Abstract
There are five antigenically distinct ebolaviruses that cause hemorrhagic fever in humans or non-human primates (Ebola virus, Sudan virus, Reston virus, Taï Forest virus, and Bundibugyo virus). The small handful of antibodies known to neutralize the ebolaviruses bind to the surface glycoprotein termed GP1,2. Curiously, some antibodies against them are known to neutralize in vitro but not protect in vivo, whereas other antibodies are known to protect animal models in vivo, but not neutralize in vitro. A detailed understanding of what constitutes a neutralizing and/or protective antibody response is critical for development of novel therapeutic strategies. Here, we show that paradoxically, a lower affinity antibody with restricted access to its epitope confers better neutralization than a higher affinity antibody against a similar epitope, suggesting that either subtle differences in epitope, or different characteristics of the GP1,2 molecules themselves, confer differential neutralization susceptibility. Here, we also report the crystal structure of trimeric, prefusion GP1,2 from the original 1976 Boniface variant of Sudan virus complexed with 16F6, the first antibody known to neutralize Sudan virus, and compare the structure to that of Sudan virus, variant Gulu. We discuss new structural details of the GP1-GP2 clamp, thermal motion of various regions in GP1,2 across the two viruses visualized, details of differential interaction of the crystallized neutralizing antibodies, and their relevance for virus neutralization.
Keywords:
Filovirus; Ebola; ebolavirus; Sudan virus; neutralization: glycoprotein; antibodies; structure 1. Introduction
Ebolaviruses are filamentous viruses with a negative-sense RNA genome that cause severe hemorrhagic fever with high lethality. There are five different ebolaviruses, each named after the location of the outbreak in which it was first discovered: Ebola virus (EBOV; formerly known as Zaire ebolavirus), Sudan virus (SUDV), Reston virus (RESTV), Taï Forest virus (TAFV), and Bundibugyo virus (BDBV) [1,2]. These virions differ by 40-50% in primary amino acid sequence and are antigenically distinct. Sudan virus was the first ebolavirus to be discovered when it caused an outbreak of hemorrhagic fever among cotton factory workers in Nzara, Sudan, in 1976 [3]. Travel of these patients and their caretakers spread the disease to other villages including Maridi, where in the hospital at which several of the patients were treated, the doctor-in-charge and 61 of the 154 nursing staff became infected [3]. The variant of Sudan virus linked to this outbreak is termed Boniface (or SUDV-Bon) after the index case. This emergence of SUDV-Bon ultimately led to 284 cases of disease and 151 deaths.
A separate variant, termed Gulu (or SUDV-Gul), is a modern variant linked to a disease outbreak in the Gulu municipality of Uganda in 2000 [4]. The Gulu outbreak was the largest outbreak of Ebola virus disease yet recorded, with 425 cases and 224 deaths and a large number of cases among children and adolescents. The glycoprotein (GP1,2) of SUDV-Bon and SUDV-Gul differ in sequence by ~5% and an ideal diagnostic or immunotherapeutic for Sudan virus would be able to recognize and neutralize both the Boniface and Gulu variants. An even better diagnostic or therapeutic would be able to recognize all ebolaviruses, taking advantage of structural information of the distinct GP1,2 glycoproteins of each virus.
The trimeric, membrane-attached glycoprotein GP1,2 is the only virally encoded protein present on the virus surface and is critical for virus adhesion, entry and internalization into target cells. In infected cells, GP1,2 is cleaved by furin to yield two subunits, termed GP1 and GP2, that remain linked by a disulfide bond. GP1 is responsible for recognition and engagement of new target cells, while GP2 drives fusion of the viral membrane with the endosomal membrane of the target cell. Antibodies against EBOV GP1,2 have been tested for protection against EBOV challenge in various models with mixed outcomes [5,6,7,8,9,10]; although recent studies are quite promising [11].
A crystal structure of trimeric GP1,2 from Ebola virus variant Mayinga (EBOV-May) in its prefusion form (PDB code 3CSY) [12] revealed that the three GP1 subunits are gathered and tethered into a bowl/chalice shape in the viral surface trimer by the three GP2 subunits that wrap around the GP1s. In crystal structures of the post-fusion six-helix bundle conformation of GP2, heptad repeat 1 (HR1) adopts a single long helix [13,14,15]. However, in the prefusion structure, the hydrophobic fusion peptides of GP2 (residues 529-535) wrap around the outside of the trimer, packing against the GP1 subunits of adjacent monomers, and only the first of two heptad repeats in GP2 is visible. This single helix is broken into four separate sections in the prefusion structure: HR1A-D. The crystal structure also illustrated that within GP1, putative receptor-binding regions are somewhat sequestered inside the bowl of the chalice at the top of GP1,2. A subsequent crystal structure of SUDV-Gul GP1,2 (PDB code 3S88) [16] illustrates that its GP1,2 has a similar fold but curiously, different electrostatic characteristics than EBOV-May GP1,2. EBOV-May GP1,2 is neutral/basic while SUDV-Gul GP1,2 is acidic at the base where the GP1 and GP2 subunits meet. In addition, the ectodomain of SUDV GP1,2 is more susceptible to proteolysis than EBOV GP1,2. Treatment of SUDV GP1,2 with either thermolysin or cathepsin L/B results in significant degradation of GP1,2 [17]. It is possible that the different electrostatics may contribute to the differing behavior of the GP1,2 in the endosome [16]. Recent studies also show that the GP1,2 of filoviruses exhibit distinct protease preferences. The GP1,2 of EBOV is strongly dependent on the endosomal protease cathepsin B while the GP1,2 of SUDV is not dependent on the protease [18,19].
Both EBOV-May GP1,2 and SUDV-Gul GP1,2 structures were determined in complex with antibodies. EBOV-May GP1,2 was crystallized in complex with an EBOV-specific human antibody termed KZ52, which was derived from a human survivor of a natural infection in Kikwit, Zaire (EBOV-May), in 1995 [5]. SUDV-Gul GP1,2 was crystallized in complex with a novel Fab termed 16F6 that was raised by immunization of mice with irradiated SUDV-Bon virions [16]. Unexpectedly, the epitopes of KZ52 and 16F6 overlap, at the base of the GP1,2 spike where GP2 meets GP1[16], suggesting that the two antibodies may function in the same way and that the shared site may be a hot spot for neutralization of ebolaviruses.
Here we report new molecular details of neutralization of the different GP1,2s by respective antibodies. Surface Plasmon Resonance and neutralization assays reveal that paradoxically, the antibody with lower affinity for recombinant GP1,2, with hindered access to GP1,2 epitope, has better neutralizing capacity. Here, we also report a new crystal structure, to 3.35 Å resolution, of SUDV-Bon GP1,2 from the historical 1976 Boniface variant, in complex with mAb 16F6. This SUDV-Bon GP1,2 structure allows better visualization of key contacts between GP1 and GP2 and the “chain reversal regions” of GP2 involved in conformational changes during fusion (a step likely blocked by both 16F6 and KZ52) [20]. Comparison of the now three available structures of ebolavirus GP1,2s (SUDV-Bon, SUDV-Gul and EBOV-May) provides new insights into particular components of the two neutralizing epitopes and the puzzle of what constitutes an effective neutralizing antibody response against ebolaviruses.
2. Results and Discussion
2.1. Crystal Structure of Sudan Virus Boniface GP1,2
The crystal structure of SUDV-Bon GP1,2 bound to antibody 16F6 was determined to a resolution of 3.35 Å using molecular replacement. The crystallographic asymmetric unit contains one GP1,2 monomer bound to one 16F6 Fab fragment. The final model contains residues 32-191, 213-285, 300-311 and 510-614 of GP1,2 and residues 1-220 and 1-212 of the heavy and light chains of 16F6, respectively. A disulfide bond between C53 and C609 covalently links the GP1 and GP2 subunits together, and is only visible in structures of SUDV, but not EBOV GP1,2. In all three ebolavirus GP1,2 structures, GP1 and GP2 bury a surface area of ~2400 Å2 on each other and are held together by numerous hydrogen bonds and non-bonded contacts. Although GP1,2 was treated with peptide-N-glycosidase F (PNGase F), clear electron density is still observed for the first two monosaccharides of glycans attached to N257 (NAG 850) in GP1 and N563 (NAG 901) in GP2 indicating that these sites (in both SUDV and EBOV GP1,2) are resistant to PNGaseF digestion (Figure 1). The trimeric form of the GP1,2 is obtained by application of 3-fold crystal symmetry, and has an approximate dimension of 95 Å × 95 Å × 90 Å.
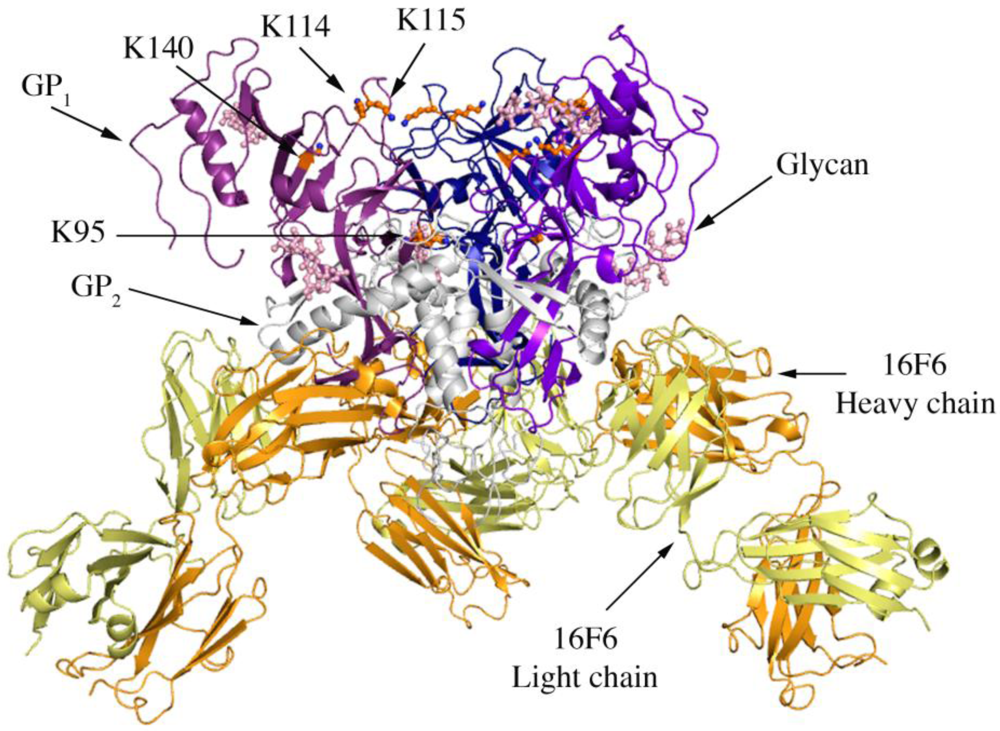
Figure 1.
Cartoon representation of trimer of SUDV-Bon GP1,2 bound to 16F6. The GP1 chains are colored in various shades of blue and GP2 chains are colored in white. The light chain of 16F6 is shown in pale yellow and the heavy chain of 16F6 is colored bright orange. The sugar residues on the glycoprotein are shown in ball and colored cyan. Lysine residues critical for receptor binding are shown in ball and stick and colored orange.
Four lysines (95, 114, 115, and 140) in the putative receptor-binding head of GP1 have been identified as critical for attachment of EBOV to target cells [21]. Three of these (114, 115, and 140) are strictly conserved among all known ebolaviruses, while position 95 is occupied by glutamine in some variants of SUDV (but is lysine in the SUDV-Bon and SUDV-Gul sequences crystallized). In all three ebolavirus GP1,2 structures, K95 is buried deep in the chalice bowl of GP1 and projects into GP2 rather than out toward the target cell and receptor (Figure 1). Inside GP1,2, K95 forms a hydrogen bond with the main-chain carbonyl of T576 and forms van der Waals interactions with L573, which is located in the loop between HR1B and HR1C of GP2. Hence the crystal structures support biochemical predictions [21] that the observed functional importance of K95 may not be in direct receptor engagement, but rather in proper maintenance or springing of the prefusion GP1,2 assembly. Alternatively, K95 may become better exposed for interaction with host factor(s) in the endosome if any conformational changes occur as a result of receptor binding. K115 also points down toward the viral membrane, but is solvent exposed and could potentially reorient upon receptor engagement. K114 and K140 project into solvent towards the target cell and may serve as direct contacts for receptor.
In the prefusion conformation of ebolavirus GP1,2, the first heptad repeat of GP2 is broken into four structural segments (HR1A-D) that wrap around GP1. We observe in all three structures (SUDV-Gul, SUDV-Bon, and EBOV-May) that residues R580-T581 of HR1C form a short β strand that assembles into a continuous β sheet with the K95-S99 β strand of the GP1 receptor-binding head. These residues are completely conserved among all ebolaviruses and marburgviruses.
Other structural details not previously reported include the noncovalent assembly by which GP1 is anchored to GP2. Five β strands of the GP1 base and two antiparallel β strands of the GP2 internal fusion loop combine to form a continuous seven-stranded twisted β sheet. The GP1 component of that sheet and a second small β sheet of the GP1 base together assemble a semicircular GP1 horseshoe surface that clamps the first heptad repeat of GP2 in its metastable, prefusion structure (Figure 2 A,B). In order for GP2 to rearrange into its post-fusion six-helix bundle structure [13,15] both the continuous GP1-GP2β sheet and the horseshoe clamp must be broken. The C-terminal heptad repeat 2 is disordered in all three SUDV-Bon, SUDV-Gul and EBOV-May structures, which may result from the functional mobility of this region as well as the lack of the transmembrane regions that tether GP1,2 on the viral surface.
2.2. Comparison of SUDV-Bon and SUDV-Gul Glycoproteins
The Boniface and Gulu variants of Sudan GP1,2 differ in sequence by ~ 5%. The clones of SUDV-Bon and SUDV-Gulu GP1,2 used for crystallization contain 449 residues, of which 7 differ (position 237 in the glycan cap - N/D for Bon/Gul; position 243 in the glycan cap - L/R; position 272 in the glycan cap -K/R; position 310 in the glycan cap - T/A; position 503 near the furin cleavage site - V/T; position 506 near the furin cleavage site -R/K; and position 631 close to the transmembrane region - I/V). Overall, structures of SUDV-Bon and SUDV-Gul GP1,2 align with an r.m.s.d. of 1.0 Å, and the entire SUDV GP1,2-antibody complexes align with an r.m.s.d. of 0.74 Å, because the Fab components of the complexes are better ordered and better superimpose than the glycoproteins to which they are bound.
2.3 Implications for fusion
A CX6CC motif in GP2 that is conserved among all ebolaviruses is visible in both structures of SUDV GP1,2. This motif contains the disulfide bond that anchors GP1 to GP2 (C53-C609) and a second disulfide bond within GP2 (C601-C608). The glycoproteins of some retroviruses have a CXXC motif within the receptor-binding subunit that isomerizes to release the disulfide bond between the receptor-binding and fusion subunits [20]. Filoviruses, however, lack such a motif in GP1, and it is currently unknown if the GP1-GP2 disulfide link, which is encoded by a different motif, is retained or released during fusion. It has been speculated that endosomal thiolreductases could reduce the C53-C609 GP1-GP2 bond [22], and mild reduction of the enzymatically processed 19 kDa GP1,2 does confer binding to liposomes (an ability presumably attained via conformational change, as unreduced 19 kDa GP1,2 does not bind liposomes) [23]. The neighboring, C601-C608 intra-GP2 disulfide bond however, is observed in pre-fusion as well as post-fusion crystal structures and thus, probably remains intact during fusion. Given the close proximity of both the GP1-GP2 and GP2-GP2 disulfide bonds, reduction is either very specific for the GP1-GP2 disulfide bond or plays no role during fusion. Separate possibilities are that GP1 simply rotates out of the way, or that continual enzymatic processing of GP1 by cathepsins digests enough of GP1 to remove steric hindrance to GP2 conformational rearrangement [22].
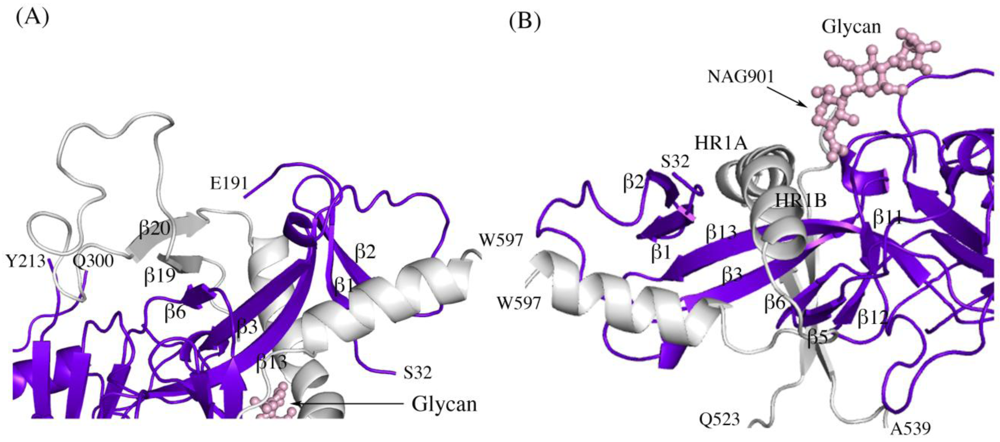
Figure 2.
Secondary structure interactions between SUDV-Bon GP1 (colored blue and purple) and GP2 (colored light grey). (A) Close up of the continuous seven-stranded twisted β sheet formed by five β strands of the GP1 base (β2, β1, β13, β3, and β6 in blue) and two antiparallel β strands of the GP2 internal fusion loop (β19 and β20 in grey). (B) The first heptad repeat of GP2 (which contains the grey coils labeled HR1A and HR1B and the loop afterwards) is clamped by a horseshoe surface formed by five β strands of GP1 base (β12, β5, β6, β3, β13), a second small β sheet of the GP1 base (β1 and β2) and a glycan (pink ball-and-stick). To orient the reader, key residues are labeled.
A recent study by Delos et al. has revealed residues in the “chain reversal region” of GP2 critical for fusion of Ebola virions [20]. This region comprises a short stretch of hydrophobic residues, a glycine-glycine pair, a CX6CC motif, and a bulky hydrophobic residue after the motif. Mutational analysis in EBOV GP1,2 has shown that residues R587, F592, W597, G598, H602, and I610 are critical for viral infectivity. All the above residues except H602 (which is R in SUDV and RESTV) are conserved in SUDV. The better ordered structures of SUDV provide us with newer insights into the role of the conserved residues in the prefusion conformation, and are reported here. Interactions made by the side chains of these residues in the prefusion-form in SUDV and the post-fusion form in analogous EBOV (strain Mayinga, PDB code 1EBO) are shown in Table 1. In the pre-fusion state, the conserved residues make interactions with GP1 (hydrogen bonding to T60′ and stacking interaction against L57 and I185 (Figure 3A); the ′ denotes residues from a 3-fold related monomer). W597 is involved in a stacking interaction with other W597 residues from two 3-fold related monomers, suggesting its role in stabilizing the trimeric form in the heptad repeat region. Definitive density was not seen for the side chains of R602 and I610 in either SUDV-Bon or SUDV-Gul GP1,2 and we could not assign any interactions of these residues; positions 602 and 610 are thus modeled as alanine. However, in the post-fusion state, the conserved residues make interactions solely with residues in GP2 (Figure 3B). In addition, the conserved residues make interactions with different residues in the pre-fusion and post-fusion forms suggesting a greater role of these residues during the fusion process.
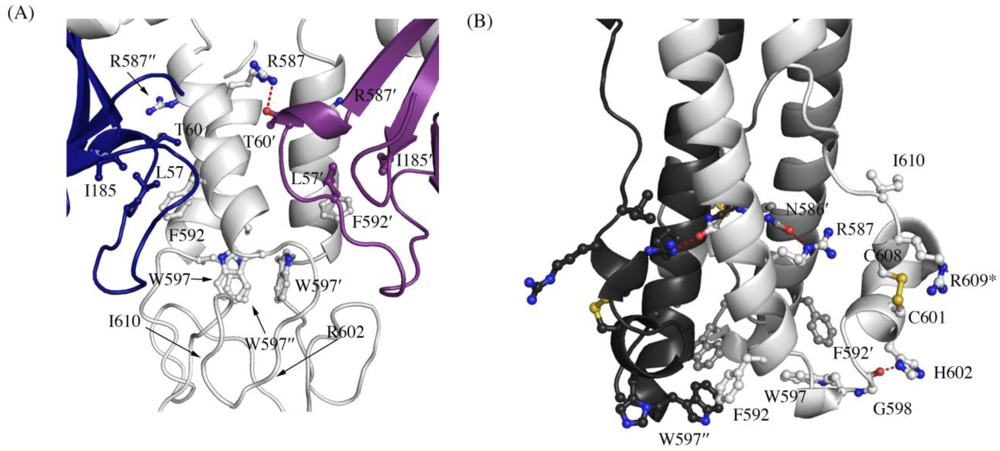
Figure 3.
Interaction of residues in the chain reversal region in (A) the prefusion SUDV-Bon GP1,2 and (B) the post fusion EBOV-May GP2. Prefusion SUDV-Bon is used here as it is better ordered than prefusion EBOV-May. Interacting residues are shown in ball and stick. In (A), different monomers of GP1 are colored blue and purple and the three copies of GP2 are all colored light grey. In (B), the three copies of GP2 are colored different shades of grey. Equivalent residues in the 3-fold related protomers are labeled with ´ and ´´ respectively. Hydrogen bonds are shown as red dashed lines. The residue R609* in the postfusion form is an engineered mutation to replace the cysteine residue (C609) in the native protein that is involved in a disulfide bond with C53 of GP1.
2.4 Interactions between 16F6 and SUDV GP
The complementarity determining regions (CDRs) H1 and H3 of 16F6 form a network of hydrogen bonds, van der Waals interactions and one salt bridge to the GP1 base. CDR L2 also hydrogen bonds to the GP1 base and forms additional hydrophobic interactions to the stem region of the internal fusion loop of GP2 (Figure 4). Specific interactions between 16F6 and the glycoprotein have not been previously reported and are shown in Table 2. The heavy chain and light chain of 16F6 bury a surface area of ~1630 Å2 between them. The antibody 16F6 interacts with GP1,2 primarily using its heavy chain, burying an area of ~350 Å2 with GP1 and ~200 Å2 with GP2. The interface between GP1,2 and 16F6 is predominantly hydrophobic with the exception of four hydrogen bonds.
Figure 4.
Residues at the interface of SUDV-Bon GP1,2 and 16F6 (cutoff distance of 3.5 Å). GP1 is colored purple, GP2 is colored white, the 16F6 heavy chain is colored orange and the light chain is colored pale yellow. Hydrogen bonds are shown as red dashed lines.
2.5 Thermal Motion in GP
Comparison of B-factor values (an atomic displacement parameter arising from thermal vibration of atoms and static disorder of atoms in different unit cells of the protein crystal) of key portions of GP1 and GP2 in SUDV GP1,2 reveals that motion predominates in the glycan cap regions, the C-terminal half of the fusion loop, and the visible C-terminal regions of GP2 (Figure 5). How does SUDV compare to EBOV GP1,2 in this regard? Deuterium Exchange Mass Spectrometry (DXMS) reveals that although GP1 of SUDV and EBOV exhibit nearly identical rates of exchange of amide hydrogens with solvent deuterium, all regions of GP2 of SUDV, including the fusion loop, heptad repeats, disulfide-containing linker and C-terminal regions, are fundamentally more mobile than those of EBOV GP1,2 [17] (Figure 6). Interestingly, the disulfide-containing linker regions of GP2 are only visible in crystals of SUDV GP1,2, not EBOV GP1,2. The unique crystal packing environment of the SUDV I23 unit cell and the acute angle made by the bound 16F6 antibody may have constrained this region.
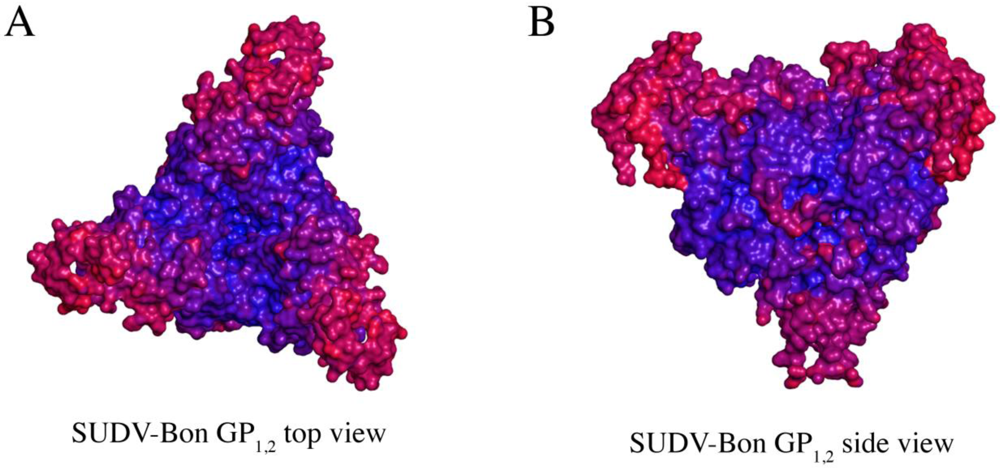
Figure 5.
Comparison of thermal factors (B-factors) of various regions of SUDV GP1,2 (Panels A and B). Regions of low thermal mobility are deep blue (deep blue color set at ~ 80 Å2), whereas regions of high thermal mobility are red (deep red color set at ~ 220 Å2). The glycan cap and C-terminal “stem” regions of SUDV GP1,2 have higher B-values than the core region, indicating higher motion in those regions.
Another difference in mobility between SUDV and EBOV GP1,2 relevant for antibody binding is the N terminus of GP2, which is released from GP1 by furin cleavage. In EBOV GP1,2, this peptide is hydrogen bonded to the GP1,2 core, is bound between heavy and light chains in the central region of the KZ52 paratope and forms a significant portion of its epitope. By contrast, in the SUDV structure, the same peptide is disordered and is not bound by 16F6. Instead, 16F6 binds the underlying GP1,2 core beneath the peptide (Figure 7).
One explanation could be that the GP2 N terminus is simply tacked down in the EBOV GP1,2 crystal structure by KZ52 binding. A more likely explanation, supported by DXMS, is that the mobility of this region fundamentally differs between SUDV and EBOV, even in the absence of antibody binding. Key differences in sequence between SUDV and EBOV at this site may explain this observation. The anchor point of the GP2 N terminus to the GP1,2 core is residue G509. At position 509, EBOV contains a proline, which may restrict mobility, while SUDV contains a glycine, which may enhance mobility. Further, residues N506 and Q508 of EBOV use both their terminal oxygen and nitrogen atoms to form a network of hydrogen bonds to the EBOV GP1,2 core; specifically, to amino acids as well as the attached glycan of heptad repeat 1. By contrast, position 506 is K in SUDV-Gul and Q in SUDV-Bon, and position 508 is T in both SUDV GP1,2 (Gulu and Boniface). None of those hydrogen bonds made by EBOV are observed for SUDV, and the specific hydrogen bonds made by the side-chain oxygens of N506 and Q508 in EBOV are not possible for the corresponding residues in SUDV. The key differences in sequence and inherent mobility of N-term region of SUDV GP1,2 reflects in a marginally varied epitope for binding of 16F6.
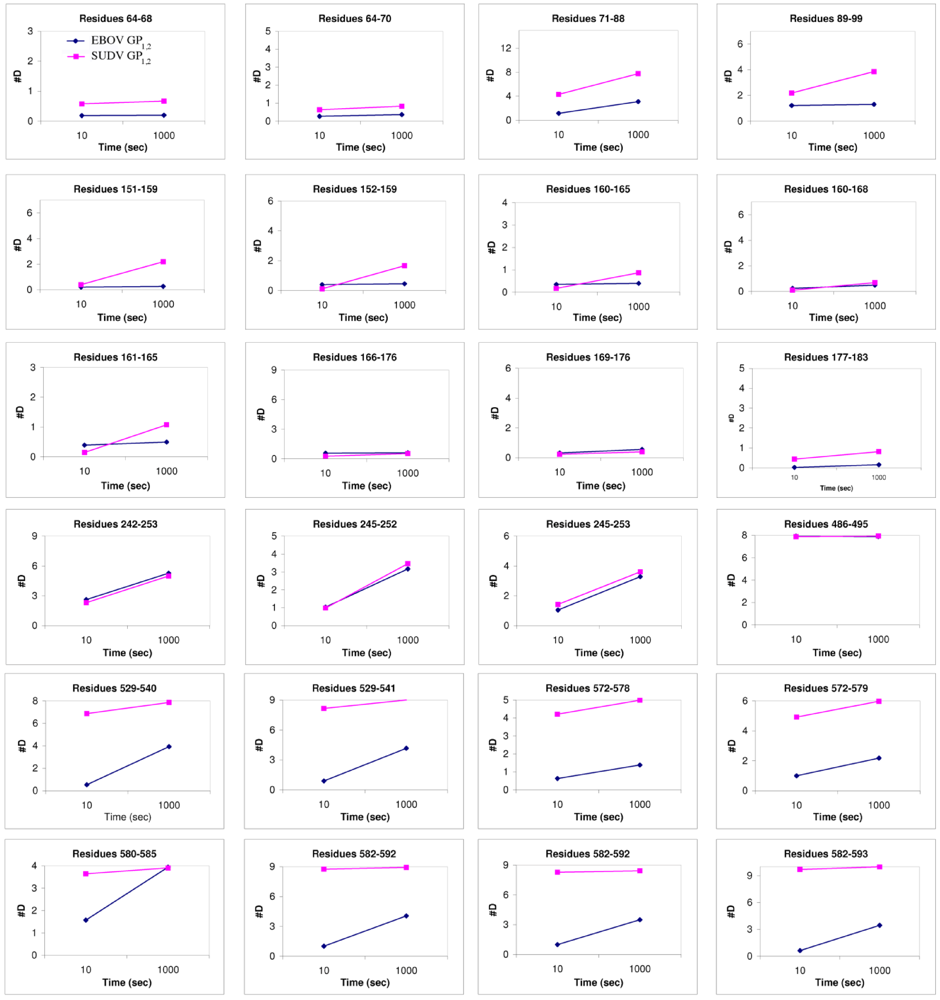
Figure 6.
Plots of number of deuterons vs. time. These compare the deuteration levels of key peptides of EBOV GP1,2 (shown in blue) to their counterparts in SUDV-Gul GP1,2 (shown in magenta), measured over a time scale of 10-1000 sec using deuterium exchange mass spectrometry [17].
2.6 Comparison of Antibody-GP complexes
The crystal structures reveal that the 16F6 and KZ52 antibodies bind at overlapping epitopes that contain residues from both GP1 and GP2. However, the number of interactions made by each antibody with its respective antigen differs (Table 3). 16F6 interacts with SUDV GP1,2 via 7 hydrogen bonds and 19 non-bonded contacts. KZ52 interacts with EBOV GP1,2 via 13 hydrogen bonds and 29 non-bonded contacts. Hence, the binding of KZ52 to EBOV GP1,2 is expected to be stronger than the binding of SUDV GP1,2 to 16F6. Accordingly, Surface Plasmon Resonance studies reveal that the Kd of KZ52 binding to recombinant, soluble EBOV GP1,2 ectodomain is 6.31 ± 3.70 nM while the binding of 16F6 to recombinant, soluble SUDV GP1,2 ectodomain is 1.94 ± 1.40 µM to SUDV-Bon, and 1.59 ± 1.0 µM to SUDV-Gul. Further, the angle the antibodies make when binding against the core of the GP1,2 differs dramatically. KZ52 binds GP1,2 at an angle more parallel to the viral membrane, while 16F6 is angled 50° downward toward the membrane, such that the C-terminal, constant regions of 16F6 are 50 Å closer to the viral membrane than those of KZ52 (Figure 8). The hinge regions attached to the membrane-proximal end of the 16F6 Fab must therefore adopt acute angles so that the other Fab and Fc of the IgG do not penetrate the viral membrane. It is a bit surprising that the 16F6 IgG can reach down to access the epitope from that angle.
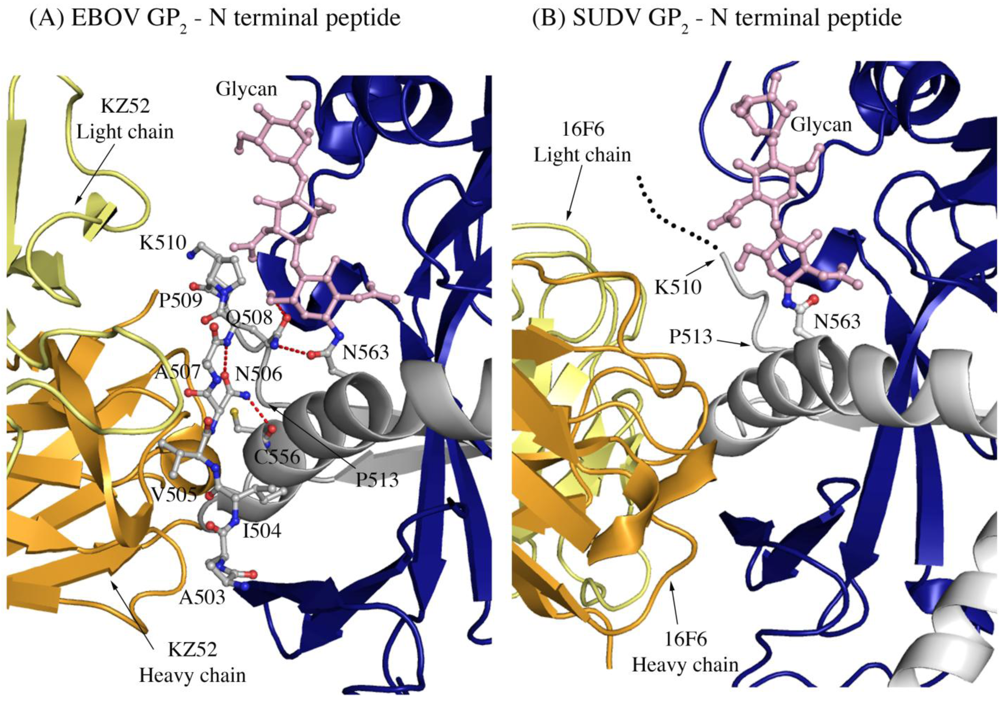
Figure 7.
The N-terminal peptide of GP2 in (A) EBOV-May GP1,2 and (B) SUDV-Bon GP1,2. In both panels, GP2 is shown in ball-and-stick and colored grey. GP1 is colored purple and the heavy and light chains of the antibody are colored bright orange and pale yellow, respectively. Note that the first eight residues of GP2 are disordered in SUDV-Bon/-Gul GP2 and are shown as black dots. Hydrogen bonds in (A) are shown as dashed red lines. There are no hydrogen bonds observed in (B) as the N-terminal peptide is disordered. The glycan residue connected to N563 is shown in ball and stick and colored light pink.
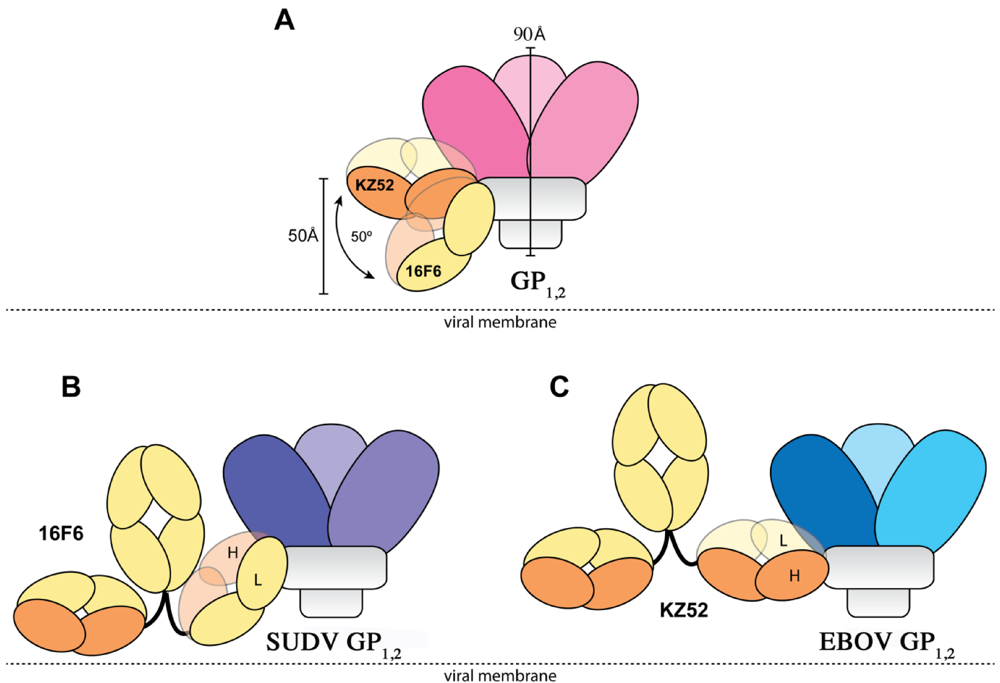
Figure 8.
Cartoon illustration of the different modes of antibody binding. (A) KZ52 binds perpendicular to the central axis of GP1,2 (parallel to the viral membrane), while 16F6 recognizes the overlapping epitope from an acute angle. The central axes of the two Fabs make a 50° angle with each other, and the constant portion of the 16F6 Fab is 50 Å lower than that of KZ52. Fab heavy chains are colored orange, while light chains are colored yellow. Approximate location of the viral membrane is indicated by a dotted line. C terminal portions of GP2 that anchor the visualized trimer to the viral membrane are disordered and not drawn here. (B) Model of a 16F6 IgG binding SUDV GP1,2 (purple) based on the 16F6 Fab-SUDV-Bon GP1,2 complex (PDB: 3VE0). (C) Model of a KZ52 IgG binding EBOV-May GP1,2 (blue) based on the KZ52 Fab-EBOV-May GP1,2 complex structure (PDB: 3CSY). The IgG and GP1,2 fragments are drawn to scale in the models.
As a result of the higher affinity for the GP1,2 ectodomain, greater number of contacts, and presumably easier access to its epitope, one might expect KZ52 to neutralize better than 16F6. Surprisingly, however, 16F6 neutralizes SUDV GP1,2-bearing VSIV better than KZ52 neutralizes EBOV GP1,2-bearing vesicular stomatitis Indiana virus (VSIV) [16] (data replotted here as Figure 9). At all IgG concentrations, a greater portion of virus remains un-neutralized by KZ52 than by 16F6. Even small differences in neutralization capacity may confer significant functional variation in vivo for filoviruses because a single p.f.u. can be a lethal dose for primates.
Our data suggests that binding affinity and access to the epitope alone do not account completely for antibody neutralization efficacy; the composition of the particular epitopes and the susceptibility of the different GP1,2, themselves, to be neutralized are also important. For example, although the epitopes overlap, KZ52 is shifted more towards GP2, burying mostly GP2 while 16F6 buries more of GP1 on SUDV GP1,2 [16]. Affinity for transmembrane-anchored GP1,2 could differ somewhat from affinity for the GP1,2 ectodomain as well, although the epitopes of these antibodies are quite some distance (>50 Å) from the transmembrane anchor.
In addition, the 16F6 and KZ52 Fabs are rotated relative to each other when bound to GP1,2. KZ52 binds with its heavy chain “down” toward the membrane, while 16F6 binds with its heavy chain “up” toward GP1. In addition, the crystal structures reveal that the heavy chain of 16F6 interacts directly with both heptad repeat 1 of GP2 (specifically HR1B) and the β2 strand of GP1. By contrast, KZ52 interacts primarily with the N-terminal peptide of GP2 and the turn region prior to the HR1 helix. KZ52 is unable to make the same contacts as 16F6 because (1) its heavy and light chain are switched in space relative to those of 16F6 and (2) the N-terminal peptide of GP2 is nestled into the center of the paratope in EBOV (Figure 7). The N-terminal peptide of GP2 is probably less important for GP1,2 function than are the HR1 helix and GP1 base strand. Hence, an antibody that anchors a less-important terminal peptide might be expected to be less effective than one that anchors a heptad repeat critical for conformational change to its prefusion GP1β-sheet clamp. Hence, 16F6 binds more weakly, but may bind more effectively. These antibodies are thought to neutralize by blocking conformational changes during fusion. Perhaps better bridging of GP1 and GP2 by 16F6 more effectively anchors the pair in a pre-fusion conformation.
Alternatively, key differences between EBOV and SUDV GP1,2 may render them differently susceptible to neutralization. GP2 of SUDV is less stably ordered than GP2 of EBOV. Perhaps 16F6 has recognized a greater portion of GP1 because its GP2 is a “moving target”. Certainly, the increased mobility of the GP2 N-terminal peptide in SUDV means that more of underlying GP1,2 structure is available for antibody interaction, antibody anchoring of the prefusion conformation and greater neutralization. Another difference is that EBOV GP1,2 has a greater requirement for cathepsin cleavage than does SUDV GP1,2 [18], and crystal structures reveal that the two GP1,2 have different electrostatics at their trimer interface. Electrostatics at the trimer interface may influence how the low pH environment of the endosomes determines susceptibility of GP1,2 to triggering [16].
In summary, we find by structural and functional analysis, that the antibody of higher affinity and presumably easier access to a shared site is not necessarily the most effective at viral neutralization. Other characteristics of the epitope bound, including which amino acids are contained in the individual epitopes and the functional importance of these amino acids, may confer neutralization susceptibility. A conundrum for filoviruses has been that antibody efficacy in vitro and in vivo do not always correlate. Here, we demonstrate that certain in vitro characteristics of antibody binding strength, epitope access, and in vitro neutralization also do not necessarily correlate. In addition to binding tightly, an effective antibody may need to bind correctly.
Development of effective immunotherapeutics against filoviruses will thus require detailed and coupled structural and functional characterization of candidate antibodies. It will also require production and analysis of larger panels of antibodies against the filoviruses than have been currently described so that we may compare several different antibodies within the same competition group, in order to determine what the most effective antibodies are and why this is so. This knowledge base will also clarify what would constitute an ideal antibody response elicited by vaccination, and will aid in development of vaccines designed to elicit these types of antibodies.

Table 1.
Interactions made by key residues in the “chain reversal region” in pre-fusion and post-fusion forms of ebolavirus GP1,2.
| Residue | Prefusion (SUDV-Bon) | Prefusion (EBOV-May) | Postfusion (EBOV-May) |
|---|---|---|---|
| R587 | Hydrogen bond to T60′ | No interactions | Hydrogen bond to N586′ |
| F592 | Stacks against L57, I185, L594′, R596 | Stacks against V48, L57, L593 | Stacks against L594′, W597′, I603′, |
| W597 | Stacks against W597′, W597′′ | Side chain disordered | Stacks against F592′, L593′, L594, I603, R596 |
| G598 | No interactions | No interactions | Hydrogen bond to H602 |
| H602 (R in SUDV) | Side chain disordered | Residue disordered | Hydrogen bond to G598 |
| I610 | Side chain disordered | Residue disordered | No interactions |
Note that ′ and ′′ denote residues from the other two monomers related by the three-fold axis
Table 2.
Interactions between SUDV GP1,2 and Fab 16F6.

GP1 to antibody heavy chain*
| 16F6-H | GP1 | Interaction | ||
| Y31 | OH | O | S32 | H-bond |
| Y31 | OH | OE2 | E47 | H-bond |
| Y32 | OH | OE1 | E44 | H-bond |
| R98 | NH2 | OE1 | E44 | H-bond |
| Y31 | OH | C | S32 | Nonbonded |
| Y31 | CE1 | CG | P34 | Nonbonded |
| R98 | NH2 | CD | E44 | Nonbonded |
| R98 | NH2 | OE2 | E44 | Nonbonded |
| Y32 | OH | N | V45 | Nonbonded |
| A28 | CB | O | V45 | Nonbonded |
| Y31 | OH | CG | E47 | Nonbonded |

GP2 to antibody heavy chain
| 16F6-H | GP2 | Interaction | ||
| N103 | ND2 | O | A553 | H-bond |
| Y101 | OH | OE1 | E564 | H-bond |
| Y101 | OH | OE2 | E564 | H-bond |
| Y101 | O | CA | G557 | Nonbonded |
| Y101 | CE1 | CB | Q560 | Nonbonded |
| Y101 | CE1 | N | L561 | Nonbonded |
| Y101 | OH | CA | L561 | Nonbonded |
| Y101 | OH | CD | E564 | Nonbonded |
| Y101 | OH | N | L561 | Nonbonded |

GP1 to antibody light chain
| 16F6-L | GP1 | Interaction | ||
| T56 | CG2 | ND2 | N40 | Nonbonded |

GP2 to antibody light chain
| 16F6-L | GP2 | Interaction | ||
| W50 | NE1 | C | N552 | Nonbonded |
| W50 | CD2 | CB | N552 | Nonbonded |
| W50 | NE1 | CB | N552 | Nonbonded |
| W50 | CE2 | CB | N552 | Nonbonded |
| W50 | CD1 | N | N553 | Nonbonded |
*Interactions were calculated with the PDBsum server and confirmed by visual inspection with COOT. The cutoff distance for non-bonded interactions listed above is 3.5 Å.

Table 3.
Comparison of the 16F6 and KZ52 epitopes on SUDV and EBOV GP1,2, respectively.Residues at the shared intersection of the two epitopes are colored orange. Residues listed are conserved between SUDV and EBOV, unless specified.
| Residues of SUDV GP1,2 bound by 16F6 | Residues of EBOV GP1,2 bound by KZ52 |
|---|---|
| S32 | |
| P34 | |
| T39 (note: residue 39 is H in EBOV) | |
| N40 | |
| S41 | |
| T42 | V42 |
| L43 | L43 |
| E44 | Q44 |
| V45 | |
| T46 (is S in EBOV) | |
| E47 (is D in EBOV) | |
| Q50 (is K in EBOV) | |
| V52 | |
| A503 (is T in SUDV) | |
| I504 (is N in SUDV) | |
| V505 (is T in SUDV) | |
| N506 (is K in SUDV) | |
| A507 | |
| Q508 (is T in SUDV) | |
| P509 (is G in SUDV) | |
| K510 | |
| C511 | |
| P513 | P513 |
| N514 | |
| H549 | |
| N550 | N550 |
| Q551 | Q551 |
| N552 | D552 |
| A553 | G553 |
| C556 | C556 |
| G557 | |
| Q560 | |
| L561 | |
| E564 |

Table 4.
Data collection and refinement statistics of SUDV-Bon GP1,2 bound to 16F6.
| Beamline | ALS 8.3.1 |
| Space group | I23 |
| Cell parameters (Å) | a=b=c=194.86 |
| Wavelength (Å) | 1.1159 |
| Resolution (Å) (last shell) | 50.00-3.35 (3.47-3.35)a |
| Total observations | 149425 |
| Unique reflections | 17818 |
| I/σ | 13.4 (2.8) |
| Rsym (%)b | 7.9 (77.7) |
| Completeness (%) | 99.9 (100.0) |
| Redundancy | 8.4 |
| Matthews coefficient (VM, A3Da-1) | 3.5, 65% solvent content |
| Rwork (%)c | 21.98 |
| Rfree (%) | 28.44 |
| Number of protein residues/atoms | 782/6003 |
| Number of glycan residues/atoms | 9/111 |
| r.m.s.d. bond length (Å) | 0.012 |
| r.m.s.d. bond angles (°) | 1.561 |
| Average B values (Å2) | |
| Fab 16F6 (chains heavy/light) | 116/124 |
| GP1 | 175 |
| GP2 | 148 |
| Overall | 141 |
| Ramachandran plot (%) | |
| Most favored | 78.8 |
| Allowed | 18.4 |
| Generously allowed | 2.2 |
| Disallowed | 0.6 |
aValues in parenthesis are for the highest resolution shell.bRsym = ΣΣi| Ii - <I> | /Σ<I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h,k,l.cR factor = Σhkl||Fobs|-k|Fcal|/Σhkl|Fobs|, where Fobs and Fcal are observed and calculated structure factors respectively.
Figure 9.
The ability of 16F6 and KZ52 IgGs to neutralize SUDV GP1,2-bearing VSIV or EBOV GP1,2-bearing VSIV, plotted in a log (A) and linear (B) scales. VSIV particles were pre-incubated with the indicated concentrations of antibody for one hour at room temperature, and then exposed to Vero cells at 37° C. Viral infectivity was scored at 16 hours post-infection. The starting titers were 9×107 IU/ml for EBOV and 3×107 IU/ml for SUDV on Vero cells [16].
3. Experimental Section
3.1. Molecular Biology
Sudan virus Boniface (SUDV-Bon) glycoprotein (GP1,2) was cloned into the pDISPLAY vector (Invitrogen) for expression in mammalian cells with an Ig K-chain leader sequence in place of its natural signal sequence, and a Factor Xa-cleavable N-terminal HA tag for purification. The construct includes residues 33-313 and 473-637, with deletion of the mucin-like domain (residues 314–472) and C-terminal transmembrane domain (residues 638–676). The GP1,2 gene was amplified by PCR using oligonucleotides containing BglII restriction sites at their 5′ ends and SalI restriction sites at the 3′ end. The PCR fragment was then cloned into pDISPLAY using BglII and SalI DNA restriction and T4 ligase DNA ligation to produce the IgGK secretion leader-HA-FactorXa-ARS-GPdelta mucin deltaTM plasmid. GP1,2 is composed of more than 50% oligosaccharides by weight, primarily because of a heavily glycosylated mucin-like domain that is non-essential for cellular attachment or entry [24].
3.2. Protein Expression
Recombinant SUDV GP1,2 was transiently expressed in HEK293T cells by calcium phosphate precipitation in ten-layer CellStacks (Corning). The DNA–calcium phosphate mixture was added to 70% confluent cells grown in DMEM plus 1x penicillin-streptomycin and 5% (v/v) fetal bovine serum. The supernatant was harvested four days post-transfection, concentrated using a Centramate tangential flow system and purified by anti-HA immunoaffinity chromatography. GP1,2 was natively deglycosylated using Peptide:N-glycosidase F (PNGaseF) in PBS, overnight at room temperature. The extent of the digestion was followed by SDS-PAGE.
3.3. Structural Determination of SUDV GP–Fab 16F6 Complex.
For crystallization, deglycosylated SUDV GP1,2 was mixed with excess Fab 16F6 and incubated for 1 hour at 4° C and subequently purified on a Superdex-200GL 10/300 column equilibrated with 10 mM Tris-HCl, pH 7.5 and 150 mM NaCl. Fractions containing trimeric SUDV GP1,2-Fab16F6 complex were concentrated to 10 mg/ml and initial crystallization conditions were screened using a Topaz Fluidigm system at 20° C. Initial crystal hits were obtained in the presence of 20% PEG 3350 and 0.2 M lithium citrate and were translated and optimized by hanging-drop vapor diffusion. Initial crystals grew over a period of one month at room temperature up to a maximum size of 0.2 × 0.2 × 0.2 mm3 and have a cubic morphology. Initial crystals diffracted up to 4.5 Å at the Beamline 8.2.2 of the Advanced Light Source (ALS, Berkeley). Improved crystals were obtained by adding a mixture of 3 μL of the SUDV GP1,2-Fab16F6 complex at 10 mg/mL in 10 mM Tris HCl pH 7.5 and 0.5 μL of 20% benzamidine hydrochloride to 3 μL of mother liquor crystallization solution 15% (w/v) PEG 3350, and 0.2 M lithium citrate. These crystals grew in 3-4 weeks at room temperature to a maximum size of 0.2 × 0.3 × 0.3 mm3 and have a star-shaped morphology. The crystals were cryoprotected with 17.5% (v/v) glycerol plus mother liquor before flash cooling in liquid nitrogen. The crystals diffracted beyond 3.2 Å resolution, but due to radiation damage, a complete dataset was collected with good statistics up to 3.35 Å, at ALS beamline 8.3.1. The data were indexed, integrated and scaled using HKL2000 [25]. The SUDV GP1,2–Fab16F6 crystals belong to space group I23, with unit cell dimensions of a=b=c=194 Å. The solvent content is approximately 65% [26] with one monomer of the GP1,2Fab complex in the asymmetric unit. Data collection statistics are shown in Table 4.
The structure of SUDV GP1,2 was determined by molecular replacement using an edited poly-alanine model based on the sequence alignment and on the conservation of structure elements between SUDV GP1,2 and the previously determined EBOV-May GP1,2 structure [12]. The edited poly-alanine search model comprised residues 62-113, 129-186, 215-260 from GP1 and 503-595 from GP2. An initial molecular replacement solution was found using the first 4.8 Å data set using PHASER [26]. A partial solution for the glycoprotein model was obtained and revealed unambiguous extra electron density for key parts that were absent from the search model. This partial solution was fixed and the Fab position was determined using poly-alanine models of separate variable and constant domains of a different Fab (PDB 1JLP) [27], with CDRs deleted. Interpretable electron density maps with clear secondary structural elements and solvent boundaries were obtained, with the biologically relevant trimer generated by the crystallographic three-fold axis that extends through the diagonal vertices of the cubic spacegroup I23. This initial solution was assembled into a complete search model and used with the subsequent 3.35 Å data set in PHASER (LL-gain=604, TFZ=33.8, RFZ=10.4). After an initial step of rigid body refinement, the structure was refined with CNS [28] using torsion-angle simulated-annealing refinement with a maximum-likelihood amplitude target. The initial model was iteratively improved by visual inspection and model building using COOT [29] and refinement in CNS [28]. In the final rounds of refinement, riding hydrogens, grouped atomic displacement and TLS parameters were implemented using PHENIX [30]. The structure was refined to final Rwork and Rfree values of 21.98% and 28.44% and contains SUDV-Bon GP1,2 residues 32–191, 213–285, 300–311 and 510–614, and Fab 16F6 residues 1–220 (heavy chain) and 1–212 (light chain). Electron density was missing for GP1,2 residues 192–212, 286-299, 312–313, 473–509 and 615–637. Weak or discontinuous electron density is observed in the outer regions of the glycan cap (residues 261–285 and 300–307) and these regions were tentatively assigned as poly-alanine fragments. Despite the treatment of the GP1,2 with PNGaseF, two glycan chains resistant to deglycosylation at N257 on GP1 and N563 on GP2 were preserved. These glycans show a high B-factor (Baverage= 242.5 Å2), but there is unambiguous electron density for the chitobiose core and up to the first mannose residues of these glycans. Final refinement statistics and analysis are shown in Table 4. Atomic coordinates and structure factors are deposited in the RCSB Protein Data Bank under accession number 3VE0. The biologically relevant trimer was generated by the crystallographic three-fold axis that extends through the diagonal vertices of the cubic spacegroup I23.
3.4. Antibody binding studies
The binding affinity of KZ52 to EBOV-May GP1,2 and 16F6 to SUDV-Gul and -Bon GP1,2 was measured using BIAcore-2000 Surface Plasmon Resonance spectrophotomer. All the samples were buffer exchanged to HBS-EP buffer (10 mM Hepes, pH 7.4, 150 mM Sodium Chloride, 3 mM EDTA, and 0.005% Polysorbate 20) prior to complexation. KZ52 and 16F6 were immobilized at 25 °C on various channels of carboxymethylated dextran CM5 sensor chips to ~200 response units using standard amine coupling protocol. EBOV GP1,2, SUDV-Bon GP1,2 and SUDV-Gul GP1,2 were injected over channels coated with KZ52 and 16F6 respectively. The glycoproteins were injected sequentially at increasing concentrations of 1, 2.5, 5, 10 and 20 µM respectively at a flow rate of 10 µL/min for 250 sec and dissociation of the complexes were measured over a period of ~600 sec. The chromatogram data were analyzed using a 1:1 Langmuir binding model to determine the ka (on), kd (off) and Kd (dissociation) values using the BIAevaluation software.
3.5. Antibody Neutralization Studies
VSIV pseudotypes bearing EBOV or SUDV GP and expressing eGFP were generated and concentrated by ultracentrifugation as described previously [31,32]. Virus-antibody incubations were carried out as follows: Concentrated virus (1-3 µL, ~1×105 IU) was mixed with PBS containing antibody at the indicated concentration (0-40 µg/mL final; total reaction volume, 20 µL). Reactions were incubated at room temperature for 1 hour, and viral infectivity was determined by titration on Vero cell monolayers. Briefly, Vero cells were exposed to samples for 1 hour at 37° C and infectivity was scored at 16 hours post-infection by manually enumerating GFP-positive cells, as described previously [32].
3.6. Deuterium Exchange Mass Spectrometry Experiments
A detailed description of DXMS methodology was described previously [17]. Briefly, quench conditions that produced an optimal pepsin fragmentation pattern were established for both EBOV and SUDV GP1,2. Then, the non-deuterated samples, functional deuterated samples and the equilibrium-deuterated back-exchange control samples were prepared by mixing the GP1,2 proteins with non-deuterated (8.3 mM Tris, 150 mM NaCl, in H2O, pH 7.15), deuterated (8.3 mM Tris, 150 mM NaCl, in D2O, pDREAD 7.2) and equilibrium-deuterated (1% formic acid in 99.9% D2O) buffers, separately. Later, optimized quench buffer was added to stop the reactions. Finally, the samples were passed over pepsin column, and the resulting peptides were collected on a C18 trap and separated using a C18 reversed phase column. Data were acquired in both data-dependent MS/MS mode and MS1 profile mode by a LCQ mass spectrometer, and the data analyzed by SEQUEST (Thermo Finnigan Inc.) and DXMS explorer (Sierra Analytics Inc., Modesto, CA).
4. Conclusions
We report the crystal structure of SUDV-Bon GP1,2 bound to 16F6 and describe new structural details of the complex including thermal motion of GP2, approach angle and access to the antibody epitope and molecular interactions between antibody and GP1,2. Surface Plasmon Resonance studies, also presented here, reveal that 16F6 binds SUDV-Gul/Bon GP1,2 with a lower affinity than KZ52 binding to EBOV-May GP1,2. Paradoxically, however, 16F6 confers better neutralization than KZ52. Together, these studies suggest that the key to effective neutralization might lie in binding the epitope correctly rather than in affinity alone.
Acknowledgments
We thank Dennis Burton’s lab at TSRI for use of the BIAcore spectrophotometer and assistance with experiments. We thank Dafna Abelson and Michelle Zandonatti for technical assistance, and Christina Corbaci for preparation of Figure 8. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U. S. Army. The work is supported by NIH grants AI081982, AI072106, AI068730, AI2008031, GM020501, GM066170, NS070899, GM093325, and RR029388 (V.L.W.); AI088027, AI082437 (K.C.); AI067927, AI081982, AI070530, The Skaggs Institute for Chemical Biology, and an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (E. O. S.). J. M. D. kindly thanks DTRA project CBM.THRV.01.RD.11.001 for their generous support. This is manuscript #21503 from The Scripps Research Institute.
Conflict of Interest
The authors declare no conflicts of interest.
References and Notes
- Sanchez, A.; Khan, A.S.; Zaki, S.R.; Nabel, G.J.; Ksiazek, T.G.; Peters, C.J. Filoviridae: Marburg and Ebola viruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams, and Wilkins: Philadelphia, 2001; pp. 1279–1304. [Google Scholar]
- Towner, J.S.; Sealy, T.K.; Khristova, M.L.; Albarino, C.G.; Conlan, S.; Reeder, S.A.; Quan, P.L.; Lipkin, W.I.; Downing, R.; Tappero, J.W.; Okware, S.; Lutwama, J.; Bakamutumaho, B.; Kayiwa, J.; Comer, J.A.; Rollin, P.E.; Ksiazek, T.G.; Nichol, S.T. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008, 4, e1000212. [Google Scholar]
- World Health Organization, International Commission to Sudan, Ebola Hemorrhagic Fever in Sudan, 1976. In Bulletin of the World Health Organization; 1978; 56, pp. 247–270.
- Sanchez, A.; Rollin, P.E. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005, 113, 16–25. [Google Scholar]
- Maruyama, T.; Parren, P.W.; Sanchez, A.; Rensink, I.; Rodriguez, L.L.; Khan, A.S.; Peters, C.J.; Burton, D.R. Recombinant human monoclonal antibodies to Ebola virus. J. Infect. Dis. 1999, 179 Suppl 1, S235–S239. [Google Scholar]
- Gupta, M.; Mahanty, S.; Bray, M.; Ahmed, R.; Rollin, P.E. Passive transfer of antibodies protects immunocompetent and imunodeficient mice against lethal Ebola virus infection without complete inhibition of viral replication. J. Virol. 2001, 75, 4649–4654. [Google Scholar]
- Wilson, J.A.; Hevey, M.; Bakken, R.; Guest, S.; Bray, M.; Schmaljohn, A.L.; Hart, M.K. Epitopes involved in antibody-mediated protection from Ebola virus. Science 2000, 287, 1664–1666. [Google Scholar]
- Parren, P.W.H.I.; Geisbert, T.W.; Maruyama, T.; Jahrling, P.B.; Burton, D.R. Pre- and post-exposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J. Virol. 2002, 76, 6408–6412. [Google Scholar]
- Oswald, W.B.; Geisbert, T.W.; Davis, K.J.; Geisbert, J.B.; Sullivan, N.J.; Jahrling, P.B.; Parren, P.W.; Burton, D.R. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007, 3, 62–66. [Google Scholar]
- Jahrling, P.B.; Geisbert, J.B.; Swearengen, J.R.; Larsen, T.; Geisbert, T.W. Ebola Hemorrhagic Fever: Evaluation of Passive Immunotherapy in Nonhuman Primates. J. Infect. Dis. 2007, 196, 400–403. [Google Scholar]
- Dye, J.M.; Herbert, A.S.; Kuehne, A.I.; Barth, J.F.; Muhammad, M.A.; Zak, S.E.; Ortiz, R.A.; Prugar, L.I.; Pratt, W.D. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc. Natl. Acad. Sci. U S A 2012. [Google Scholar]
- Lee, J.E.; Fusco, M.H.; Hessell, A.J.; Oswald, W.B.; Burton, D.R.; Saphire, E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008, 454, 177–183. [Google Scholar]
- Weissenhorn, W.; Calder, L.J.; Wharton, S.A.; Skehel, J.J.; Wiley, D.C. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 1998, 95, 6032–6036. [Google Scholar]
- Weissenhorn, W.; Carfi, A.; Lee, K.H.; Skehel, J.J.; Wiley, D.C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 1998, 2, 605–616. [Google Scholar]
- Malashkevich, V.N.; Schneider, B.J.; McNally, M.L.; Milhollen, M.A.; Pang, J.X.; Kim, P.S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl. Acad. Sci. USA 1999, 96, 2662–2667. [Google Scholar]
- Dias, J.M.; Kuehne, A.I.; Abelson, D.M.; Bale, S.; Wong, A.C.; Halfmann, P.; Muhammad, M.A.; Fusco, M.L.; Zak, S.E.; Kang, E.; Kawaoka, Y.; Chandran, K.; Dye, J.M.; Saphire, E.O. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 2011, 18, 1424–1427. [Google Scholar]
- Bale, S.; Liu, T.; Li, S.; Wang, Y.; Abelson, D.; Fusco, M.; Woods, V.L., Jr.; Ollmann Saphire, E. , Ebola Virus Glycoprotein Needs an Additional Trigger, beyond Proteolytic Priming for Membrane Fusion. PLoS Negl. Trop. Dis. 2011, 5, e1395. [Google Scholar]
- Misasi, J.; Chandran, K.; Yang, J.Y.; Considine, B.; Filone, C.M.; Cote, M.; Sullivan, N.; Fabozzi, G.; Hensley, L.; Cunningham, J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J. Virol. 2012, 86, 3284–3292. [Google Scholar]
- Gnirss, K.; Kuhl, A.; Karsten, C.; Glowacka, I.; Bertram, S.; Kaup, F.; Hofmann, H.; Pohlmann, S. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology 2012, 424, 3–10. [Google Scholar]
- Delos, S.E.; La, B.; Gilmartin, A.; White, J.M. Studies of the "chain reversal regions" of the avian sarcoma/leukosis virus (ASLV) and ebolavirus fusion proteins: analogous residues are important, and a His residue unique to EnvA affects the pH dependence of ASLV entry. J. Virol. 2010, 84, 5687–5694. [Google Scholar]
- Dube, D.; Brecher, M.B.; Delos, S.E.; Rose, S.C.; Park, E.W.; Schornberg, K.L.; Kuhn, J.H.; White, J.M. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 2009, 83, 2883–2891. [Google Scholar]
- Schornberg, K.; Matsuyama, S.; Kabsch, K.; Delos, S.; Bouton, A.; White, J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006, 80, 4174–4178. [Google Scholar]
- Brecher, M.B.; Schornberg, K.L.; Delos, S.E.; Fusco, M.L.; Saphire, E.O.; White, J.M. Cathepsin Cleavage Potentiates the Ebola Virus Glycoprotein to Undergo a Fusion Relevant Conformational Change. J. Virol. 2011, in press.. [Google Scholar]
- Yang, Z.Y.; Duckers, H.J.; Sullivan, N.J.; Sanchez, A.; Nabel, E.G.; Nabel, G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000, 6, 886–889. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Meth. Enzymol. 1997, 276A, 307–326. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Storoni, L.C.; Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 458–464. [Google Scholar]
- Yokota, A.; Tsumoto, K.; Shiroishi, M.; Kondo, H.; Kumagai, I. The role of hydrogen bonding via interfacial water molecules in antigen-antibody complexation. The HyHEL-10-HEL interaction. J. Biol. Chem. 2003, 278, 5410–5418. [Google Scholar]
- Brünger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; Read, R.J.; Rice, L.M.; Simonson, T.; Warren, G.L. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998, D54, 905–921. [Google Scholar]
- Emsley, P.; Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004, D60, 2126–2132. [Google Scholar]
- Adams, P.D.; Grosse-Kunstleve, R.W.; Hung, L.W.; Ioerger, T.R.; McCoy, A.J.; Moriarty, N.W.; Read, R.J.; Sacchettini, J.C.; Sauter, N.K.; Terwilliger, T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002, D58, 1948–1954. [Google Scholar]
- Takada, A.; Robison, C.; Goto, H.; Sanchez, A.; Murti, K.G.; Whitt, M.A.; Kawaoka, Y. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 1997, 94, 14764–14769. [Google Scholar]
- Wong, A.C.; Sandesara, R.G.; Mulherkar, N.; Whelan, S.P.; Chandran, K. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 2010, 84, 163–175. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).