Like Diseases of Pome and Stone Fruits; Hadidi, A., Barba, M., Candresse, T., Jelkmann, W., Eds.; APS Press: St. Paul, MN, USA, 2011; pp. 281–287. [Google Scholar]
- Conners, I.L. Twentieth Annual Report of the Canadian Plant Disease Survey 1940; Department of Agriculture: Ottawa, Canada, 1941; p. 98. [Google Scholar]
- Crowe, F.J. Witches’ broom of rose: A new outbreak in several Central States. Plant Dis. 1983, 67, 544–546. [Google Scholar] [CrossRef]
- Epstein, A.H.; Hill, J.H.; Nutter, F.W., Jr. Augmentation of rose rosette disease for biocontrol of multiflora rose (Rosa multiflora). Weed Sci. 1997, 45, 172–178. [Google Scholar]
- Gergerich, R.C.; Kim, K.S.; Kitajima, E.W. A particle of unique morphology associated with the disease of rose in northwest Arkansas. Phytopathology 1983, 73, 500–501. [Google Scholar]
- Amrine, J.W., Jr.; Hindal, D.F.; Stasny, T.A.; Williams, R.L.; Coffman, C.C. Transmission of the rose rosette disease agent to Rosa multiflora by Phyllocoptes fructiphylus (Acari: Eriophyidae). Entomol. News 1988, 99, 239–252. [Google Scholar]
- Di, R.; Hill, J.H.; Epstein, A.H. Double-stranded RNA associated with the rose rosette disease of multiflora rose. Plant Dis. 1990, 74, 56–58. [Google Scholar] [CrossRef]
- Gordon, S.C.; Taylor, C.E. Some aspects of the biology of the raspberry leaf and bud mite (Phyllocoptes (Eriophyes) gracilis Nal.) Eriophyidae in Scotland. J. Hortic. Sci. 1976, 51, 501–508. [Google Scholar]
- Jones, A.T.; Gordon, S.C.; Jennings, D.L. A leaf-blotch disorder of tayberry associated with the leaf and bud mite (Phyllocoptes gracilis) and some effects of three aphid-borne viruses. J. Hortic. Sci. 1984, 59, 523–528. [Google Scholar]
- Jensen, S.G.; Lane, L.C. A new virus disease of corn and wheat in the High Plains. Phytopathology 1994, 84, 1158. [Google Scholar]
- Slykhuis, J.T. Wheat spot mosaic, caused by a mite-transmitted virus associated with Wheat streak mosaic virus. Phytopathology 1956, 46, 682–687. [Google Scholar]
- Jensen, S.G.; Lane, L.C.; Seifers, D.L. A new disease of maize and wheat in the high plains. Plant Dis. 1996, 80, 1387–1390. [Google Scholar] [CrossRef]
- Mahmood, T.; Hein, G.L.; Jensen, S.G. Mixed infection of hard red winter wheat with High Plains virus and Wheat streak mosaic virus from wheat curl mites in Nebraska. Plant Dis. 1998, 82, 311–315. [Google Scholar] [CrossRef]
- Seifers, D.L.; Harvey, T.L.; Martin, T.J.; Jensen, S.G. A partial host range of the High Plains virus of corn and wheat. Plant Dis. 1998, 82, 875–879. [Google Scholar] [CrossRef]
- Jensen, S.G. High plains virus: A new twist to an old story. Phytopathology 1999, 89, S84. [Google Scholar] [CrossRef]
- Lebas, B.S.M.; Ochoa-Corona, F.M.; Elliott, D.R.; Tang, Z.; Alexander, B.J.R. Development of an RT-PCR for High Plains virus indexing scheme in New Zealand post-entry quarantine. Plant Dis. 2005, 89, 1103–1108. [Google Scholar] [CrossRef]
- Jones, A.T.; Kumar, P.L.; Saxena, K.B.; Kulkarni, N.K.; Muniyappa, V.; Waliyar, F. Sterility mosaic disease: The ‘green plague’ of pigeonpea. Plant Dis. 2004, 88, 436–445. [Google Scholar] [CrossRef]
- Mitra, M. Report of the imperial mycologist. Sci. Rep. Agric. Res. Inst. 1931, 19, 58–71. [Google Scholar]
- Saxena, K.B. Genetic improvement of pigeonpea. Trop. Plant Biol. 2008, 1, 159–178. [Google Scholar] [CrossRef]
- Kumar, P.L.; Latha, T.K.S.; Kulkarni, N.K.; Raghavendra, N.; Saxena, K.B.; Waliyar, F.; Rangaswamy, K.T.; Muniyappa, V.; Doriswamy, S.; Jones, A.T. Broad-Based resistance to pigeonpea sterility mosaic disease in wild relatives of pigeonpea (Cajanus: Phaseolae). Ann. Appl. Biol. 2005, 146, 371–379. [Google Scholar] [CrossRef]
- Kannaiyan, J.; Nene, Y.L.; Reddy, M.V.; Ryan, J.G.; Raju, T.N. Prevalence of pigeonpea disease and associated crop losses in Asia, Africa and the Americas. Trop. Pest Manag. 1984, 30, 62–71. [Google Scholar] [CrossRef]
- Reddy, M.V.; Kannaiyan, J.; Nene, Y.L. Increased susceptibility of sterility mosaic infected pigeonpea to powdery mildew. Int. J. Trop. Plant Dis. 1984, 2, 35–40. [Google Scholar]
- Sithanantham, S.; Reddy, M.V.; Rameshwar, R.V. Increased damage by the spider mite Schizotetranychus cajani in Pigeonpea Plants affected by sterility mosaic. In Progress in Acarology; Channabasavanna, G.P., Viraktamath, C.A., Eds.; Oxford & IBH Publishing Co: New Delhi, India, 1989; Volume 2, pp. 11–14. [Google Scholar]
- Kumar, P.L.; Duncan, G.H.; Roberts, A.M.; Jones, A.T.; Reddy, D.V.R. Cytopathology of pigeonpea sterility mosaic virus in pigeonpea and Nicotiana benthamiana: Similarities with those of eriophyid mite-borne agents of undefinded aetiology. Ann. Appl. Biol. 2002, 140, 87–96. [Google Scholar] [CrossRef]
- Bradfute, O.E.; Whitmoyer, R.E.; Nault, R.L. Ultrastructure of plant leaf tissue infected with mite-borne viral-like particles. Proc. Electron Microsc. Soc. Am. 1970, 258, 178–179. [Google Scholar]
- Ebrahim-Nesbat, F.; Izadpanah, K. Virus-Like particles associated with ringfleck mosaic of mountain ash and a mosaic disease of raspberry in the Bavarian forest. Eur. J. For. Path. 1992, 22, 1–10. [Google Scholar] [CrossRef]
- Kikkert, M.; van Poelwijk, F.; Storms, M.; Kassies, W.; Bloksma, H.; van Lent, J.; Kormelink, R.; Goldbach, R. A protoplast system for studying Tomato spotted wilt virus infection. J. Gen. Virol. 1997, 78, 1755–1763. [Google Scholar]
- Plavsic, B.; Milicic, D. Intracellular changes in trees infected with fig mosaic. Acta Hort. 1980, 110, 281–286. [Google Scholar]
- Appiano, A.; Conti, M.; Zini, N. Cytopathological study of the double-membrane bodies in fig plants affected by fig mosaic disease. Acta Hort. 1995, 386, 585–592. [Google Scholar]
- Ammar, E.D.; Gingery, R.E.; Nault, L.R. Two types of inclusions in maize infected with maize stripe virus. Phytopathology 1985, 75, 84–89. [Google Scholar] [CrossRef]
- Espinoza, A.M.; Pereira, R.; Macaya-Lizano, A.V.; Hernandez, M.; Goulden, M.; Rivera, C. Comparative light and electron microscopic analyses of tenuivirus major noncapsid protein (NCP) inclusion bodies in infected plants, and of the NCP in vitro. Virology 1991, 195, 156–166. [Google Scholar]
- Ahn, K.K.; Kim, K.S.; Gergerich, R.C.; Jensen, S.G. High Plains disease of corn and wheat: Ultrastructural and serological aspects. J. Submicrosc. Cytol. Pathol. 1998, 30, 563–571. [Google Scholar]
- Louie, R.; Seifers, D.L.; Bradfute, O.E. Isolation, transmission and purification of the High Plains virus. J. Virol. Meth. 2006, 135, 214–222. [Google Scholar] [CrossRef]
- Black, L.M.; Brakke, M.K.; Vatter, A.E. Purification and electron microscopy of tomato spotted-wilt virus. Virology 1963, 20, 120–130. [Google Scholar] [CrossRef]
- Kitajima, E.W.; de Avila, A.C.; Resende, R.D.O.; Goldbach, R.W.; Peters, D. Comparative cytological and immunogold labelling studies on different isolates of tomato spotted wilt virus. J. Submicrosc. Cytol. Pathol. 1992, 24, 1–14. [Google Scholar]
- Silvestro, S.R.; Chapman, G.B. A transmission electron microscope study of “New Dawn” climber rose (Rosa wichuraiana x safrano) exhibiting rose rosette disease. Plant Cell Rep. 2004, 23, 345–351. [Google Scholar] [CrossRef]
- Kikkert, M.; van Lent, J.; Storms, M.; Bodegom, P.; Kormelink, R.; Goldbach, R. Tomato spotted wilt virus particle morphogenesis in plant cells. J. Virol. 1999, 73, 2288–2297. [Google Scholar]
- Duijsings, D.; Kormelink, R.; Goldbach, R. In vivo analysis of the TSWV cap-snatching mechanism: Single base complementarity and primer length requirements. EMBO J. 2001, 20, 2535–2552. [Google Scholar]
- Rao, P.; Yuan, W.; Krug, R.M. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 2003, 22, 1188–1198. [Google Scholar] [CrossRef]
- Walia, J.J.; Falk, B.W. Fig mosaic virus mRNAs show generation by cap-snatching. Virology 2012, 426, 162–166. [Google Scholar] [CrossRef]
- Liu, D.Y.; Tesh, R.B.; Travassos da Rosa, A.P.A.; Peters, C.J.; Yang, Z.; Guzman, H.; Xiao, S.Y. Phylogenetic relationships among members of the genus Phlebovirus (Bunyaviridae) based on partial M segment sequence analyses. J. Gen. Virol. 2003, 84, 465–473. [Google Scholar] [CrossRef]
- Fodor, E.; Pritlove, D.C.; Brownlee, G.G. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994, 68, 4092–4096. [Google Scholar]
- Barr, J.N.; Elliott, R.M.; Dunn, E.F.; Wertz, G.W. Segment specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology 2003, 311, 326–228. [Google Scholar] [CrossRef]
- Kohl, A.; Lowen, A.C.; Leonard, V.H.J.; Elliott, R.M. Genetic elements regulating packaging of the Bunyamwera orthobunyavirus genome. J. Gen. Virol. 2006, 87, 177–187. [Google Scholar] [CrossRef]
- Elbeaino, T.; Digiaro, M.; Martelli, G.P. RNA-5 and -6, two additional negative-sense RNA segments associated with fig mosaic virus. J. Plant Pathol. 2012, 1. [Google Scholar]
- Ishikawa, K.; Maejima, K.; Komatsu, K.; Kitazawa, Y.; Hashimoto, M.; Takata, D.; Yamaji, Y.; Namba, S. Identification and characterization of two novel genomic RNA segments of fig mosaic virus, RNA5 and RNA6. J. Gen. Virol. 2012, 93, 1612–1619. [Google Scholar] [CrossRef]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar]
- Seifers, D.L.; Harvey, T.L.; Martin, T.J.; Jensen, S.G. Identification of the wheat curl mite as the vector of the High Plains virus of corn and wheat. Plant Dis. 1997, 81, 1161–1166. [Google Scholar] [CrossRef]
- She, Y.-M.; Seifers, D.L.; Haber, S.; Ens, W.; Standing, K.G. Characterization of the agent of ‘High Plains disease’. J. Biol. Chem. 2004, 279, 488–494. [Google Scholar]
- Kulkarni, N.K.; Kumar, P.L.; Muniyappa, V.; Jones, A.T.; Reddy, D.V.R. Transmission of pigeon pea sterility mosaic by the eriophyid mite, Aceria cajani (Acari: Arthropoda). Plant Dis. 2002, 86, 1297–1302. [Google Scholar] [CrossRef]
- Seifers, D.L.; Harvey, T.L.; Louie, R.; Gordon, D.T.; Martin, T.J. Differential transmission of isolates of the High Plains virus by different sources of wheat curl mites. Plant Dis. 2002, 86, 138–142. [Google Scholar] [CrossRef]
- Skare, J.M.; Wijkamp, I.; Rezende, J.A.M.; Michels, G.; Rush, C.; Scholthof, K.-B.G.; Scholthof, H.B. Colony establishment and maintenance of the eriophyid wheat curl mite Aceria tosichella for controlled transmission studies on a new virus-like pathogen. J. Virol. Methods 2003, 108, 133–137. [Google Scholar] [CrossRef]
- Mielke-Ehret, N.; Thoma, J.; Schlatermund, N.; Mühlbach, H.-P. Detection of European mountain ash ringspot-associated virus-specific RNA and protein P3 in the pear leaf blister mite Phytoptus pyri (Eriophyidae). Arch. Virol. 2010, 155, 987–991. [Google Scholar] [CrossRef]
- Elbeshehy, E.K.; Elbeaino, T. Viruses infecting figs in Egypt. Phytopathol. Mediterr. 2011, 50, 327–332. [Google Scholar]
- El Air, M.; Mahfoudhi, N.; Elbeaino, T.; Dhouibi, M.H.; Digiaro, M. Occurrence of fig mosaic virus in Tunisian fig orchards. J. Plant Pathol. 2012, 1. [Google Scholar]
- Alhudaib, K. Incidence of fig leaf mottle-associated virus and fig mosaic virus in Eastern Province of Saudi Arabia. Int. J. Virol. 2012, 8, 128–132. [Google Scholar] [CrossRef]
- Shahmirzaie, M.; Rakhshanderoo, F.; Zamanizadeh, H.R.; Elbeaino, T. Current status of fig mosaic disease in Iran. J. Phytopathol. 2012, 160, 324–330. [Google Scholar] [CrossRef]
- Ishikawa, K.; Maejima, K.; Nagashima, S.; Sawamura, N.; Takinami, Y.; Komatsu, K.; Hashimoto, M.; Yamaji, Y.; Yamamoto, J.; Namba, S. First report of fig mosaic virus infecting common fig (Ficus carica) in Japan. J. Gen. Plant Pathol. 2012, 78, 136–139. [Google Scholar] [CrossRef]
- Flock, R.A.; Wallace, J.M. Transmission of fig mosaic by the eriophyid mite Aceria ficus. Phytopathology 1955, 45, 52–54. [Google Scholar]
- Seth, M.L. Transmission of pigeonpea sterility by an eriophyid mite. Indian Phytopathol. 1962, 15, 225–227. [Google Scholar]
- Proeseler, G. Beziehungen zwischen Virus, Vektor und Wirtspflanze am Beispiel des Feigenmosaik-Virus und Aceria ficus Cotte (Eriophyoidea) (in German). Acta Phytopathol. Acad. Sci. Hung. 1972, 7, 179–186. [Google Scholar]
- Capoor, S.P. Observations on pigeonpea sterility disease in Bombay. Indian J. Agric. Sci. 1952, 22, 271–274. [Google Scholar]
- Alfieri, S.A., Jr. Fig mosaic. In Plant Pathology Circular No. 64; Florida Department of Agriculture, Division of Plant Industry: Gainesville, FL, USA, 1967. [Google Scholar]
- Führling, M.; Büttner, C. Transmission experiments of viruses to woody seedlings (Quercus robur L. and Sorbus aucuparia L.) by grafting and mechanical inoculation. Eur. J. For. Path. 1995, 25, 129–135. [Google Scholar] [CrossRef]
- Forster, R.L.; Seifers, D.L.; Strausbaugh, C.A.; Jensen, S.G.; Ball, E.M.; Harvey, T.L. Seed transmission of the High Plains virus in sweet corn. Plant Dis. 2001, 85, 696–699. [Google Scholar] [CrossRef]
- Divya, P.; Kumar, L.L.; Rangaswamy, K.T.; Muniyappa, V. Detection of pigonpea sterility mosaic virus in floral parts and seeds. Indian J. Virol. 2005, 16, 36. [Google Scholar]
- Kim, K.S.; Martin, E.M. Virus-like particles associated with yellow ringspot of redbud. Phytopathol. News 1978, 12, 119. [Google Scholar]
- Ahn, K.K.; Kim, K.S.; Gergerich, R.C.; Jensen, S.G.; Anderson, E.J. Comparative ultrastructure of double-membrane-bound particles and inclusions associated with eriophyid mite-borne plant diseases of unknown etiology: A potentially new group of plant viruses. J. Submicrosc. Cytol. Pathol. 1996, 28, 345–355. [Google Scholar]
- Laney, A.G.; Gergerich, R.; Keller, K.; Martin, R.; Tzanetakis, I. Rose rosette and redbud yellow ringspot are caused by two new emaraviruses. Phytopathology 2010, 100, S67. [Google Scholar]
- Robertson, N.L.; Brown, K.L. Ringspot leaf symptoms on Sorbus scopulina greene associated with virus-like particles. Phytopathology 2009, 99, S185–S186. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mielke-Ehret, N.; Mühlbach, H.-P. Emaravirus: A Novel Genus of Multipartite, Negative Strand RNA Plant Viruses. Viruses 2012, 4, 1515-1536. https://doi.org/10.3390/v4091515
Mielke-Ehret N, Mühlbach H-P. Emaravirus: A Novel Genus of Multipartite, Negative Strand RNA Plant Viruses. Viruses. 2012; 4(9):1515-1536. https://doi.org/10.3390/v4091515
Chicago/Turabian StyleMielke-Ehret, Nicole, and Hans-Peter Mühlbach. 2012. "Emaravirus: A Novel Genus of Multipartite, Negative Strand RNA Plant Viruses" Viruses 4, no. 9: 1515-1536. https://doi.org/10.3390/v4091515
APA StyleMielke-Ehret, N., & Mühlbach, H.-P. (2012). Emaravirus: A Novel Genus of Multipartite, Negative Strand RNA Plant Viruses. Viruses, 4(9), 1515-1536. https://doi.org/10.3390/v4091515




