Abstract
Bacteriophages have been a model system to study assembly processes for over half a century. Formation of infectious phage particles involves specific protein-protein and protein-nucleic acid interactions, as well as large conformational changes of assembly precursors. The sequence and molecular mechanisms of phage assembly have been elucidated by a variety of methods. Differences and similarities of assembly processes in several different groups of bacteriophages are discussed in this review. The general principles of phage assembly are applicable to many macromolecular complexes.
1. Introduction
How proteins and nucleic acids assemble, often spontaneously, into structurally well-defined three-dimensional objects is an intriguing question. The limited size of the phage genome and the multicomponent composition of bacteriophages make them well suited for assembly investigations. Genetic manipulation of phages has made it easy to observe the effects of gene inactivation on protein-protein association, providing information on the sequence of assembly processes (Figure 1; [1]). Over the past fifty years, mutational, biochemical and biophysical analyses, X-ray crystallography, NMR, cryo-electron microscopy (cryo-EM), thin sectioning and single molecule methods have been used to study bacteriophages [2,3,4,5,6,7,8,9,10,11,12,13,14,15]. This review will describe what has been achieved and will contemplate what still needs to be accomplished, focusing mostly on dsDNA tailed phages (Table 1).
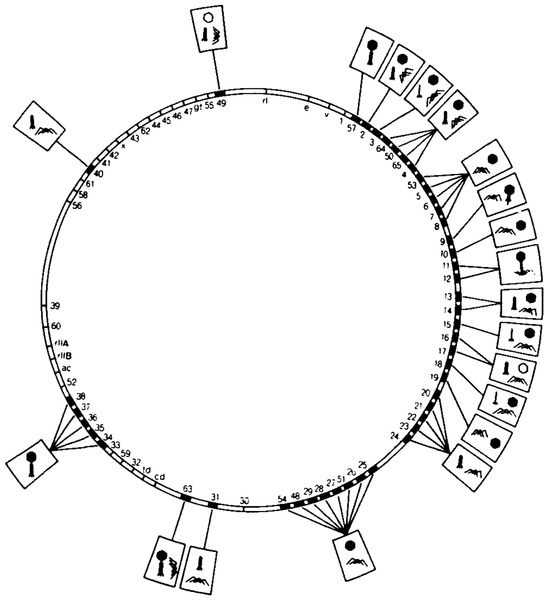
Figure 1.
A simplified bacteriophage T4 genome map showing the effect of mutations in morphological genes on virion assembly. Boxed symbols attached to a particular gene show the phenotype, arising from the defects in that gene. Reprinted from Wood et al. [1]; with permission. Copyright for this figure is owned by the Federation of American Societies for Experimental Biology. This image may not be reproduced in any way without explicit permission from the copyright owner.

Table 1.
Phages discussed in the current review.
There are substantial similarities in the assembly processes of all types of viruses, and of various cellular complexes. For example, most bacteriophages as well as herpesviruses, adenoviruses, poxviruses and the giant mimivirus make an empty protein shell that is subsequently packaged with the viral genome [4,5,16,17,18,19,20,21]. The majority of viruses undergo proteolytic cleavages during assembly that are often essential to trigger the next assembly step [22,23]. Additionally, the capsid protein fold common to dsDNA tailed phages also occurs in herpesviruses [24], as well as in bacterial molecular compartments [25]. Moreover, the bacterial injectosome, the hook of flagella and phage tails all use molecular ruler proteins in order to assemble a complex of correct length [26]. Nevertheless, even within the same group of phages, there are notable differences in the assembly pathway, size and symmetry of the capsid and tail, positions of individual genes in the genome, the number of structural proteins and whether or not they are cleaved during assembly [27]. In this review we discuss only those groups of phages for which a significant amount of knowledge on assembly has been accumulated. Thus, for instance, we make no mention of Corticoviridae and Plasmaviridae families of phages.
2. dsDNA Tailed Phages
All tailed bacterial viruses are distinguished by a special organelle, called a tail, which contains cell recognition proteins and triggers DNA release from the head. Tailed phages form the Caudovirales family, which is subdivided into phages with short tails (Podoviridae), phages with long non-contractile tails (Siphoviridae) and phage with contractile tails (Myoviridae). Viruses with similar morphology are found in archaea, suggesting that tailed phages appeared before the separation of the domains of life [28,29]. The size of the tail ranges from a small protein complex of about 40 proteins in Podoviridae phages to about 300 proteins in Myoviridae tails. The length of the tail varies from about 300 Å in P22 to about 4550 Å for bacteriophage G [30,31,32]. Although tails are not commonly observed in eukaryotic viruses, algal viruses have been reported to have similar structures [33] and a tail-like unique vertex was demonstrated for PBCV-1 [34].
2.1. Head Organization
Many bacterial and eukaryotic viral capsids have similar organization. Multiple copies of viral capsid proteins are frequently assembled into icosahedral shells, as was predicted by Crick and Watson [35] and later confirmed by Caspar [36]. A minimum of 60 copies of the capsid protein is necessary to make an icosahedral particle, in which case all the subunits have identical environments. A bigger capsid can be made by placing several monomers into the icosahedral asymmetric unit with “quasi-equivalent” environments, making the total number of capsid proteins a multiple of 60. For example, if there are T (the triangulation number) quasi-equivalent positions, there are T × 60 capsid proteins in a capsid [37]. A T = 1 particle consists of twelve pentameric capsomers, whereas a capsid with a higher T number contains both pentameric and hexameric capsomers. Not all T values are permissible (although some disallowed T numbers can occur in practice [38]) as some T numbers cannot be arranged with quasi-symmetric environments. However, for each virus there is usually a dominant capsid organization. This implies that virus assembly must ensure that the capsid proteins form a unique structure with a specific T number. A different way of increasing capsid size is assembling an elongated or prolate head, by inserting an additional cylindrical section between the two icosahedral end-caps. Such a structure is described by two triangulation numbers: a T number is used for the terminal caps and a Q number for the cylindrical section. Prolate heads occur in several genera of plant, fungal and animal viruses [39], but are more common among bacteriophages, where they are observed in about 15% of studied viruses [28]. As was pointed out by Moody [40], elongation of the head in one direction does not affect receptor binding sites in bacteriophages, because only the vertex where the tail is attached to the head is functional for cell binding. In a prolate head there are two types of five-fold vertices, the ten surrounding the cylindrical section and the two five-folds at the center of the caps. These differences would affect receptor attachment and uncoating in viruses that otherwise use any of the five-fold vertices during infection (e.g., picornaviruses) perhaps explaining the rare occurrence of prolate heads among eukaryotic viruses [40].
Most dsDNA tailed phages assemble their capsid from multiple copies of one capsid protein. However, bacteriophage T4 encodes two capsid proteins, one forming the pentameric capsomers, consisting of gp24, and the other hexameric capsomers, consisting of gp23. Although both are present in the wild-type T4 phage, a mutant gp23 can form both hexameric and pentameric capsomers, producing a capsid that contains only one type of capsid protein [41,42,43]. These two proteins share 21% sequence similarity and probably occurred as the result of gene duplication [42,44]. Many eukaryotic viruses, like herpesvirus, adenovirus, picornaviruses, PBCV-1 and others, code for more than one capsid protein [45]. Although the presence of several different proteins requires a larger viral genome, it might provide some evolutionary advantages. For example, the problem of accommodating quasi-equivalent environments in the vicinity of pentameric vertices is mitigated. Additionally, mutations would be easier to accommodate, as they would affect only certain positions in the capsid [23].
2.2. Formation of the Prohead
An empty shell, called the prohead or procapsid, is usually a required stable assembly intermediate which is then packaged with DNA. The difference in the structure of proheads and mature heads is discussed in Section 2.4. Prohead assembly is not unique to bacteriophages, but also occurs in many other viruses such as adenoviruses, herpesviruses, poxviruses and the giant mimivirus [18,19,20,21]. The similarities between tailed phages and herpesviruses are even more striking as they were shown to share the same capsid protein fold [24], termed a HK97-like fold, which was first found in bacteriophage HK97 [46]. However, some phages have domains in the capsid protein structure that are not part of the HK97 fold. Such domains are structurally distinct in different phages, represented by BIG2-like domain in phi29 [47], a chitin-binding-like domain in T4 [44] and an insertion domain in P22 [48,49,50]. In T4 and phi29 these insertion domains stabilize the capsid, by bridging the neighboring molecules within one capsomer (T4) or between neighboring capsomers (phi29). Although the capsid protein of HK97 does not have an insertion domain, the capsid is stabilized through a network of covalent cross-links [46]. Bacteriophage T7 does not have an extra capsid domain, but 10% of its capsid proteins have C-terminal extensions of 52 residues, which arise from a frameshift during translation [51]. Although complementation showed that T7 capsid without the larger protein is as stable as the wild-type virus, this frameshift is conserved in the related phage T3 [51].
A capsid protein by itself is not capable of ensuring the correct geometry of the capsid shell, requiring an additional scaffolding protein for prohead assembly [8,52,53]. Most phages have a separate scaffolding protein gene, with the exception of HK97 and T5 that have scaffolding domains, or delta-domains, within capsid protein sequences [54,55,56]. In assembled procapsids, scaffolding proteins form a core inside the prohead, which does not have the icosahedral symmetry of the outer capsid shell. Many phage scaffolding proteins, including those of P22, phi29 and SPP1, form dimers and tetramers [57,58,59], increasing the local concentration of capsid proteins and therefore acting as an entropic sink and promoting association of coat proteins [8,53]. Without scaffolding most capsid proteins form aberrant structures. In the case of P22, the assembly of aberrant particles proceeds ten-times slower than head assembly in the presence of the scaffolding protein [60]. A mutation in the scaffolding protein of phi29 leads to the formation of isometric particles instead of prolate [61]. In the absence of scaffolding proteins, capsid proteins of T4 and lambda can assemble into long cylindrical structures [62,63,64]. Similarly, bacteriophage T5 capsid protein can form open tubular assemblies under certain conditions, even though the scaffolding domain is a part of the capsid protein sequence [56]. Assembly of bacteriophage T7 into polycapsids occurs in the presence of scaffolding protein, but only when the ratio of scaffolding protein to the capsid protein is 0.11:1 [65,66]. The resulting polycapsids consist almost entirely of head protein [66].
Both the phi29 scaffolding protein and the C-terminal portion of the P22 scaffolding protein have a helix-loop-helix structure [58,67]. Moreover, the sequence alignment suggests that the N-terminal delta domains of HK97 and T5 would have a similar motif [58]. The binding sites of P22 scaffolding protein on the capsid were determined by electron microscopy, but the arrangement of the scaffolding protein inside the capsid could not be visualized due to icosahedral averaging [57,68,69]. The reconstruction of the phi29 prohead assuming no symmetry showed a cage-like scaffolding protein density inside the capsid, organized in several shells with different symmetry [58]. In contrast to the majority of phages that code for one scaffolding protein, the bacteriophage T4 capsid assembly requires six different scaffolding, or core, proteins [70,71]. Co-expression of these proteins results in formation of tubular polycores which are unique to T4 [72]. Although it was previously shown that the core of T4 has six-fold symmetry, a later re-examination indicated that polycores could have six-, eight- or ten-fold symmetry [73]. How exactly scaffolding proteins influence the shape of the head is unknown.
The third essential component of head assembly is a portal protein, also called a connector, which is situated on one pentameric vertex of the capsid. As these names suggest, portal or connector is required for DNA entry and release, as well as for attachment of the neck proteins to the head. Additionally, the phage head assembly is probably initiated from the portal vertex by copolymerization of the scaffolding and capsid proteins [3,15,74]. In bacteriophage T4, the ellipsoidal cores are formed in the absence of the capsid protein [75], whereas in many other phages the scaffolding proteins cannot assemble without the presence of capsid proteins. Assembly initiation in bacteriophage T4 additionally requires a membrane scaffolding protein, which interacts with the connector protein [76]. The membrane association of proheads during the assembly was not demonstrated for other phages.
The recombinantly expressed portal proteins were shown to assemble into different oligomers, but in all structurally characterized phages the portal protein forms a dodecameric ring [10]. Therefore, to ensure proper prohead assembly the portal has to co-assemble with the capsid and scaffolding proteins [77,78]. The outer diameter of portal protein rings ranges from 140 to 170 Å and the protein molecular weight ranges from 36 kDa (phi29) to 83 kDa (P22) [74]. Electron microscopy and X-ray crystallography have shown that the basic morphology of the portal ring is similar in all known tailed phages [79,80,81,82]. Some phages, like P22, HK97 and T7, can form prohead-like structures in the absence of portal proteins. However, these virus-like particles cannot package DNA and represent a dead end assembly. It was noted earlier that aberrant particles often form slower than the viral precursors, indicating that connector and scaffolding affect not only the accuracy but the kinetics of the assembly process [60,83,84].
2.3. dsDNA Packaging
Once the head is assembled, the packaging complex binds and utilizes ATP hydrolysis to translocate the genome into the head [5,13,85,86]. Because of a symmetry mismatch between the dodecameric portal ring and the five-fold capsid vertex, it was suggested that the portal ring is rotating during packaging [87]. Later single-molecule spectroscopy of the phi29 connector [88] and studies on T4 connector in which its motion was restricted by binding to the capsid [89] showed that connector rotation does not accompany packaging.
The initiation of DNA packaging depends on how viral DNA was replicated. Many phages produce head-to-tail multimers, or concatemers of DNA, and these are used as a packaging substrate in T4, lambda, and P22. In that case, the ATPase packages the length of the genome (or more in some phages) and cuts the concatemer. Hence, the term “terminase” is used for the packaging ATPase. This enzyme is also called a large terminase to distinguish it from the small terminase, which does not cut the DNA. The small terminase modulates the activity of the large terminase and is involved in packaging initiation [5,90]. The crystal structure of a small terminase from a Podoviridae phage Sf61989 was solved by X-ray crystallography, suggesting it could form a ring below the large terminase [91]. Bacteriophage phi29 does not have a small terminase, but it utilizes a unique viral encoded structural RNA, called p-RNA [92,93]. Some phages, like Mu, N4 and phi29, do not generate a concatemeric DNA [5]. Phages that utilize concatemeric DNA package more than one length of the genome, or a headful. The determination of when the head is full is not well understood. There are known mutations in T4, P22 and SPP1 that demonstrate the interplay between the terminase and portal protein ring [74,94,95,96,97]. Therefore, conformational changes in the connector are thought to provide the signal for the terminase to cut the DNA and dissociate from the head [5,74,82]. Single molecule studies of the phi29 packaging motor [98,99] and the crystal structure of T4 terminase [100], as well as the structure of the dsRNA packaging enzyme from phage phi12 [101], have formed the basis for several packaging mechanisms [13,102,103,104].
The packaged genome inside the phage head is wound into a spool-like structure such that several layers of dsDNA are visible in electron micrographs of individual virions as well as in cryo-EM image reconstructions of phage heads [10,105]. There are several proposed models for the formation of the spool structure [5,12,106,107]. The DNA end that is packaged first is likely associated with the inside surface of the capsid, whereas the end that is packaged last is probably the first to be ejected [5]. Strong binding of DNA to the connector was shown for phage SPP1 [97] and both ends of DNA were localized within or near the portal in bacteriophage T4 [108].
2.4. Head Maturation
During or before packaging the scaffolding proteins either exit from the capsid (P22, phi29) or are proteolytically cleaved by phage encoded protease (HK97, T4). The recycled P22 scaffolding protein can be reused four more times in the assembly process [109]. Lambda scaffolding protein can also exit from the capsid without being cleaved, as was shown for the phage with a genetically inactivated protease [110]. The release of scaffolding proteins could be induced by their interaction with DNA, possibly through a leucine-zipper motif, identified in the scaffolding protein of phi29 [58]. The exit of the scaffolding proteins and the packaging of DNA initiate maturation, involving a large structural transition of the prohead and resulting in a bigger, angular and more stable head with a thinner shell [3,6,8,9,14,40,64]. In contrast to mature heads, proheads can dissociate into subunits at low concentration [111], which could allow proofreading and correction of misassembled intermediates. Additionally, the thick shell of the prohead might make it easier to control the curvature during assembly [40]. In some phages, like HK97 and T4, cleavage of the capsid and scaffolding proteins precedes the expansion, but the cleaved-unexpanded intermediates are short-lived. The viral protease of T4 cleaves about 3,000 peptide bonds per virion [70]. Cleavages of capsid protein can affect the thermodynamic stability of the capsid through changes of quaternary interactions [23] and could also influence the kinetic stability of capsid protein as shown for self-cleaving enzymes [112,113]. In bacteriophage lambda, in addition to the capsid protein cleavage, the connector protein is also cleaved [114]. Although this cleavage is not essential for assembly, it might play a role during DNA ejection [23]. In vitro treatment of proheads with denaturants can also trigger maturation, probably through unfolding of the domains that are cleaved by protease in vivo. Maturation intermediates of HK97 proheads and T4 polyheads have been trapped in vitro [115,116,117,118,119]. Head expansion is probably initiated at one end by the portal protein [120] and is then propagated through the prohead. Such a wave was captured in a giant capsid of phage T4, for which several different maturation states were observed along the axis of the head [121]. The head expansion leads to a 50% increase of the head volume and a change in the appearance of the hexameric capsomers, which in most phages have two-fold rather than six-fold symmetry in the prohead [105,118,122,123]. In phage P22 skewed capsomers have central holes, through which the scaffolding proteins could exit [49,123]. Although detailed information about the head expansion during maturation is derived mostly from the work on HK97, it is likely to be applicable to other tailed phages, all of which have the same capsid protein fold. Comparison of the prohead and the mature head crystal structures of HK97 showed that the capsid proteins are probably trapped in a distorted form. During maturation of HK97, the cross-linking reaction drives the procapsid into a metastable state, where the capsid protein refolds into its lower energy conformation after the delta-domain has been cleaved [14]. Although the cross-linking is unique for HK97, the binding of capsid stabilization proteins, represented by gp soc in T4 and gpD in lambda, could act similarly during maturation and promote the head expansion [14,124,125,126,127]. Both gp soc and gpD can only attach to matured capsids, but have different effects on capsid stability. In the absence of gpD, lambda capsids cannot package the full genome [128], whereas gp soc is only required in the extremes of pH and temperature [124].
In the course of head assembly many phages incorporate minor or pilot proteins into the head. These are usually present in low copy numbers (less than 12 subunits) and are nonessential for the formation of the structure, but crucial for infectivity of the virion. There are three minor proteins in P22, which modulate DNA ejection [52,129] and were thought to localize in the shaft above the connector [82]. However, the shaft density was later reassigned to be part of the connector [50]. A more elaborate shaft structure, called the inner core, is present in the capsid of bacteriophage T7 [65,105,130] and consists of a ring of proteins with twelve-fold symmetry immediately above the connector, followed by an eight-fold and a four-fold symmetric protein rings. A difference in the core structure before and after DNA packaging is perhaps important for the release of terminase after packaging. A similar signal could be propagated by a conformational change of the connector [82,131].
2.5. Head Assembly Completion
After the head is packaged with the DNA, the terminase complex is substituted by neck proteins, which together with the connector form the “gatekeeper” complex [81], preventing premature DNA leakage from the head and possibly initiating DNA exit upon attachment to the host bacteria [132]. In Siphoviridae and Myoviridae phages there are two types of neck protein, each making a ring below the portal. The neck proteins that form a ring closest to the connector have similar structure in Siphoviridae phages SPP1 and HK97, but have a different fold in lambda. However, the neck proteins forming the second ring have similar folds in SPP1 and lambda [132,133,134,135,136]. Additionally, a Myoviridae prophage was identified that has structurally similar neck proteins to those of Siphoviridae [135]. A related structure of the tail-binding platform among Sipho- and Myoviridae phages indicates a common tail binding mechanism and suggests an evolutionary relationship, as well as a possibility that there once existed a phage which could attach two different tail structures.
Phage neck proteins, as well as several other structural proteins, like gpD of lambda, have large (more than 60%) unstructured regions. Naturally disordered proteins that participate in protein-protein interactions are abundant in cells and by becoming folded can influence the sequence of assembly [137]. Additionally, it has been shown that a small disordered protein, which does not have a hydrophobic core, can donate a large surface area to the binding interface. On the other hand, an ordered protein has to be much larger to donate an equivalent surface area to complex. The presence of unstructured proteins inside the cell was suggested to prevent overcrowding [138], whereas for the virus it could play a role in keeping the genome size small.
2.6. Tail Assembly
After the completion of the head assembly, the tail proteins of Podoviridae phages are sequentially attached to the capsid [139,140]. However, for Sipho- and Myoviridae phages, there is a separate tail assembly branch to the assembly pathway, allowing the preformed tail to bind the head via the neck proteins [15]. In the case of T4, this is followed by an attachment of the preassembled fibers [71,141]. Although the tail and the head assemblies occur independently, within each pathway protein association follows a strict order, implying that the third component does not bind until the first two proteins form a complex. Likewise, if an assembly component is absent, the assembly is stalled, and all the proteins that would be added after the missing component should have assembled will remain free in solution. Such a sequential assembly was shown, for example, for the distal part of the T4 contractile tail, called the baseplate [142,143,144]. In such processes the monomers are added to a growing complex and are not wasted on incomplete intermediates [15].
The sequential attachment of proteins can be controlled by different mechanisms. One of the mechanisms, very common in protein assemblies, is called conformational switching. Such a process occurs when a protein structure changes upon attachment to an initiator complex and often involves refolding of some part of the structure, for example a loop-to-helix. Conformational switching is observed in viral capsids with T numbers higher than one, when a capsid protein has to adopt several quasi-equivalent conformations [145,146,147]. The sequence of assembly can also be controlled though formation of composite binding surfaces, created by more than one protein. In such a situation the portion of the surface donated by a single protein is insufficient for stable attachment of an assembly component [15,148].
The tail assembly in Siphoviridae and Myoviridae phages starts from the initiator complex, which forms the absorption device of the phage at the distal end of the tail. The size of this complex ranges from six proteins in Siphoviridae phage lambda or eight in Myoviridae phage Mu [149,150] to about 150 in the baseplate of Myoviridae phage T4 [71,151]. In addition to priming the tail assembly, baseplate complexes undergo structural changes during infection that involve large motions of the component proteins, as was shown for baseteriophage T4 [152] and Lactococcal Siphoviridae phage p2 [153]. During tail assembly, baseplate initiates polymerization of the cylindrical section of the tail, which contributes to the majority of the tail mass. In Siphoviridae the cylinder of the tail is composed of multiple copies of the tail tube [154,155], whereas in Myoviridae phages the tail tube is covered by an outer contractile sheath [152,156,157]. Podoviridae phage N4 also has a two-layered tail, albeit short with a non-contractile “sheath” [158]. Initiation of the tail tube polymerization in Sipho- and Myoviridae phages probably occurs via conformational switching. Without the initiator complex the lambda tail tube protein, gpV, cannot form a tubular structure and exists as a monomer. One of the structural proteins of the bacterial complex, the type VI secretion system, is structurally homologous to lambda gpV. In contrast to gpV, the secretion system homolog does not require an initiator to form the tube. Structural comparison of these homologous tube proteins suggested that a loop-to-helix transition is required to initiate polymerization of gpV [154]. Similarly to lambda gpV, the tail tube protein of Myoviridae phage T4 does not assemble into tubes without baseplates [159]. Nevertheless, disassembled tubes of T4 can repolymerize without baseplates [160], probably because of an irreversible conformational switching that occurred during the initial binding of the tube protein to the baseplate. In Myoviridae phages after the tail tube is assembled, the tail sheath wraps around it. Siphoviridae phage SPP1 has two tail proteins in the ratio 3:1, forming a tail tube, with one of them arising from a translational frame shift [161]. The larger tail protein is predicted to have an additional immunoglobulin-like domain. Although some immunoglobulin-like folds, such as BIG2 domain of phi29, have a function, the roles of others remain unknown. Such domains may have been acquired by phages from hosts [162]. The cylinder of the Sipho and Myoviridae tail is almost exclusively six-fold symmetric. The two exceptions are the three-fold symmetric tail of Siphoviridae bacteriophages phiCbK [163] and T5 [55]. The six-fold symmetry of the tail might be functionally advantageous due to the interaction of the phages with the oligosaccharides which form hexagonal arrays on the outer surface of some bacterial strains [40].
After polymerization of the cylindrical part of the tail, binding of terminator proteins completes the tail assembly [164,165,166]. Terminator proteins, in turn, interact with the neck proteins attached to the head and mediate the association of the tail with the head. The tail-terminator protein of Siphoviridae phage lambda is structurally similar to a Myoviridae prophage protein, providing further evidence of the evolutionary relationship of these tails [164]. Moreover, structural and functional comparison of neck, tail tube and tail completion proteins suggest that these proteins evolved from a single ancestral gene [136]. The length of Sipho- and Myoviridae tails is determined by a tape-measure or ruler protein, also found in cellular complexes such as an injectesome (also called a type III secretion system) and the hook of flagellum [26]. The presence of the ruler protein was first identified in bacteriophage lambda and later in bacteriophage T4 and T5 [167,168,169]. Presumably, the ruler protein is stretched the entire length of the tail and acts as a scaffold for the polymerization of the tail tube [170]. If a ruler protein extends inside the tail cylinder, it would be about 1000 Å long in lambda or T4 and 5000 Å in bacteriophage G. The bacteriophage T4 tape measure protein consists of 590 residues and, thus, cannot have a helical structure in order to be able to stretch the entire length of the tail. The cryo-EM reconstruction of the T4 and SPP1 tails showed density inside the tail tube, which could be attributed to either the ruler protein or DNA [152,155,156]. The copy number of ruler proteins per phage is unknown, but for the type III secretion system it was determined that only one ruler protein is present per complex [171].
The length of the tail tube in Myoviridae phages determines the length of the sheath. In contrast to the tail tube proteins, the tail sheath protein can polymerize into polysheaths in the absence of the tail tube, although 50-fold less efficiently [172,173]. Similarly, capsid protein forms polyheads in the absence of scaffolding proteins. In both cases formation of aberrant structures occurs slower than formation of the viral precursors due to a nucleation barrier. The presence of the tail tube-baseplate complex for the tail sheath or the scaffolding protein for the capsid lowers the nucleation barrier, resulting in a faster assembly. A similar requirement is observed in many viruses, with some viral assemblies primed by nucleic acid [174]. Nucleation complexes are ubiquitous among cellular complexes, like microtubules and actin [175,176]. During the formation of an oligomer, reversible interactions between two subunits sharing one interaction site are unstable. However, if a third subunit binds to the two subunits before they dissociate, the probability of dissociation would be reduced [3]. Hence, after the nucleation barrier is overcome, the polymerization occurs very fast. Likewise, during the formation of the tail sheath no intermediates were observed even when the tube-baseplate complexes were in excess, showing that either complete tail sheaths or only naked tubes were present [172,173]. In addition to nucleating the polymerization of the sheath, the tail tube-baseplate complex induces assembly of the sheath subunits into a helical arrangement that is different to the helical symmetry observed in polysheaths [177]. When assembled on the tail tube, the tail sheath is in its extended conformation with the sheath proteins making contacts to the tail tube as well as to each other. In polysheaths the subunits only make contact with each other and have an arrangement similar to the contracted sheath that occurs after infection. In the contracted form the tail sheath subunits are detached from the inner tube, similarly to polysheaths. The binding of the sheath to the tube does not require nucleation in contrast to the assembly of polysheath and therefore acts as a kinetic trap, inducing assembly of the sheath into the extended higher free-energy structure [157,178].
2.7. Assembly Completion
In Sipho- and Myoviridae phage assembly the completed head and tail are joined spontaneously. In the case of phage T4, the head to tail association is followed by binding of gp wac to the neck region, forming whiskers. The whiskers are essential for attachment of the preassembled long tail fibers [141]. This aligns the fibers along the length of the tail, positioning the proximal part at the baseplate attachment site. Additionally, attachment of fibers through whiskers insures that no fibers would bind to the free tails. The fibers are known to stay in such a retracted position if the conditions (pH and ionic strength) are unfavorable for phage growth, preventing infection [179].
In summary, there are several distinct steps of dsDNA tailed phage assembly: 1. Assembly of a prohead, or a spherical shell of capsid protein filled with scaffolding protein that contains a dodecameric portal. 2. Packaging of DNA using the energy of ATP. 3. Maturation of proheads into angular mature heads. 4. Attachment of the neck and tail proteins or a preassembled tail (Figure 2).
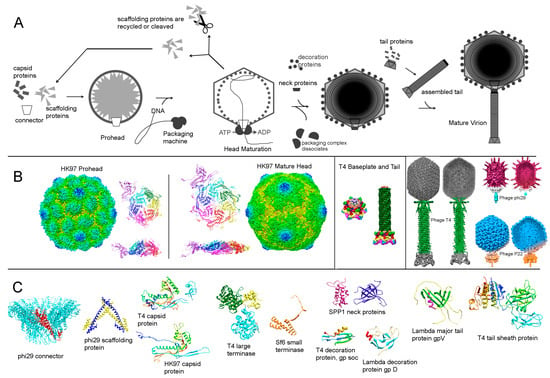
Figure 2.
Assembly of dsDNA tailed phages. (A) Schematic of the assembly steps. (B) Examples of assembly intermediates and mature virions. The left panel shows HK97 prohead and mature head, displaying the whole structure, as well as side and top views of the icosahedral asymmetric units. The middle panel shows the T4 baseplate and tail. The right panel shows T4, phi29 and P22 mature virions, as a surface rendered view and a slab view, displaying the DNA structure. (C) Structural proteins involved in the assembly of several phages. The figure was created using the program CHIMERA [180]. The relevant structures were downloaded from the PDB and EMDB databases.
3. ssDNA Phages
3.1. Microviridae: phiX174
The most studied member of the Microviridae family is bacteriophage phiX174. The isometric T = 1 capsid of phiX174 assembles via an empty procapsid intermediate, similarly to dsDNA tailed phages discussed above. In contrast to most dsDNA phages, phiX174 utilizes two scaffolding proteins during assembly: an inner protein B and an outer protein D [53]. The phage assembly is initiated by the association of five capsid proteins (F) with five copies of the internal scaffolding protein (B) into a 9S particle. In turn, the 9S intermediate binds a spike pentamer (G) and a pilot protein (H), forming a 12S particle. Subsequently, binding of twenty external D proteins to a 12S particle results in the formation of an 18S particle, which is a capsomer of a phiX174 procapsid. Association of twelve 18S particles leads to the formation of an icosahedral procapsid [8]. The outer procapsid shell, formed by protein D, is more elaborate than the actual T = 1 capsid underneath. Another bacteriophage that requires an external scaffolding protein during assembly is the satellite phage P4 [181]. In the external scaffolding shell of phiX174, D proteins adopt four different conformations [182], making two types of asymmetric dimers. In a crystal structure D proteins also form asymmetric dimers, which are similar to the two dimers present in the procapsid [183]. The formation of such an asymmetric dimer represents the first step in the assembly of D and is accomplished through conformational switching [183]. Asymmetric dimers are common among cellular enzymes, for example two identical hexokinase molecules form an asymmetric dimer where one of the two subunits binds substrate more tightly than the other [184]. The dimers of phiX174 scaffolding protein D undergo another conformational switch upon tetramerization. The subsequent binding of the D tetramer to the 12S particle induces yet another structural change, allowing twelve 18S subunits to associate. The specific residues involved in the conformational switching of the D protein were identified using structural and mutational investigations [183,185,186].
The structure of the phiX174 procapsid was determined by cryo-EM and image reconstruction [187,188]. This structure has 30Å pores at the three-fold axes of the icosahedron and 10Å gaps between the F pentamers, which are bridged together by the external scaffolding D protein shell. This “open” procapsid structure probably represents a true viral precursor. When the same complex was crystallized, it changed into a “closed” procapsid that has a different conformation of F and G proteins, similar to that in a mature virus [182,188]. The “closed” procapsid lacks the three-fold holes and is thought to be an aberrant particle, not a true intermediate state. After assembly, the “open” procapsid is packaged with the ssDNA genome, probably through the three-fold holes, resulting in the exit of the internal scaffolding protein B. Similarly, the internal scaffolding proteins of some dsDNA tailed phages exit from the procapsid though holes in skewed hexameric capsomers, as discussed above. The intermediate assembly state of phiX174, that does not contain B but still contains the external scaffolding protein D, is called “provirion”, which subsequently loses D proteins and matures into a virion [189]. During maturation, instead of the expansion observed in dsDNA tailed phages, the head collapses, as the holes between the F pentamers and the holes at the three-fold axes close and the scaffolding protein dissociates (Figure 3).
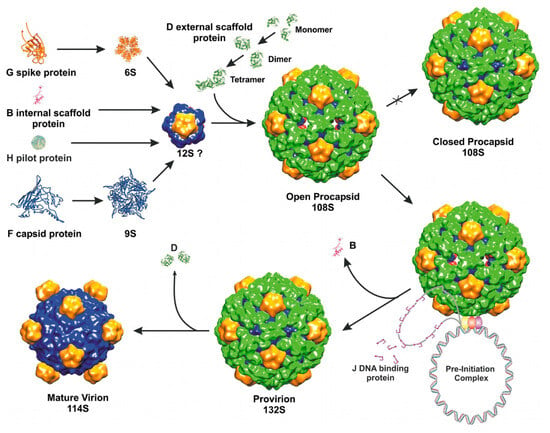
Figure 3.
Microviridae assembly pathway (reprinted from Bernal et al. [189] with permission from Elsevier).
3.2. Filamentous Phages
Filamentous phages have been widely used as a molecular biology tool since the development of the phage display method [190], which proved indispensable to study protein-protein interactions, modify proteins and select antibodies [191]. The assembly and structure of filamentous phages have been extensively studied [192,193,194,195,196]. The filamentous virion has a 9000 Å-long rod-like structure with a ~65 Å diameter that contains ssDNA genome. In contrast to DNA bacteriophages discussed above, filamentous phages assemble on the internal membrane of the host cell. The head of the tailed bacteriophage T4 also assembles on the internal membrane of E. coli, anchored by a connector chaperon protein, as discussed above. In filamentous bacteriophages, all structural proteins are anchored on the inner membrane of E. coli. Moreover, the major capsid protein, pVIII, of filamentous phages, forms a transmembrane helix, but the final virion does not contain any lipids [194]. In contrast to the majority of phages that lyse the infected cell, filamentous phages form adhesion zones and are extruded from the host without killing it.
Filamentous phages belong to Inoviridae family, represented by M13, fd and f1 as most studied members that infect E. coli. Infectious virions are assembled from five different structural proteins. There are about 2,700 copies of the major capsid protein that form a five-start helical rod [197,198,199,200], capped by pentamers of pIII and pVI on one side and pVII and pIX on the other [201,202]. In some filamentous phages, e.g., f1, the capsid protein has a leader sequence that is cleaved by a cellular peptidase after membrane insertion [203]. Three non-structural proteins, pI, pIV and pXI, mediate assembly and release of the phage from the host cell. The assembly is initiated when the ssDNA binding protein pV displaces the DNA replication machinery. Next, the pI protein interacts with the 32 bp DNA leader sequence and mediates the DNA interaction with capsid protein, removing pV. It has been suggested that this process requires ATP hydrolyses. Because pI has a nucleotide-binding motif, it was proposed to be a motor [193]. If this is indeed the case this would be the second use of an ATP-driven motor employed in bacteriophage assembly, with the first motor being the phage packaging machine. Prior to DNA binding, the capsid protein is probably associated with minor proteins pVII and pIX, which are required for assembly initiation [202]. It is possible that they trigger a conformational switch in the capsid protein, inducing it to polymerize around DNA. How the capsid protein transitions from the lipid membrane into the rod that does not contain any lipids is unknown. The length of the rod is determined by the length of DNA. In contrast to TMV, where three bases fit into a defined groove of the capsid protein, there is no specific protein/DNA association in filamentous phages. The length of packaged DNA can be altered by the number of charged residues of the capsid protein [204,205]. The terminal cap proteins pIII and pVI bind last and terminate the assembly. Similar to the tail assembly termination proteins of dsDNA phages, the absence of pIII and pVI results in formation of polyphages. The virions exit through the adhesion zones, formed by two non-structural proteins pI and pXI (Figure 4). Another viral protein, pIV, was shown to assemble into 10–12 subunit oligomers that probably form pores in the outer membrane of the cell [206,207]. Homologs of pIV were found in both type II and type III bacterial secretion systems. The location of gene IV in the genomes of several different filamentous phages is not conserved, in comparison with other phage proteins, suggesting that filamentous phages could have acquired the pIV gene from a host [193].
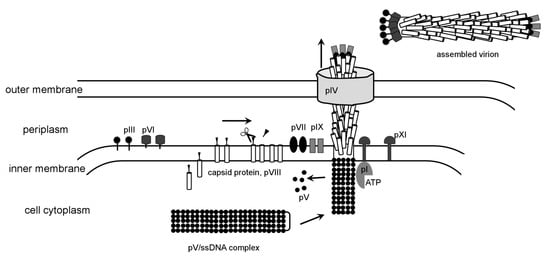
Figure 4.
Model for the assembly and extrusion of filamentous phages. The figure was created based on a figure by Russel and Model [196].
4. Lipid Containing Phages
The two groups, Tectiviridae and Cystoviridae, described below assemble via an empty capsid shell, similar to dsDNA tailed phages and ssDNA isometric phages.
The most studied member of the Tectiviridae family is bacteriophage PRD1. The crystal structure of PRD1 was determined by X-ray crystallography, showing the arrangement of capsid proteins that surround the membrane sack and dsDNA genome [208,209]. Four of eighteen viral structural proteins are icosahedrally arranged and, therefore, visible in the X-ray crystal structure. Although phages from the Tectiviridae group do not have a permanent or stable tail structure, a non-permanent tail is produced upon infection [210,211]. Furthermore, PRD1 has a special five-fold vertex with a different protein composition to the proteins at other five-fold axes [212] and carries the packaging ATPase [213]. However, this unique vertex has not been shown to be the same as the vertex used for infection, but is utilized during packaging [214]. Therefore, in contrast to the tailed dsDNA phages, the PRD1 packaging enzyme is a structural protein and does not dissociate after the assembly is complete [17,212]. The rate of PRD1 packaging in vitro was calculated to be more than 340 bp/s [215], which is comparable with that of some dsDNA tailed phages (e.g., 350 bp/s for SPP1 [216]). Bacteriophage T4 can package with a rate of up to 2000 bp/s [217]), whereas dsRNA phages translocate RNA at a much slower rate of about 30 bp/s [218]. Although PRD1 has no temporary scaffolding, its membrane is coated by the structural protein p30. This glue protein mediates the assembly of the coat protein p3 into correct pseudo T = 25 quasi-symmetry and cements capsomers together (Figure 5) [208]. A similar, but external, network of scaffolding proteins is formed by protein Sid of bacteriophage P4, a satellite phage of P2 [181,219]. No expansion of the PRD1 capsid was detected after DNA packaging, although expansion of the viral membrane was observed [220]. There are many similarities between phage PRD1 and adenovirus, including the topology of the capsid protein jelly-roll fold, capsomer arrangement and replication priming by a protein. These similarities suggest that although these viruses infect two different domains of life they might be evolutionary related [221,222]. Additionally, recent structures of adenovirus by X-ray crystallography [223] and atomic resolution cryo-EM [224] showed that one of the adenovirus cement proteins, although structurally different from PRD1 p30, also forms a net underneath the capsid protein [224]. This adenoviral cement protein affects the amount of DNA packaged by the virus [225,226] and might be a functional equivalent of the PRD1 tape measure protein p30.
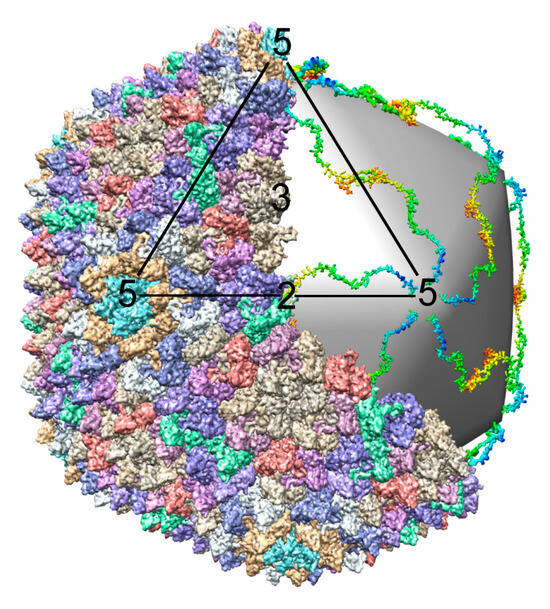
Figure 5.
Structure of PRD1 virion. A portion of the capsid protein was removed to show the mesh underneath made by 60 copies of tape measure protein p30. The figure was created using the program CHIMERA [180]. The relevant PDB entry 1W8X was downloaded from the PDB databank.
Another group of lipid-containing viruses that package their nucleic acid into the preformed capsid shell belong to the Cystoviridae family of phages and have a segmented dsRNA genome. The most studied members of this group are bacteriophages phi6 and phi12. The procapsid of Cystoviridae consists of 120 copies of the capsid protein (p1), about 14 copies of RNA polymerase (p2), located at the five-fold vertices [227], a packaging enzyme p4 and its cofactor p7 (Figure 6A). The procapsid is not spherical and has deeply recessed vertexes [220]. The genome is packaged into the procapsid, somewhat similarly to the DNA phages discussed above. Because the genome is segmented, packaging is sequential. The (+) strands of the S, M and L segments are packaged in strict order, probably determined by the binding site for the RNA strand on the capsid surface. Once S, the first segment, is packaged another binding site appears for the next strand to be packaged, which continues until the third segment is internalized. After all (+) stands are packaged the (−) strands are synthesized inside the capsid by the polymerase (Figure 6B) [16,228]. The packaging is accompanied by capsid expansion resulting in elimination of surface depressions. Both RNA polymerase and the packaging enzyme are structural proteins [229]. The phi6 packaging enzyme can use any nucleotide for packaging in contrast to dsDNA tailed phages, which have specificity for ATP. A mechanism of the RNA translocation was proposed based on the X-ray structure of the NTPase [101]. In contrast to PRD1, where ATPase is located only on one special vertex, the phi6 ATPase (p4) is present on more than one vertex [229]. Upon completion of assembly and packaging of phi6, the procapsid is coated with another protein, p8, that forms a T = 13 icosahedral shell. This outer shell has a different arrangement in related phages, such as phi12 and phi8 [230,231]. The assembly is completed when the virus has been surrounded by a lipid membrane containing four integral membrane proteins [228]. The formation of the lipid envelope does not involve budding and is not understood.
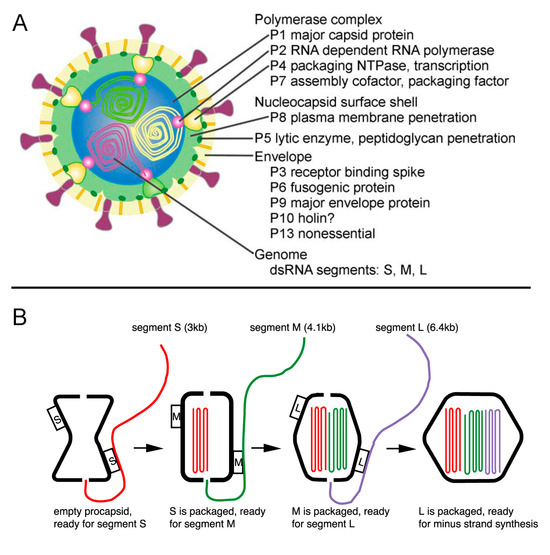
Figure 6.
(A) A diagram showing the phi6 virion architecture (reprinted from [228] with permission from Elsevier). (B) Genome packaging in dsRNA phages (reprinted from Figure 6 in Mindich et al. [16] with permission from Elsevier).
5. ssRNA Icosahedral Phages
The single-stranded RNA phages such as MS2, Qβ, fr, GA, R17, f2 and phiCb5 form the Leviviridae family. The capsid protein of ssRNA phages has a unique fold, specific only to this phage group. Ninety dimers of the capsid protein are arranged into a T = 3 icosahedral lattice [232,233,234,235,236]. Additionally, the virion contains one copy of a maturation protein, or A protein and viral ssRNA genome. Similar to some ssRNA plant viruses, a large proportion of the ssRNA is icosahedrally ordered [174]. The A protein mediates phage attachment to pili and was suggested to initiate assembly of the coat protein around the folded RNA [237]. Even without the genome, the coat protein assembles into virus-like particles, although this requires higher coat protein concentration. In the T = 3 capsid there are two quasi-equivalent coat protein dimers, both of which are required for efficient assembly. The ssRNA induces the conformational switching of the capsid protein dimer during assembly [238,239]. In the assembly of Leviviridae phiCb5 phage, three RNA bases interact with the two capsid subunits from one dimer as well as one subunit from a neighboring dimer, facilitating capsid formation and stabilizing the final structure [240]. Co-interaction of capsid protein with the RNA is critical during assembly, similar to the interactions between scaffold and capsid proteins in dsDNA phages.
6. Concluding Remarks
The majority of structural information about phage assembly has been accumulated by studying individual viral components. Although the dynamics of the viral assembly processes has been investigated with biophysical methods [241,242,243,244], most of current models of dynamic processes are based on investigations of stable intermediates, captured by slowing the assembly reactions in vitro. Analysis of transient dynamic intermediates will provide new insights into molecular mechanisms of phage assembly. Visualization of the conformational changes, like capturing the rearrangement of the capsid proteins during the capsid maturation, might soon be possible with the development of live high-speed imaging techniques [245].
Despite great morphological and genetic diversity of phages, there are extensive structural similarities in the component proteins from which the whole virus is assembled and that govern the viral life cycle. As has frequently been observed, the three-dimensional structure of proteins is perhaps the most conserved aspect of evolution [246]. The most highly conserved structures relate to the most basic biological functions, such as ATP driven molecular motors and the capsid structures of many phage families. The implication of the conserved capsomer structure with either a HK97 or a jelly-roll fold is that phages and, hence, bacteria have co-evolved and that this coexistence has provided the means for gene transfer and the subsequent evolutionary modifications in all kingdoms of life.
Acknowledgments
We are grateful to Sheryl Kelly for assistance in preparation of the manuscript. This work was supported by National Science Foundation grant MCB-1014547 and National Institutes of Health grant AI081726, both awarded to MGR.
References and Notes
- Wood, W.B.; Edgar, R.S.; King, J.; Lielausis, I.; Henninger, M. Bacteriophage assembly. Fed. Proc. 1968, 27, 1160–1166. [Google Scholar] [PubMed]
- Casjens, S.; King, J. Virus assembly. Annu. Rev. Biochem. 1975, 44, 555–611. [Google Scholar] [CrossRef] [PubMed]
- Murialdo, H.; Becker, A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol. Rev. 1978, 42, 529–576. [Google Scholar] [CrossRef] [PubMed]
- Bazinet, C.; King, J. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 1985, 39, 109–129. [Google Scholar] [CrossRef]
- Black, L.W. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 1989, 43, 267–292. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Garcea, R.L. Capsid assembly of dsDNA viruses. Sem. Virol. 1994, 5, 15–26. [Google Scholar] [CrossRef]
- Kellenberger, E.; Wunderli-Allenspach, H. Electron microscopic studies on intracellular phage development—History and perspectives. Micron 1995, 26, 213–245. [Google Scholar] [CrossRef]
- Fane, B.A.; Prevelige, P.E., Jr. Mechanism of scaffolding-assisted viral assembly. Adv. Prot. Chem. 2003, 64, 259–299. [Google Scholar]
- Steven, A.C.; Heymann, J.B.; Cheng, N.; Trus, B.L.; Conway, J.F. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 2005, 15, 227–236. [Google Scholar] [CrossRef]
- Johnson, J.E.; Chiu, W. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 2007, 17, 237–243. [Google Scholar] [CrossRef]
- Papapostolou, D.; Howorka, S. Engineering and exploiting protein assemblies in synthetic biology. Mol. Biosyst. 2009, 5, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C.; Petrov, A.S.; Devkota, B.; Box, M.B. Viral assembly: A molecular modeling perspective. Phys. Chem. Chem. Phys. 2009, 11, 10553–10564. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Rao, V.B.; Rossmann, M.G. Genome packaging in viruses. Curr. Opin. Struct. Biol. 2010, 20, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E. Virus particle maturation: Insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 2010, 20, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Hendrix, R. Control mechanisms in dsDNA bacteriophage assembly. In The Bacteriophages; Calendar, R.L., Ed.; Plenum Press: New York, NY, USA, 1988; pp. 15–75. [Google Scholar]
- Mindich, L. Packaging, replication and recombination of the segmented genome of bacteriophage f6 and its relatives. Virus Res. 2004, 101, 83–92. [Google Scholar] [CrossRef]
- Mindich, L.; Bamford, D.; McGraw, T.; Mackenzie, G. Assembly of bacteriophage PRD1: Particle formation with wild-type and mutant viruses. J. Virol. 1982, 44, 1021–1030. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: A tale of two membranes. Curr. Opin. Microbiol. 2006, 9, 423–429. [Google Scholar] [CrossRef]
- Ostapchuk, P.; Hearing, P. Regulation of adenovirus packaging. Curr. Top. Microbiol. 2003, 272, 165–185. [Google Scholar]
- Cassetti, M.C.; Merchlinsky, M.; Wolffe, E.J.; Weisberg, A.S.; Moss, B. DNA packaging mutant: Repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 1998, 72, 5769–5780. [Google Scholar] [CrossRef]
- Zauberman, N.; Mutsafi, Y.; Halevy, D.B.; Shimoni, E.; Klein, E.; Xiao, C.; Sun, S.; Minsky, A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008, 6, e114. [Google Scholar] [CrossRef]
- Hershko, A.; Fry, M. Post-translational cleavage of polypeptide chains: Role in assembly. Annu. Rev. Biochem. 1975, 44, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Hellen, C.U.; Wimmer, E. The role of proteolytic processing in the morphogenesis of virus particles. Experientia 1992, 48, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.L.; Jiang, W.; Rixon, F.J.; Chiu, W. Common ancestry of herpesvirus and tailed DNA bacteriophages. J. Virol. 2005, 79, 14967–14970. [Google Scholar] [CrossRef] [PubMed]
- Sutter, M.; Boehringer, D.; Futmann, S.; Günther, S.; Prangishvili, D.; Loessner, M.J.; Stetter, K.O.; Weber-Ban, E.; Ban, N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 2008, 15, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.R.; Agrain, C.; Sorg, I. Length control of extended protein structures in bacteria and bacteriophages. Curr. Opin. Microbiol 2006, 9, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res. Microbiol. 2008, 159, 340–348. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Tailed bacteriophages: The order Caudovirales. Adv. Virus Res. 1998, 51, 135–201. [Google Scholar]
- Bamford, D.H.; Grimes, J.M.; Stuart, D.I. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005, 15, 655–663. [Google Scholar] [CrossRef]
- Teschke, C.M.; Parent, K.N. ‘Let the phage do the work’: using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 2010, 401, 119–130. [Google Scholar] [CrossRef]
- Donelli, G.; Guglielmi, F.; Paoletti, L. Structure and physico-chemical properties of bacteriophage G. I. Arrangement of protein subunits and contraction process of tail sheath. J. Mol. Biol. 1972, 71, 113–125. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Classification of Bacteriophages. In The Bacteriophages, 2nd ed.; Calendar, R.L., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 8–16. [Google Scholar]
- Dodds, J.A.; Cole, A. Microscopy and biology of Uronema gigas, a filamentous eucaryotic green alga, and its associated tailed virus-like particle. Virology 1980, 100, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Cherrier, M.V.; Kostyuchenko, V.A.; Xiao, C.; Bowman, V.D.; Battisti, A.J.; Yan, X.; Chipman, P.R.; Baker, T.S.; Van Etten, J.L.; Rossmann, M.G. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 11085–11089. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H.C.; Watson, J.D. Structure of small viruses. Nature 1956, 177, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Caspar, D.L.D. Structure of bushy stunt virus. Nature 1956, 177, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Caspar, D.L.D.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962, 27, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T. Freedom and restraint: Themes in virus capsid assembly. Structure 2000, 8, R157–R162. [Google Scholar] [CrossRef]
- Luque, A.; Reguera, D. The structure of elongated viral capsids. Biophys. J. 2010, 98, 2993–3003. [Google Scholar] [CrossRef]
- Moody, M.F. Geometry of phage head construction. J. Mol. Biol. 1999, 293, 401–433. [Google Scholar] [CrossRef]
- McNicol, L.A.; Simon, L.D.; Black, L.W. A mutation which bypasses the requirement for p24 in bacteriophage T4 capsid morphogenesis. J. Mol. Biol. 1977, 116, 261–283. [Google Scholar] [CrossRef]
- Fokine, A.; Battisti, A.J.; Kostyuchenko, V.A.; Black, L.W.; Rossmann, M.G. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: The evolution of pentameric vertex proteins in icosahedral viruses. J. Struct. Biol. 2006, 154, 255–259. [Google Scholar] [CrossRef]
- Johnson, K.; Condie, B.; Mooney, D.T.; Doermann, A.H. Mutations that eliminate the requirement for the vertex protein in bacteriophage T4 capsid assembly. J. Mol. Biol. 1992, 224, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Fokine, A.; Leiman, P.G.; Shneider, M.M.; Ahvazi, B.; Boeshans, K.M.; Steven, A.C.; Black, L.W.; Mesyanzhinov, V.V.; Rossmann, M.G. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 7163–7168. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989, 58, 533–573. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef]
- Morais, M.C.; Choi, K.H.; Koti, J.S.; Chipman, P.R.; Anderson, D.L.; Rossmann, M.G. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of j29. Mol. Cell 2005, 18, 149–159. [Google Scholar] [CrossRef]
- Jiang, W.; Li, Z.; Zhang, Z.; Baker, M.L.; Prevelige Jr., P. E.; Chiu, W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 2003, 10, 131–135. [Google Scholar] [CrossRef]
- Parent, K.N.; Khayat, R.; Tu, L.H.; Suhanovsky, M.M.; Cortines, J.R.; Teschke, C.M.; Johnson, J.E.; Baker, T.S. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure 2010, 18, 390–401. [Google Scholar] [CrossRef]
- Chen, D.H.; Baker, M.L.; Hryc, C.F.; Dimaio, F.; Jakana, J.; Wu, W.; Dougherty, M.; Haase-Pettingell, C.; Schmid, M.F.; Jiang, W.; Baker, D.; King, J.A.; Chiu, W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 1355–1360. [Google Scholar] [CrossRef]
- Condron, B.G.; Atkins, J.F.; Gesteland, R.F. Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol. 1991, 173, 6998–7003. [Google Scholar] [CrossRef]
- King, J.; Lenk, E.V.; Botstein, D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J. Mol. Biol. 1973, 80, 697–731. [Google Scholar] [CrossRef]
- Dokland, T. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 1999, 56, 580–603. [Google Scholar] [CrossRef] [PubMed]
- Duda, R.L.; Martincic, K.; Hendrix, R.W. Genetic basis of bacteriophage HK97 prohead assembly. J. Mol. Biol. 1995, 247, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Effantin, G.; Boulanger, P.; Neumann, E.; Letellier, L.; Conway, J.F. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 2006, 361, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Huet, A.; Conway, J.F.; Letellier, L.; Boulanger, P. In vitro assembly of the T = 13 procapsid of bacteriophage T5 with its scaffolding domain. J. Virol. 2010, 84, 9350–9358. [Google Scholar] [CrossRef]
- Thuman-Commike, P.A.; Greene, B.; Jakana, J.; Prasad, B.V.V.; King, J.; Prevelige, P.E., Jr.; Chiu, W. Three-dimensional structure of scaffolding-containing phage P22 procapsids by electron cryo-microscopy. J. Mol. Biol. 1996, 260, 85–98. [Google Scholar] [CrossRef]
- Morais, M.C.; Kanamaru, S.; Badasso, M.O.; Koti, J.S.; Owen, B.A.L.; McMurray, C.T.; Anderson, D.L.; Rossmann, M.G. Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Biol. 2003, 10, 572–576. [Google Scholar] [CrossRef]
- Poh, S.L.; el-Khadali, F.; Berrier, C.; Lurz, R.; Melki, R.; Tavares, P. Oligomerization of the SPP1 scaffolding protein. J. Mol. Biol. 2008, 378, 551–564. [Google Scholar] [CrossRef]
- Casjens, S.; King, J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J. Supromol. Struct. 1974, 2, 202–224. [Google Scholar] [CrossRef]
- Choi, K.H.; Morais, M.C.; Anderson, D.L.; Rossmann, M.G. Determinants of bacteriophage f29 head morphology. Structure 2006, 14, 1723–1727. [Google Scholar] [CrossRef]
- Epstein, C.J.; Goldberg, R.F.; Anfinsen, C.B. The genetic control of tertiary protein structure: Studies with model systems. Cold Spring Harbor Symp. Quant. Biol. 1963, 28, 439–449. [Google Scholar] [CrossRef]
- Kemp, C.L.; Howatson, A.F.; Siminovitch, L. Electron microscopy studies of mutants of lambda bacteriophage. I. General description and quantitation of viral products. Virology 1968, 36, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, E. Form determination of the heads of bacteriophages. Eur. J. Biochem. 1990, 190, 233–248. [Google Scholar] [CrossRef]
- Serwer, P. Internal proteins of bacteriophage T7. J. Mol. Biol. 1976, 107, 271–291. [Google Scholar] [CrossRef] [PubMed]
- Cerritelli, M.E.; Studier, F.W. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J. Mol. Biol. 1996, 258, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Parker, M.H.; Weigele, P.; Casjens, S.; Prevelige, P.E., Jr.; Krishna, N.R. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 2000, 297, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Thuman-Commike, P.A.; Greene, B.; Malinski, J.A.; Burbea, M.; McGough, A.; Chiu, W.; Prevelige, P.E., Jr. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids Biophys. J. 1999, 76, 3267–3277. [Google Scholar]
- Thuman-Commike, P.A.; Greene, B.; Jakana, J.; McGough, A.; Prevelige, P.E., Jr; Chiu, W. Interactions in a bacteriophage P22 mutant defective in maturation. J. Virol. 2000, 74, 3871–3873. [Google Scholar] [CrossRef]
- Black, L.W.; Showe, M.K.; Steven, A.C. Morphogenesis of the T4 head. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 218–258. [Google Scholar]
- Rossmann, M.G.; Mesyanzhinov, V.V.; Arisaka, F.; Leiman, P.G. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 2004, 14, 171–180. [Google Scholar] [CrossRef]
- Caldentey, J.; Lepault, J.; Kellenberger, E. Isolation and reassembly of bacteriophage T4 core proteins. J. Mol. Biol. 1987, 295, 637–647. [Google Scholar] [CrossRef]
- Berger, B.; Hoest, G.W.; Paulson, J.R.; Shor, P.W. On the structure of the scaffolding core of bacteriophage T4. J. Comput. Biol. 1999, 6, 1–12. [Google Scholar] [CrossRef]
- Valpuesta, J.M.; Carrascosa, J.L. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Quart. Rev. Biophys. 1994, 27, 107–155. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.; van Driel, R.; Driedonks, R. A proposed structure of the prolate phage T4 prehead core. An electron microscopic study. J. Ultrastruct. Res. 1982, 80, 12–22. [Google Scholar] [CrossRef]
- Michaud, G.; Zachary, A.; Rao, V.B.; Black, L.W. Membrane-associated assembly of a phage T4 DNA entrance vertex structure studied with expression vectors. J. Mol. Biol. 1989, 209, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Dröge, A.; Santos, M.A.; Stiege, A.C.; Alonso, J.C.; Lurz, R.; Trautner, T.A.; Tavares, P. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: Protein components and interactions during assembly. J. Mol. Biol. 2000, 296, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Lurz, R.; Orlova, E.V.; Günther, D.; Dube, P.; Dröge, A.; Weise, F.; van Heel, M.; Tavares, P. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 2001, 310, 1027–1037. [Google Scholar] [CrossRef]
- Simpson, A.A.; Tao, Y.; Leiman, P.G.; Badasso, M.O.; He, Y.; Jardine, P.J.; Olson, N.H.; Morais, M.C.; Grimes, S.; Anderson, D.L.; Baker, T.S.; Rossmann, M.G. Structure of the bacteriophage f29 DNA packaging motor. Nature 2000, 408, 745–750. [Google Scholar] [CrossRef]
- Lebedev, A.A.; Krause, M.H.; Isidro, A.L.; Vagin, A.A.; Orlova, E.V.; Turner, J.; Dodson, E.J.; Tavares, P.; Antson, A.A. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007, 26, 1984–1994. [Google Scholar] [CrossRef]
- Orlova, E.V.; Gowen, B.; Droge, A.; Stiege, A.; Weise, F.; Lurz, R.; van Heel, M.; Tavares, P. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003, 22, 1255–1262. [Google Scholar] [CrossRef]
- Lander, G.C.; Tang, L.; Casjens, S.R.; Gilcrease, E.B.; Prevelige, P.; Poliakov, A.; Potter, C.S.; Carragher, B.; Johnson, J.E. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 2006, 312, 1791–1795. [Google Scholar] [CrossRef]
- Fu, C.-Y.; Prevelige, P.E., Jr. In vitro incorporation of the phage phi29 connector complex. Virology 2009, 394, 149–153. [Google Scholar] [CrossRef]
- Serwer, P.; Watson, R.H. Function of an internal bacteriophage T7 core during assembly of a T7 procapsid. J. Virol. 1982, 42, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, H.; Morita, M. Phage DNA packaging. Genes Cells 2003, 2, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Casjens, S.R. DNA packaging by double-stranded DNA bacteriophages. Cell 1980, 21, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 4779–4783. [Google Scholar] [CrossRef] [PubMed]
- Hügel, T.; Michaelis, J.; Hetherington, C.L.; Jardine, P.J.; Grimes, S.; Walter, J.M.; Falk, W.; Anderson, D.L.; Bustamante, C. Experimental test of connector rotation during DNA packaging into bacteriophage f29 capsids. PLoS Biology 2007, 5, e59. [Google Scholar] [CrossRef]
- Baumann, R.G.; Mullaney, J.; Black, L.W. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 2006, 61, 16–32. [Google Scholar] [CrossRef]
- Al-Zahrani, A.S.; Kondabagil, K.; Gao, S.; Kelly, N.; Ghosh-Kumar, M.; Rao, V.B. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. J. Biol. Chem. 2009, 284, 24490–24500. [Google Scholar] [CrossRef]
- Zhao, H.; Finch, C.J.; Sequeira, R.D.; Johnson, B.A.; Johnson, J.E.; Casjens, S.R.; Tang, L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1971–1976. [Google Scholar] [CrossRef]
- Bjornsti, M.A.; Reilly, B.E.; Anderson, D.L. Morphogenesis of bacteriophage f29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA-protein gp3. J. Virol. 1983, 45, 383–396. [Google Scholar] [CrossRef]
- Morais, M.C.; Koti, J.S.; Bowman, V.D.; Reyes-Aldrete, E.; Anderson, D.L.; Rossmann, M.G. Defining molecular and domain boundaries in the bacteriophage f29 DNA packaging motor. Structure 2008, 16, 1267–1274. [Google Scholar] [CrossRef]
- Hsiao, C.L.; Black, L.W. DNA packaging and pathway of bacteriophage T4 head assembly. Proc. Natl. Acad. Sci. U. S. A. 1977, 74, 3652–3656. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Wyckoff, E.; Heyden, M.; Sampson, L.; Eppler, K.; Randall, S.; Moreno, E.T.; Serwer, P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 1992, 224, 1055–1074. [Google Scholar] [CrossRef]
- Tavares, P.; Santos, M.A.; Lurz, R.; Morelli, G.; de Lencastre, H.; Trautner, T.A. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 1992, 225, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Tavares, P.; Lurz, R.; Stiege, A.; Rückert, B.; Trautner, T.A. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol. 1996, 264, 954–967. [Google Scholar] [CrossRef]
- Moffitt, J.R.; Chemla, Y.R.; Aathavan, K.; Grimes, S.; Jardine, P.J.; Anderson, D.L.; Bustamante, C. Intersubunit coordination in a homomeric ring ATPase. Nature 2009, 457, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Aathavan, K.; Politzer, A.T.; Kaplan, A.; Moffitt, J.R.; Chemla, Y.R.; Grimes, S.; Jardine, P.J.; Anderson, D.L.; Bustamante, C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 2009, 461, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Kondabagil, K.; Draper, B.; Alam, T.I.; Bowman, V.D.; Zhang, Z.; Hegde, S.; Fokine, A.; Rossmann, M.G.; Rao, V.B. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 2008, 135, 1251–1262. [Google Scholar] [CrossRef]
- Mancini, E.J.; Kainov, D.E.; Grimes, J.M.; Tuma, R.; Bamford, D.H.; Stuart, D.I. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 2004, 118, 743–755. [Google Scholar] [CrossRef]
- Oram, M.; Sabanayagam, C.; Black, L.W. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J. Mol. Biol. 2008, 381, 61–72. [Google Scholar] [CrossRef]
- Ray, K.; Sabanayagam, C.R.; Lakowicz, J.R.; Black, L.W. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology 2010, 398, 224–232. [Google Scholar] [CrossRef]
- Rao, V.B.; Black, L.W. Structure and assembly of bacteriophage T4 head. Virol. J. 2010, 7, 356. [Google Scholar] [CrossRef] [PubMed]
- Cerritelli, M.E.; Conway, J.F.; Chang, N.; Trus, B.L.; Steven, A.C. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv. Protein Chem. 2003, 64, 301–323. [Google Scholar] [PubMed]
- Jardine, P.J.; Anderson, D.L. DNA Packaging in DS DNA Phages. In The Bacteriophages, 2nd ed.; Calendar, R.L., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 49–65. [Google Scholar]
- Fang, P.-A.; Wright, E.T.; Weintraub, S.T.; Hakala, K.; Wu, W.; Serwer, P.; Jiang, W. Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J. Mol. Biol. 2008, 384, 1384–1399. [Google Scholar] [CrossRef]
- Ray, K.; Ma, J.; Oram, M.; Lakowicz, J.R.; Black, L.W. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: Colocalization of packaged phage T4 DNA ends within the capsid. J. Mol. Biol. 2010, 395, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- King, J.; Casjens, S. Catalytic head assembling protein in virus morphogenesis. Nature 1974, 251, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Medina, E.; Wieczorek, D.; Medina, E.M.; Yang, Q.; Feiss, M.; Catalano, C.E. Assembly and maturation of the bacteriophage lambda procapsid: gpC is the viral protease. J. Mol. Biol. 2010, 401, 813–830. [Google Scholar] [CrossRef]
- van Driel, R. Assembly of bacteriophage T4 head-related structures. Assembly of polyheads in vitro. J. Mol. Biol. 1977, 114, 61–72. [Google Scholar] [CrossRef]
- Sohl, J.L.; Jaswal, S.S.; Agard, D.A. Unfolded conformations of a-lytic protease are more stable than its native state. Nature 1998, 395, 817–819. [Google Scholar] [CrossRef]
- Jaswal, S.S.; Sohl, J.L.; Davis, J.H.; Agard, D.A. Energetic landscape of a-lytic protease optimizes longevity through kinetic stability. Nature 2002, 415, 343–346. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Casjens, S.R. Assembly of bacteriophage lambda heads: Protein processing and its genetic control in petit l assembly. J. Mol. Biol. 1975, 91, 187–199. [Google Scholar] [CrossRef]
- Duda, R.L.; Hempel, J.; Michel, H.; Shabanowitz, J.; Hunt, D.; Hendrix, R.W. Structural transitions during bacteriophage KH97 head assembly. J. Mol. Biol. 1995, 247, 618–635. [Google Scholar] [CrossRef] [PubMed]
- Lata, R.; Conway, J.F.; Cheng, N.; Duda, R.L.; Hendrix, R.W.; Wikoff, W.R.; Johnson, J.E.; Tsuruta, H.; Steven, A.C. Maturation dynamics of a viral capsid: Visualization of transitional intermediate states. Cell 2000, 100, 253–263. [Google Scholar] [CrossRef]
- Gan, L.; Speir, J.A.; Conway, J.F.; Lander, G.; Cheng, N.; Firek, B.A.; Hendrix, R.W.; Duda, R.L.; Liljas, L.; Johnson, J.E. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure 2006, 14, 1655–1665. [Google Scholar] [CrossRef]
- Gertsman, I.; Gan, L.; Guttman, M.; Lee, K.; Speir, J.A.; Duda, R.L.; Hendrix, R.W.; Komives, E.A.; Johnson, J.E. An unexpected twist in viral capsid maturation. Nature 2009, 458, 646–650. [Google Scholar] [CrossRef]
- Kocsis, E.; Greenstone, H.L.; Locke, E.G.; Kessel, M.; Steven, A.C. Multiple conformational states of the bacteriophage T4 capsid surface lattice induced when expansion occurs without prior cleavage. J. Struct. Biol. 1997, 118, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.; Oram, M.; Ma, J.; Black, L.W. Portal control of viral prohead expansion and DNA packaging. Virology 2009, 391, 44–50. [Google Scholar] [CrossRef]
- Steven, A.C.; Carrascosa, J.L. Proteolytic cleavage and structural transformation: Their relationship in bacteriophage T4 capsid maturation. J. Supramol. Struct. 1979, 10, 1–11. [Google Scholar] [CrossRef]
- Dokland, T.; Murialdo, H. Structural transitions during maturation of bacteriophage lambda capsids. J. Mol. Biol. 1993, 233, 682–694. [Google Scholar] [CrossRef]
- Prasad, B.V.V.; Prevelige, P.E.; Marietta, E.; Chen, R.O.; Thomas, D.; King, J.; Chiu, W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 1993, 231, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Fokine, A.; O’Donnell, E.; Rao, V.B.; Rossmann, M.G. Structure of the small outer capsid protein, Soc: A clamp for stabilizing capsids of T4-like phages. J. Mol. Biol. 2010, 395, 728–741. [Google Scholar] [CrossRef]
- Yang, F.; Forrer, P.; Dauter, Z.; Conway, J.F.; Cheng, N.; Cerritelli, M.E.; Steven, A.C.; Plückthun, A.; Wlodawer, A. Novel fold and capsid-binding properties of the l-phage display platform protein gpD. Nat. Struct. Biol. 2000, 7, 230–237. [Google Scholar] [PubMed]
- Iwasi, H.; Forrer, P.; Plückthun, A.; Güntert, P. NMR solution structure of the monomeric form of the bacteriophage l capsid stabilizing protein gpD. J. Biomol. NMR 2005, 31, 351–356. [Google Scholar]
- Lander, G.C.; Evilevitch, A.; Jeembaeva, M.; Potter, C.S.; Carragher, B.; Johnson, J.E. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location and mechanism of attachment determined by cryo-EM. Structure 2008, 16, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, N.; Weisberg, R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 1977, 117, 733–759. [Google Scholar] [CrossRef]
- Botstein, D.; Waddell, C.H.; King, J. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22: I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 1973, 80, 669–695. [Google Scholar] [CrossRef]
- Agirrezabala, X.; Velázquez-Muriel, J.A.; Gómez-Puertas, P.; Scheres, S.H.W.; Carazo, J.M.; Carrascosa, J.L. Quasi-atomic model of bacteriophage T7 procapsid shell: Insights into the structure and evolution of a basic fold. Structure 2007, 15, 461–472. [Google Scholar] [CrossRef]
- Xiang, Y.; Morais, M.C.; Battisti, A.J.; Grimes, S.; Jardine, P.J.; Anderson, D.L.; Rossmann, M.G. Structural changes of bacteriophage f29 upon DNA packaging and release. EMBO J. 2006, 25, 5229–5239. [Google Scholar] [CrossRef]
- Lhuillier, S.; Gallopin, M.; Gilquin, B.; Brasilès, S.; Lancelot, N.; Letellier, G.; Gilles, M.; Dethan, G.; Orlova, E.V.; Couprie, J.; Tavares, P.; Zinn-Justin, S. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 8507–8512. [Google Scholar] [CrossRef]
- Maxwell, K.L.; Yee, A.A.; Booth, V.; Arrowsmith, C.H.; Gold, M.; Davidson, A.R. The solution structure of bacteriophage lamdba protein W, a small morphogenetic protein possessing a novel fold. J. Mol. Biol. 2001, 308, 9–14. [Google Scholar] [CrossRef]
- Maxwell, K.L.; Yee, A.A.; Arrowsmith, C.H.; Gold, M.; Davidson, A.R. The solution structure of the bacteriophage l head-tail joining protein, gpFII. J. Mol. Biol. 2002, 318, 1395–1404. [Google Scholar] [CrossRef]
- Cardarelli, L.; Lam, R.; Tuite, A.; Baker, L.A.; Sadowski, P.D.; Radford, D.R.; Rubinstein, J.L.; Battaile, K.P.; Chirgadze, N.; Maxwell, K.L.; Davidson, A.R. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J. Mol. Biol. 2010, 395, 754–768. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, L.; Pell, L.G.; Neudecker, P.; Pirani, N.; Liu, A.; Baker, L.A.; Rubinstein, J.L.; Maxwell, K.L.; Davidson, A.R. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 14384–14389. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, K.; Tsai, C.-J.; Kumar, S.; Zanuy, D.; Nussinov, R. Extended disorder proteins: Targeting function with less scaffold. Trends Biochem. Sci. 2003, 28, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Camacho, A.; Jiménez, F.; Viñuela, E.; Salas, M. Order of assembly of the lower collar and the tail proteins of Bacillus subtilis bacteriophage f29. J. Virol. 1979, 29, 540–545. [Google Scholar] [CrossRef]
- Lander, G.C.; Khayat, R.; Li, R.; Prevelige, P.E.; Potter, C.S.; Carragher, B.; Johnson, J.E. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure 2009, 17, 789–799. [Google Scholar] [CrossRef]
- Wood, W.B.; Eiserling, F.A.; Crowther, R.A. Long tail fibers: Genes, proteins, structure, and assembly. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 282–290. [Google Scholar]
- Kikuchi, Y.; King, J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J. Mol. Biol. 1975, 99, 645–672. [Google Scholar] [CrossRef]
- Kikuchi, Y.; King, J. Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J. Mol. Biol. 1975, 99, 673–694. [Google Scholar] [CrossRef]
- Kikuchi, Y.; King, J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J. Mol. Biol. 1975, 99, 695–716. [Google Scholar] [CrossRef]
- Johnson, J.E. Functional implications of protein-protein interactions in icosahedral viruses. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 27–33. [Google Scholar] [CrossRef]
- Johnson, J.E.; Speir, J.A. Quasi-equivalent viruses: A paradigm for protein assemblies. J. Mol. Biol. 1997, 269, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Johnson, J.E. Virus assembly: Imaging a molecular machine. Curr. Biol. 1999, 9, R296–R300. [Google Scholar] [CrossRef] [PubMed]
- Aksyuk, A.A.; Leiman, P.G.; Shneider, M.M.; Mesyanzhinov, V.V.; Rossmann, M.G. The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure 2009, 17, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Morgan, G.J.; Hatfull, G.F.; Casjens, S.; Hendrix, R.W. Bacteriophage Mu genome sequence: Analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 2002, 317, 337–359. [Google Scholar] [CrossRef]
- Kondou, Y.; Kitazawa, D.; Takeda, S.; Tsuchiya, Y.; Yamashita, E.; Mizuguchi, M.; Kawano, K.; Tsukihara, T. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J. Mol. Biol. 2005, 352, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Kostyuchenko, V.A.; Leiman, P.G.; Chipman, P.R.; Kanamaru, S.; van Raaij, M.J.; Arisaka, F.; Mesyanzhinov, V.V.; Rossmann, M.G. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 2003, 10, 688–693. [Google Scholar] [CrossRef]
- Leiman, P.G.; Chipman, P.R.; Kostyuchenko, V.A.; Mesyanzhinov, V.V.; Rossmann, M.G. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 2004, 118, 419–429. [Google Scholar] [CrossRef]
- Sciara, G.; Bebeacua, C.; Bron, P.; Tremblay, D.; Ortiz-Lombardia, M.; Lichière, J.; van Heel, M.; Campanacci, V.; Moineau, S.; Cambillau, C. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 6852–6857. [Google Scholar] [CrossRef]
- Pell, L.G.; Kanelis, V.; Donaldson, L.W.; Howell, P.L.; Davidson, A.R. The phage l major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 4160–4165. [Google Scholar] [CrossRef]
- Plisson, C.; White, H.E.; Auzat, I.; Zafarani, A.; São-José, C.; Lhuillier, S.; Tavares, P.; Orlova, E.V. Structure of bacteriophage SPP1 tails reveals trigger for DNA ejection. EMBO J. 2007, 26, 3720–3728. [Google Scholar] [CrossRef]
- Kostyuchenko, V.A.; Chipman, P.R.; Leiman, P.G.; Arisaka, F.; Mesyanzhinov, V.V.; Rossmann, M.G. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 2005, 12, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Aksyuk, A.A.; Leiman, P.G.; Kurochkina, L.P.; Schneider, M.M.; Kostyuchenko, V.A.; Mesyanzhinov, V.V.; Rossmann, M.G. The tail sheath structure of bacteriophage T4: A molecular machine for infecting bacteria. EMBO J. 2009, 28, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; McPartland, J.; Kaganman, I.; Bowman, V.D.; Rothman-Denes, L.B.; Rossmann, M.G. Insight into DNA and protein transport in double-stranded DNA viruses: The structure of bacteriophage N4. J. Mol. Biol. 2008, 378, 726–736. [Google Scholar] [CrossRef]
- Wagenknecht, T.; Bloomfield, V.A. In vitro polymerization of bacteriophage T4D tail core subunits. J. Mol. Biol. 1977, 116, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Poglazov, B.F.; Nikolskaya, T.I. Self-assembly of the protein of bacteriophage T2 tail cores. J. Mol. Biol. 1969, 43, 231–233. [Google Scholar] [CrossRef]
- Auzat, I.; Dröge, A.; Weise, F.; Lurz, R.; Tavares, P. Origin and function of the two major tail proteins of bacteriophage SPP1. Mol. Microbiol. 2008, 70, 557–569. [Google Scholar] [CrossRef]
- Fraser, J.S.; Maxwell, K.L.; Davidson, A.R. Immunoglobulin-like domains on bacteriophage: Weapons of modest damage? Curr. Opin. Microbiol. 2007, 10, 382–387. [Google Scholar] [CrossRef]
- Leonard, K.R.; Kleinschmidt, A.K.; Lake, J.A. Caulobacter crescentus bacteriophage fCbK: Structure and in vitro self-assembly of the tail. J. Mol. Biol. 1973, 81, 349–365. [Google Scholar] [CrossRef]
- Pell, L.G.; Liu, A.; Edmonds, L.; Donaldson, L.W.; Howell, P.L.; Davidson, A.R. The X-ray crystal structure of the phage l tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J. Mol. Biol. 2009, 389, 938–951. [Google Scholar] [CrossRef]
- Vianelli, A.; Wang, G.R.; Gingery, M.; Duda, R.L.; Eiserling, F.A.; Goldberg, E.B. Bacteriophage T4 self-assembly: localization of gp3 and its role in determining tail length. J. Bacteriol. 2000, 182, 680–688. [Google Scholar] [CrossRef]
- Zhao, L.; Kanamaru, S.; Chaidirek, C.; Arisaka, F. P15 and P3, the tail completion proteins of bacteriophage T4, both form hexameric rings. J. Bacteriol. 2003, 185, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Katsura, I.; Hendrix, R.W. Length determination in bacteriophage lambda tails. Cell 1984, 39, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Abuladze, N.K.; Gingery, M.; Tsai, J.; Eiserling, F.A. Tail length determination in bacteriophage T4. Virology 1994, 199, 301–310. [Google Scholar] [CrossRef]
- Boulanger, P.; Jacquot, P.; Plançon, L.; Chami, M.; Engel, A.; Parquet, C.; Herbeuval, C.; Letellier, L. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J. Biol. Chem. 2008, 283, 13556–13564. [Google Scholar] [CrossRef]
- Katsura, I. Mechanism of length determination in bacteriophage lambda tails. Adv. Biophys. 1990, 26, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Königsmaier, L.; Lara-Tejero, M.; Lefebre, M.; Marlovits, T.C.; Galán, J.E. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 17745–17750. [Google Scholar] [CrossRef]
- Arisaka, F.; Tschopp, J.; van Driel, R.; Engel, J. Reassembly of the bacteriophage T4 tail from the core-baseplate and the monomeric sheath protein P18: a co-operative association process. J. Mol. Biol. 1979, 132, 369–386. [Google Scholar] [CrossRef]
- Tschopp, J.; Arisaka, F.; van Driel, R.; Engel, J. Purification, characterization, and reassembly of the bacteriophage T4D tail sheath protein P18. J. Mol. Biol. 1979, 128, 247–258. [Google Scholar] [CrossRef]
- Schneemann, A. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 2006, 60, 51–67. [Google Scholar] [CrossRef]
- Wade, R.H. On and around microtubules: an overview. Mol. Biotechnol. 2009, 43, 177–191. [Google Scholar] [CrossRef]
- Campellone, K.G.; Welch, M.D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell. Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Moody, M.F. Structure of the sheath of bacteriophage T4: I. Structure of the contracted sheath and polysheath. J. Mol. Biol. 1967, 25, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Moody, M.F. Sheath of bacteriophage T4: III. Contraction mechanism deduced from partially contracted sheaths. J. Mol. Biol. 1973, 80, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Coombs, D.H.; Arisaka, F. T4 tail structure and function. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 259–281. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T.; Wang, S.; Lindqvist, B.H. The structure of P4 procapsids produced by coexpression of capsid and external scaffolding proteins. Virology 2002, 298, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T.; McKenna, R.; Ilag, L.L.; Bowman, B.R.; Incardona, N.L.; Fane, B.A.; Rossmann, M.G. Structure of a viral procapsid with molecular scaffolding. Nature 1997, 389, 308–313. [Google Scholar] [CrossRef]
- Morais, M.C.; Fisher, M.; Kanamaru, S.; Przybyla, L.; Burgner, J.; Fane, B.A.; Rossmann, M.G. Conformational switching by the scaffolding protein D directs the assembly of bacteriophage fX174. Mol. Cell 2004, 15, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Steitz, T.A.; Anderson, W.F.; Fletterick, R.J.; Anderson, C.M. High resolution crystal structures of yeast hexokinase complexes with substrates, activators, and inhibitors. Evidence for an allosteric control site. J. Biol. Chem. 1977, 252, 4494–4500. [Google Scholar] [CrossRef]
- Cherwa, J.E., Jr.; Uchiyama, A.; Fane, B.A. Scaffolding proteins altered in the ability to perform a conformational switch confer dominant lethal assembly defects. J. Virol. 2008, 82, 5774–5780. [Google Scholar] [CrossRef]
- Cherwa, J.E., Jr.; Fane, B.A. Complete virion assembly with scaffolding proteins altered in the ability to perform a critical conformational switch. J. Virol. 2009, 83, 7391–7396. [Google Scholar] [CrossRef]
- Ilag, L.L.; Olson, N.H.; Dokland, T.; Music, C.L.; Cheng, R.H.; Bowen, Z.; McKenna, R.; Rossmann, M.G.; Baker, T.S.; Incardona, N.L. DNA packaging intermediates of bacteriophage fX174. Structure 1995, 3, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T.; Bernal, R.A.; Burch, A.; Pletnev, S.; Fane, B.A.; Rossmann, M.G. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus fX174. J. Mol. Biol. 1999, 288, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Bernal, R.A.; Hafenstein, S.; Olson, N.H.; Bowman, V.D.; Chipman, P.R.; Baker, T.S.; Fane, B.A.; Rossmann, M.G. Structural studies of bacteriophage a3 assembly. J. Mol. Biol. 2003, 325, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Bratkovič, T. Progress in phage display: evolution of the technique and its application. Cell. Mol. Life Sci. 2010, 67, 749–767. [Google Scholar] [CrossRef] [PubMed]
- Marvin, D.A. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 1998, 8, 150–158. [Google Scholar] [CrossRef]
- Russel, M.; Linderoth, N.A.; Sali, A. Filamentous phage assembly: variation on a protein export theme. Gene 1997, 192, 23–32. [Google Scholar] [CrossRef]
- Stopar, D.; Spruijt, R.B.; Wolfs, C.J.; Hemminga, M.A. Protein-lipid interactions of bacteriophage M13 major coat protein. Biochim. Biophys. Acta 2003, 1611, 5–15. [Google Scholar] [CrossRef]
- Hemminga, M.A.; Vos, W.L.; Nazarov, P.V.; Koehorst, R.B.; Wolfs, C.J.; Spruijt, R.B.; Stopar, D. Viruses: Incredible nanomachines. New advances with filamentous phages. Eur. Biophys. J. 2010, 39, 541–550. [Google Scholar] [CrossRef]
- Russel, M.; Model, P. Filamentous phage. In The Bacteriophages, 2nd ed.; Calendar, R.L., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 146–160. [Google Scholar]
- Marvin, D.A. X-ray diffraction and electron microscope studies on the structure of the small filamentous bacteriophage fd. J. Mol. Biol. 1966, 15, 8–17. [Google Scholar] [CrossRef]
- Marvin, D.A.; Hale, R.D.; Nave, C.; Helmer-Critterich, M. Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe. J. Mol. Biol. 1994, 235, 260–286. [Google Scholar] [CrossRef] [PubMed]
- Glucksman, M.J.; Bhattacharjee, S.; Makowski, L. Three-dimensional structure of a cloning vector. X-ray diffraction studies of filamentous bacteriophage M13 at 7 Å resolution. J. Mol. Biol. 1992, 226, 455–470. [Google Scholar] [CrossRef] [PubMed]
- Zeri, A.C.; Mesieh, M.F.; Nevzorov, A.A.; Opella, S.J. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 6458–6463. [Google Scholar] [CrossRef] [PubMed]
- Grant, R.A.; Lin, T.C.; Konigsberg, W.; Webster, R.E. Structure of the filamentous bacteriophage fl. Location of the A, C, and D minor coat proteins. J. Biol. Chem. 1981, 256, 539–546. [Google Scholar] [CrossRef]
- Endemann, H.; Model, P. Location of filamentous phage minor coat proteins in phage and in infected cells. J. Mol. Biol. 1995, 250, 496–506. [Google Scholar] [CrossRef]
- Chang, C.N.; Blobel, G.; Model, P. Detection of prokaryotic signal peptidase in an Escherichia coli membrane fraction: endoproteolytic cleavage of nascent fl pre-coat protein. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 361–365. [Google Scholar] [CrossRef]
- Day, L.A.; Marzec, C.J.; Reisberg, S.A.; Casadevall, A. DNA packaging in filamentous bacteriophages. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 509–539. [Google Scholar] [CrossRef]
- Hunter, G.J.; Rowitch, D.H.; Perham, R.N. Interactions between DNA and coat protein in the structure and assembly of filamentous bacteriophage fd. Nature 1987, 327, 252–254. [Google Scholar] [CrossRef]
- Kazmierczak, B.I.; Mielke, D.L.; Russel, M.; Model, P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 1994, 238, 187–198. [Google Scholar] [CrossRef]
- Russel, M. Moving through the membrane with filamentous phages. Trends Microbiol. 1995, 3, 223–228. [Google Scholar] [CrossRef]
- Abrescia, N.G.A.; Cockburn, J.J.B.; Grimes, J.M.; Sutton, G.C.; Diprose, J.M.; Butcher, S.J.; Fuller, S.D.; San Martin, C.; Burnett, R.M.; Stuart, D.I.; Bamford, D.H.; Bamford, J.K.H. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 2004, 432, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Cockburn, J.J.B.; Abrescia, N.G.A.; Grimes, J.M.; Sutton, G.C.; Diprose, J.M.; Benevides, J.M.; Thomas, G.J., Jr.; Bamford, J.K.H.; Bamford, D.H.; Stuart, D.I. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 2004, 432, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Lundström, K.H.; Bamford, D.H.; Palva, E.T.; Lounatmaa, K. Lipid-containing bacteriophage PR4: structure and life cycle. J. Gen. Virol. 1979, 43, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Bamford, D.; Mindich, L. Structure of the lipid-containing bacteriophage PRD1: Disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 1982, 44, 1031–1038. [Google Scholar] [CrossRef]
- Gowen, B.; Bamford, J.K.; Bamford, D.H.; Fuller, S.D. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J. Virol. 2003, 77, 7863–7871. [Google Scholar] [CrossRef]
- Karhu, N.J.; Ziedaite, G.; Bamford, D.H.; Bamford, J.K. Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6. J. Virol. 2007, 81, 2970–2979. [Google Scholar] [CrossRef]
- Jaatinen, S.T.; Viitanen, S.J.; Bamford, D.H.; Bamford, J.K.H. Integral membrane protein P16 of bacteriophage PRD1 stabilizes the adsorption vertex structure. J. Virol. 2004, 78, 9790–9797. [Google Scholar] [CrossRef]
- Ziedaite, G.; Kivelä, H.M.; Bamford, J.K.; Bamford, D.H. Purified membrane-containing procapsids of bacteriophage PRD1 package the viral genome. J. Mol. Biol. 2009, 386, 637–647. [Google Scholar] [CrossRef]
- Oliveira, L.; Alonso, J.C.; Tavares, P. A defined in vitro system for DNA packaging by the bacteriophage SPP1: insights into the headful packaging mechanism. J. Mol. Biol. 2005, 353, 529–539. [Google Scholar] [CrossRef]
- Fuller, D.N.; Raymer, D.M.; Kottadiel, V.I.; Rao, V.B.; Smith, D.E. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 16868–16873. [Google Scholar] [CrossRef]
- Frilander, M.; Bamford, D.H. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage f6: The three segments modulate each other’s packaging efficiency. J. Mol. Biol. 1995, 246, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Marvik, O.J.; Dokland, T.; Nøkling, R.H.; Jacobsen, E.; Larsen, T.; Lindqvist, B.H. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J. Mol. Biol. 1995, 251, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Butcher, S.J.; Bamford, D.H.; Fuller, S.D. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 1995, 14, 6078–6086. [Google Scholar] [CrossRef] [PubMed]
- Benson, S.D.; Bamford, J.K.H.; Bamford, D.H.; Burnett, R.M. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 1999, 98, 825–833. [Google Scholar] [CrossRef]
- Krupovič, M.; Bamford, D.H. Virus evolution: How far does the double b-barrel viral lineage extend? Nat. Rev. Microbiol. 2008, 6, 941–948. [Google Scholar] [CrossRef]
- Reddy, V.S.; Natchiar, S.K.; Stewart, P.L.; Nemerow, G.R. Crystal structure of human adenovirus at 3.5 Å resolution. Science 2010, 329, 1071–1075. [Google Scholar] [CrossRef]
- Liu, H.; Jin, L.; Koh, S.B.; Atanasov, I.; Schein, S.; Wu, L.; Zhou, Z.H. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 2010, 329, 1038–1043. [Google Scholar] [CrossRef]
- Ghosh-Choudhury, G.; Haj-Ahmad, Y.; Graham, F.L. Protein IX, a minor component of the human adenovirus capsid, is essential for the pakaging of full length genomes. EMBO J. 1987, 6, 1733–1739. [Google Scholar] [CrossRef]
- Sargent, K.L.; Ng, P.; Evelegh, C.; Graham, F.L.; Parks, R.J. Development of a size-restricted pIX-deleted helper virus for amplification of helper-dependent adenovirus vectors. Gene Ther. 2004, 11, 504–511. [Google Scholar] [CrossRef]
- Sen, A.; Heymann, J.B.; Cheng, N.; Qiao, J.; Mindich, L.; Steven, A.C. Initial location of the RNA-dependent RNA polymerase in the bacteriophage f6 procapsid determined by cryo-electron microscopy. J. Biol. Chem. 2008, 283, 12227–12231. [Google Scholar] [CrossRef]
- Poranen, M.M.; Tuma, R. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 2004, 101, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Nemecek, D.; Heymann, J.B.; Qiao, J.; Mindich, L.; Steven, A.C. Cryo-electron tomography of bacteriophage f6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J. Struct. Biol. 2010, 171, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Jäälinoja, H.T.; Huiskonen, J.T.; Butcher, S.J. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages f6 and f8. Structure 2007, 15, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Cheng, R.H.; Berriman, J.; Rice, W.J.; Stokes, D.L.; Katz, A.; Morgan, D.G.; Gottlieb, P. Three-dimensional structure of the enveloped bacteriophage f12: An incomplete T = 13 lattice is superposed on an enclosed T = 1 shell. PLoS ONE 2009, 4, e6850. [Google Scholar] [CrossRef] [PubMed]
- Valegård, K.; Liljas, L.; Fridborg, K.; Unge, T. The three-dimensional structure of the bacterial virus MS2. Nature 1990, 345, 36–41. [Google Scholar] [CrossRef]
- Liljas, L.; Fridborg, K.; Valegård, K.; Bundule, M.; Pumpens, P. Crystal structure of bacteriophage fr capsids at 3.5 Å resolution. J. Mol. Biol. 1994, 244, 279–290. [Google Scholar] [CrossRef]
- Golmohammadi, R.; Fridborg, K.; Bundule, M.; Valegård, K.; Liljas, L. The crystal structure of bacteriophage Qb at 3.5 Å resolution. Structure 1996, 4, 543–554. [Google Scholar] [CrossRef]
- Tars, K.; Bundule, M.; Fridborg, K.; Liljas, L. The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses. J. Mol. Biol. 1997, 271, 759–773. [Google Scholar] [CrossRef]
- Tars, K.; Fridborg, K.; Bundule, M.; Liljas, L. The three-dimensional structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7-Å resolution. Virology 2000, 272, 331–337. [Google Scholar] [CrossRef] [PubMed]
- van Duin, J.; Tsareva, N. Single-Stranded RNA Phages. In The Bacteriophages, 2nd ed.; Calendar, R.L., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 175–196. [Google Scholar]
- Beckett, D.; Wu, H.N.; Uhlenbeck, O.C. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J. Mol. Biol. 1988, 204, 939–947. [Google Scholar] [CrossRef]
- Stockley, P.G.; Rolfsson, O.; Thompson, G.S.; Basnak, G.; Francese, S.; Stonehouse, N.J.; Homans, S.W.; Ashcroft, A.E. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J. Mol. Biol. 2007, 369, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Plevka, P.; Kazaks, A.; Voronkova, T.; Kotelovica, S.; Dishlers, A.; Liljas, L.; Tars, K. The structure of bacteriophage fCb5 reveals a role of the RNA genome and metal ions in particle stability and assembly. J. Mol. Biol. 2009, 391, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Zlotnick, A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 1994, 241, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Zlotnick, A.; Aldrich, R.; Johnson, J.M.; Ceres, P.; Young, M.J. Mechanism of capsid assembly for an icosahedral plant virus. Virology 2000, 277, 450–456. [Google Scholar] [CrossRef]
- Moisant, P.; Neeman, H.; Zlotnick, A. Exploring the paths of (virus) assembly. Biophys. J. 2010, 99, 1350–1357. [Google Scholar] [CrossRef]
- Kang, S.; Prevelige, P.E., Jr. Domain study of bacteriophage P22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J. Mol. Biol. 2005, 347, 935–948. [Google Scholar] [CrossRef]
- Kodera, N.; Yamamoto, D.; Ishikawa, R.; Ando, T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 2010, 468, 72–76. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Moras, D.; Olsen, K.W. Chemical and biological evolution of a nucleotide-binding protein. Nature 1974, 250, 194–199. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2011 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).