Molecular Identification of Chronic Bee Paralysis Virus Infection in Apis mellifera Colonies in Japan
Abstract
1. Introduction
2. Results
| Sample | Virus | Microsporidia | Protist | Tracheal mite | Severe loss of workers | Use for pollination |
|---|---|---|---|---|---|---|
| Japan-1 | CBPV, BQCV, DWV | N. ceranae | − | − | + | − |
| Japan-2 | CBPV, BQCV, DWV | N. ceranae | − | − | + | + |
| Japan-3 | CBPV, BQCV, DWV, IAPV, SBV | N. ceranae | − | + | + | − |
| Japan-4 | CBPV, BQCV, DWV, IAPV | N. ceranae | − | − | + | − |
| Japan-5 | CBPV | N. ceranae | − | − | + | + |
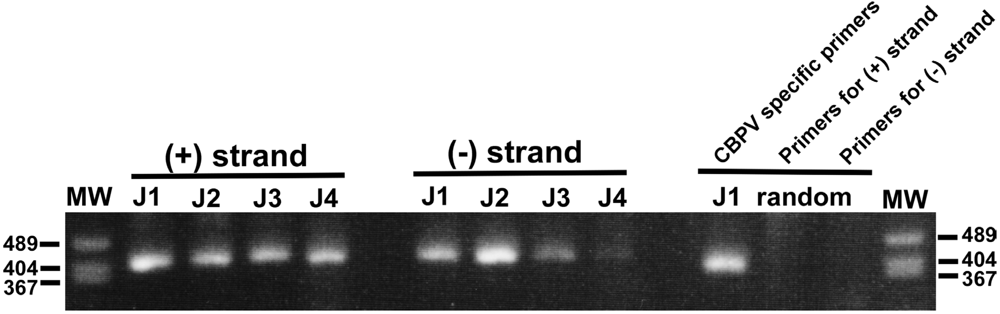

3. Discussion
4. Experimental Section
4.1. Honey Bee Sample Collection
4.2. RT-PCR Detection of Viruses
4.3. PCR Detection of N. ceranae and N. apis
4.4. PCR Detection of Tracheal Mite (A. woodi)
4.5. PCR Detection of Crithidia mellificae and Apicystis bombi
4.6. Construction of Phylogenetic Tree of CBPV
5. Conclusions
Acknowledgments
Funding
Conflict of Interest
References and Notes
- Bailey, L.; Ball, B.V.; Perry, J.N. Honeybee paralysis: Its natural spread and its diminished incidence in England and Wales. J. Apic. Res. 1983, 22, 191–195. [Google Scholar]
- Ball, B.V.; Bailey, L. Viruses. In Honey Bee Pests, Predators, & Diseases; Morse, R.A., Flottum, K., Eds.; A.I. Root Company: Medina, OH, USA, 1997; pp. 11–32. [Google Scholar]
- Blanchard, P.; Ribière, M.; Celle, O.; Lallemand, P.; Schurr, F.; Olivier, V.; Iscache, A.L.; Faucon, J.P. Evaluation of a real-time two-step RT-PCR assay for quantification of Chronic bee paralysis virus (CBPV) genome in experimentally-infected bee tissues and in life stages of a symptomatic colony. J. Virol. Methods 2007, 141, 7–13. [Google Scholar]
- Allen, M.; Ball, B.V. The incidence and world distribution of honey bee viruses. Bee World 1996, 77, 141–162. [Google Scholar]
- Olivier, V.; Blanchard, P.; Chaouch, S.; Lallemand, P.; Schurr, F.; Celle, O.; Dubois, E.; Tordo, N.; Thiéry, R.; Houlgatte, R.; et al. Molecular characterization and phylogenetic analysis of Chronic bee paralysis virus: A honey bee virus. Virus Res. 2008, 132, 59–68. [Google Scholar] [CrossRef]
- Kojima, Y.; Toki, T.; Morimoto, T.; Yoshiyama, M.; Kimur, A.K.; Kadowaki, T. Infestation of Japanese native honey bees by tracheal mite and virus from non-native European honey bees in Japan. Microbial. Ecol. 2011, 62, 895–906. [Google Scholar]
- Ribière, M.; Olivier, V.; Blanchard, P. Chronic bee paralysis: A disease and a virus like no other? J. Invertebr. Pathol. 2010, 103Suppl 1, S120–S131. [Google Scholar]
- Blanchard, P.; Schurr, F.; Olivier, V.; Celle, O.; Antùnez, K.; Bakonyi, T.; Berthoud, H.; Haubruge, E.; Higes, M.; Kasprzak, S.; et al. Phylogenetic analysis of the RNA-dependant RNA polymerase (RdRp) and a predicted structural protein (pSP) of the chronic bee paralysis virus (CBPV) isolated from various geographical regions. Virus Res. 2009, 144, 334–338. [Google Scholar] [CrossRef]
- Chen, Y.P.; Evans, J.D.; Pettis, J.S. The presence of chronic bee paralysis virus infection in honey bees (Apis mellifera L.) in the USA. J. Apic. Res. 2011, 50, 85–86. [Google Scholar] [CrossRef]
- Grabensteiner, E.; Ritter, W.; Carter, M.J.; Davison, S.; Pechhacker, H.; Kolodziejek, J.; Boecking, O.; Derakhshifar, I.; Moosbeckhofer, R.; Licek, E.; et al. Sacbrood virus of the honeybee (Apis mellifera): Rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 2001, 8, 93–104. [Google Scholar]
- Ai, H.; Yan, X.; Han, R. Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in China. J. Invertebr. Pathol. 2012, 109, 160–164. [Google Scholar] [CrossRef]
- Morimoto, T.; Kojima, Y.; Toki, T.; Komeda, Y.; Yoshiyama, M.; Kimura, K.; Nirasawa, K.; Kadowaki, T. The habitat disruption induces immune-suppression and oxidative stress in honey bees. Ecol. Evol. 2011, 1, 201–217. [Google Scholar] [CrossRef]
- Chen, Y.P.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef]
- Kohno, K.; Sokabe, T.; Tominaga, M.; Kadowaki, T. Honey bee thermal/chemical sensor, AmHsTRPA, reveals neofunctionalization and loss of transient receptor potential channel genes. J. Neurosci. 2010, 30, 12219–12229. [Google Scholar]
- Kojima, Y.; Yoshiyama, M.; Kimura, K.; Kadowaki, T. PCR-based detection of a tracheal mite of the honey bee Acarapis woodi. J. Invertebr. Pathol. 2011, 108, 135–137. [Google Scholar]
- Meeus, I.; de Graaf, D.C.; Jans, K.; Smagghe, G. Multiplex PCR detection of slowly-evolving trypanosomatids and neogregarines in bumblebees using broad-range primers. J. Appl. Microbiol. 2010, 109, 107–115. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. EGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance; and maximum parsimony methods. Mol. Biol. Evol. 2011, 10, 2731–2739. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Morimoto, T.; Kojima, Y.; Yoshiyama, M.; Kimura, K.; Yang, B.; Kadowaki, T. Molecular Identification of Chronic Bee Paralysis Virus Infection in Apis mellifera Colonies in Japan. Viruses 2012, 4, 1093-1103. https://doi.org/10.3390/v4071093
Morimoto T, Kojima Y, Yoshiyama M, Kimura K, Yang B, Kadowaki T. Molecular Identification of Chronic Bee Paralysis Virus Infection in Apis mellifera Colonies in Japan. Viruses. 2012; 4(7):1093-1103. https://doi.org/10.3390/v4071093
Chicago/Turabian StyleMorimoto, Tomomi, Yuriko Kojima, Mikio Yoshiyama, Kiyoshi Kimura, Bu Yang, and Tatsuhiko Kadowaki. 2012. "Molecular Identification of Chronic Bee Paralysis Virus Infection in Apis mellifera Colonies in Japan" Viruses 4, no. 7: 1093-1103. https://doi.org/10.3390/v4071093
APA StyleMorimoto, T., Kojima, Y., Yoshiyama, M., Kimura, K., Yang, B., & Kadowaki, T. (2012). Molecular Identification of Chronic Bee Paralysis Virus Infection in Apis mellifera Colonies in Japan. Viruses, 4(7), 1093-1103. https://doi.org/10.3390/v4071093





