Interaction of Host Cellular Proteins with Components of the Hepatitis Delta Virus
Abstract
:1. Introduction
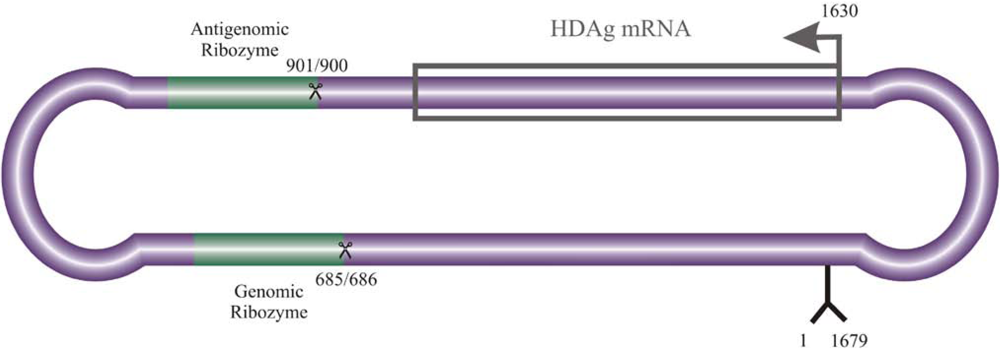
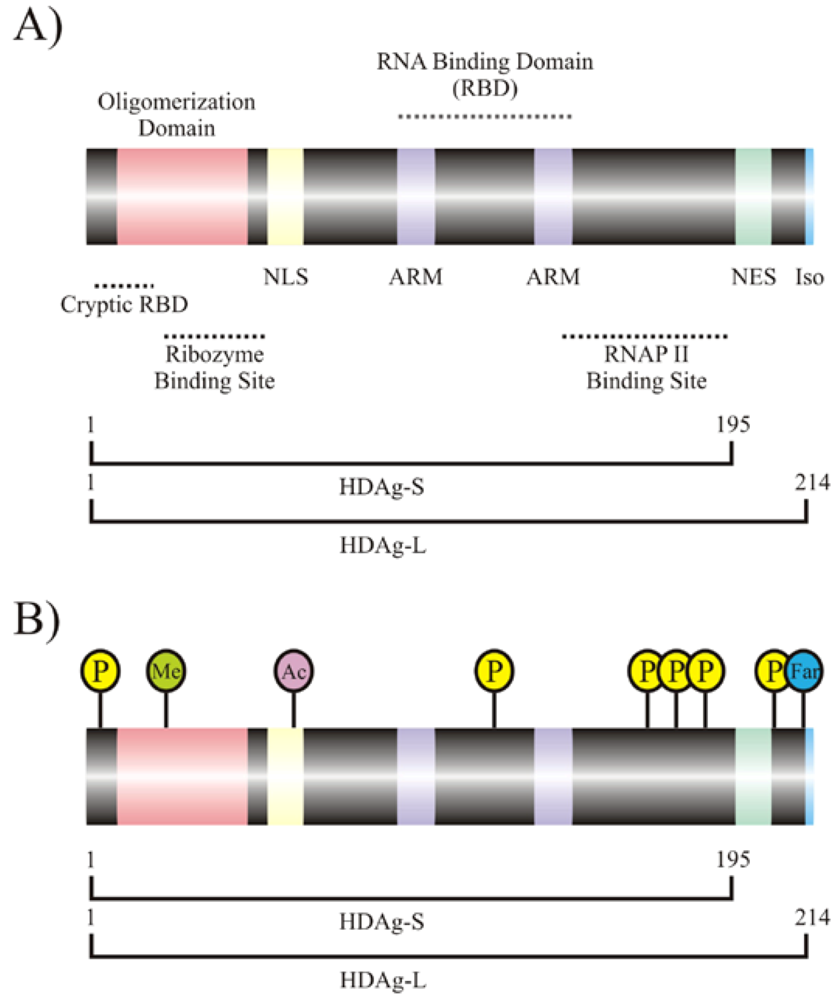
2. Interaction of host cellular proteins with HDV RNA and HDAgs
| Host protein | Proposed Function | References |
|---|---|---|
| Post translational modification | ||
| Casein Kinase II (CKII) | Phosphorylation (S2 and S213) | [22] |
| Double-stranded RNA-activated protein kinase R (PKR) | Phosphorylation (S177, S180, T182) | [23] |
| Extracellular signal-related kinases 1 and 2 (ERK1/2) | Phosphorylation (S177) | [24] |
| Protein Kinase C (PKC) a | Phosphorylation (S210) | [22] |
| Protein farnesyltransferase (FTase) a | Isoprenylation with farnesyl (C211) | [25-28] |
| Protein arginine methyltransferase 1 (PRMT1) | Methylation (R13) | [29] |
| p300 cellular acetyltransferase | Acetylation (K72) | [30,31] |
| Small ubiquitin-related modifier isoform 1 (SUMO1) b | Sumoylation of multiple lysine residues | [32] |
| Ubc9b | Sumoylation of multiple lysine residues | [32] |
| Sub-cellular localization | ||
| karyopherin (importin) 2α | Nuclear import | [33] |
| Nuclear export signal-interacting protein (NESI)a | Nuclear export | [34] |
| Clathrin heavy chaina | Exocytosis | [35,36] |
| Nucleolin (C23) | Nucleolar localization, shuttling, RNA synthesis/accumulation (?) | [37] |
| Nucleophosmin (B23) | Nucleolar localization, shuttling, RNA synthesis/accumulation (?) | [38] |
| RNA synthesis | ||
| RNAP II (Rpb1/2 mobile clamp element) | Genome and mRNA synthesis | [39,40] |
| Antigenome synthesis (?) | ||
| DRB sensitivity-inducing factor (DSIF) | Relieves transcriptional repression; stimulates elongation by RNAP II | [12] |
| delta interacting protein A | Transcriptional regulation (?) | [41] |
| Yin Yang 1 (YY1) | RNA synthesis/accumulation (?) | [30] |
| Histone H1eb | RNA synthesis/accumulation (?) | [42] |
| Other | ||
| MOV10 | RNA remodelling (?) | [43] |
| Smad3 | Alters host gene expression | [44] |
| c-Jun | Alters host gene expression | [44] |
| TRAF2 | Alters host gene expression | [45] |
2.1. Interaction of HDAgs with host proteins
| Host protein | Proposed Function | References |
|---|---|---|
| Adenosine deaminase acting on RNA (ADAR 1) | Post-transcriptional modification of the antigenome that results in production of HDAg-L | [15] |
| Glyceraldehydes 3-phosphate dehydrogenase (GAPDH) | Enhances delta ribozyme activity | [58,59] |
| Double-stranded RNA-activated protein kinase R (PKR) | Recruitment to HDAg for post-translational modification Repression of antiviral response | [60,61] |
| RNAP I | Antigenome synthesis | [62] |
| RNAP II | Genome synthesis, mRNA synthesis, Antigenome synthesis | [63-66] |
| RNAP III | Unknown | [62] |
| Polypyrimidine tract-binding protein associated splicing factor (PSF) | Suspected involvement in recruitment of HDV RNA to RNAP II | [67] |
| 54 kDa nuclear RNA-binding protein (p54nrb) | Unknown | [59] |
| Heterogeneous nuclear ribonucleoprotein L (hnRNPL) | Unknown | [59] |
| Arginine/serine-rich splicing factor (ASF) | Unknown | [59] |
| Eukaryotic elongation factor 1A1 (eEF1A1) | Unknown | [59] |
2.2. HDAg interaction with host transcription factors: A possible role for HDV in host gene regulation
2.3. HDV interacts with eukaryotic RNAPs for transcription and replication
2.4. Interaction of HDV RNA with host proteins
3. Concluding Remarks
Acknowledgments
References
- Chen, P.J.; Kalpana, G.; Goldberg, J.; Mason, W.; Werner, B.; Gerin, J.; Taylor, J. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 1986, 83, 8774–8778. [Google Scholar] [CrossRef]
- Lai, M.M. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 2005, 79, 7951–7958. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.M. Chapter 3. Replication of the hepatitis delta virus RNA genome. Adv. Virus Res. 2009, 74, 103–121. [Google Scholar] [PubMed]
- Taylor, J.M. Hepatitis delta virus. Virology 2006, 344, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Lai, M.C. Hepatitis Delta Virus RNA Replication. Viruses 2009, 1, 818–831. [Google Scholar] [CrossRef]
- Branch, A.D.; Robertson, H.D. A replication cycle for viroids and other small infectious RNA's. Science 1984, 223, 450–455. [Google Scholar] [PubMed]
- Macnaughton, T.B.; Shi, S.T.; Modahl, L.E.; Lai, M.M. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 2002, 76, 3920–3927. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.E.; Lazinski, D.W. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 2000, 97, 424–429. [Google Scholar] [CrossRef]
- Kuo, M.Y.; Goldberg, J.; Coates, L.; Mason, W.; Gerin, J.; Taylor, J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 1988, 62, 1855–1861. [Google Scholar] [PubMed]
- Weiner, A.J.; Choo, Q.L.; Wang, K.S.; Govindarajan, S.; Redeker, A.G.; Gerin, J.L.; Houghton, M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 1988, 62, 594–599. [Google Scholar] [PubMed]
- Kuo, M.Y.; Chao, M.; Taylor, J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 1989, 63, 1945–1950. [Google Scholar] [PubMed]
- Yamaguchi, Y.; Filipovska, J.; Yano, K.; Furuya, A.; Inukai, N.; Narita, T.; Wada, T.; Sugimoto, S.; Konarska, M.M.; Handa, H. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 2001, 293, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.L.; Bergmann, K.F.; Brown, T.L.; Gerin, J.L. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. USA 1992, 89, 7149–7153. [Google Scholar] [CrossRef]
- Casey, J.L.; Gerin, J.L. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 1995, 69, 7593–7600. [Google Scholar] [PubMed]
- Wong, S.K.; Lazinski, D.W. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 2002, 99, 15118–15123. [Google Scholar] [CrossRef]
- Lee, C.Z.; Chen, P.J.; Chen, D.S. Large hepatitis delta antigen in packaging and replication inhibition: role of the carboxyl-terminal 19 amino acids and amino-terminal sequences. J. Virol. 1995, 69, 5332–5336. [Google Scholar] [PubMed]
- Chang, F.L.; Chen, P.J.; Tu, S.J.; Wang, C.J.; Chen, D.S. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 1991, 88, 8490–8494. [Google Scholar] [CrossRef]
- Ryu, W.S.; Bayer, M.; Taylor, J. Assembly of hepatitis delta virus particles. J. Virol. 1992, 66, 2310–2315. [Google Scholar] [PubMed]
- Sureau, C.; Moriarty, A.M.; Thornton, G.B.; Lanford, R.E. Production of infectious hepatitis delta virus in vitro and neutralization with antibodies directed against hepatitis B virus pre-S antigens. J. Virol. 1992, 66, 1241–1245. [Google Scholar] [PubMed]
- Sureau, C. The role of the HBV envelope proteins in the HDV replication cycle. Curr. Top. Microbiol. Immunol. 2006, 307, 113–131. [Google Scholar] [PubMed]
- Chao, M.; Hsieh, S.Y.; Taylor, J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 1990, 64, 5066–5069. [Google Scholar] [PubMed]
- Yeh, T.S.; Lo, S.J.; Chen, P.J.; Lee, Y.H. Casein kinase II and protein kinase C modulate hepatitis delta virus RNA replication but not empty viral particle assembly. J. Virol. 1996, 70, 6190–6198. [Google Scholar] [PubMed]
- Chen, C.W.; Tsay, Y.G.; Wu, H.L.; Lee, C.H.; Chen, D.S.; Chen, P.J. The double-stranded RNA-activated kinase, PKR, can phosphorylate hepatitis D virus small delta antigen at functional serine and threonine residues. J. Biol. Chem. 2002, 277, 33058–33067. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Huang, W.H.; Hong, S.Y.; Tsay, Y.G.; Chen, P.J. ERK1/2-mediated phosphorylation of small hepatitis delta antigen at serine 177 enhances hepatitis delta virus antigenomic RNA replication. J. Virol. 2008, 82, 9345–9358. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.S.; Watson, J.A.; Havel, C.M.; White, J.M. Identification of a prenylation site in delta virus large antigen. Science 1992, 256, 1331–1333. [Google Scholar] [PubMed]
- Otto, J.C.; Casey, P.J. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J. Biol. Chem. 1996, 271, 4569–4572. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.B.; Lai, M.M. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J. Virol. 1994, 68, 2958–2964. [Google Scholar] [PubMed]
- Lee, C.Z.; Chen, P.J.; Lai, M.M.; Chen, D.S. Isoprenylation of large hepatitis delta antigen is necessary but not sufficient for hepatitis delta virus assembly. Virology 1994, 199, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Stallcup, M.R.; Lai, M.M. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 2004, 78, 13325–13334. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Mai, R.T.; Lee, Y.H. Transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate hepatitis delta virus RNA replication. J. Virol. 2008, 82, 7313–7324. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.J.; Tsay, Y.G.; Juan, L.J.; Fu, T.F.; Huang, W.H.; Chen, D.S.; Chen, P.J. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 2004, 319, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Cheng, T.S.; Shu, C.Y.; Jeng, K.S.; Lai, M.M. Modification of small hepatitis delta virus antigen by SUMO protein. J. Virol. 2010, 84, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Moroianu, J.; Hijikata, M.; Blobel, G.; Radu, A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 1995, 92, 6532–6536. [Google Scholar] [CrossRef]
- Wang, Y.H.; Chang, S.C.; Huang, C.; Li, Y.P.; Lee, C.H.; Chang, M.F. Novel nuclear export signal-interacting protein, NESI, critical for the assembly of hepatitis delta virus. J. Virol. 2005, 79, 8113–8120. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chang, S.C.; Yu, I.C.; Tsay, Y.G.; Chang, M.F. Large hepatitis delta antigen is a novel clathrin adaptor-like protein. J. Virol. 2007, 81, 5985–5994. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Huang, C.R.; Chao, M.; Lo, S.J. The C-terminal sequence of the large hepatitis delta antigen is variable but retains the ability to bind clathrin. Virol. J. 2009, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Chang, S.C.; Chen, C.J.; Chang, M.F. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 1998, 273, 7650–7656. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Yung, B.Y.; Syu, W.J.; Lee, Y.H. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 2001, 276, 25166–25175. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Mura, T.; Chanarat, S.; Okamoto, S.; Handa, H. Hepatitis delta antigen binds to the clamp of RNA polymerase II and affects transcriptional fidelity. Genes Cells 2007, 12, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Haussecker, D.; Huang, Y.; Kay, M.A. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA 2009, 15, 1971–1979. [Google Scholar] [CrossRef] [PubMed]
- Brazas, R.; Ganem, D. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science 1996, 274, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Z.; Sheu, J.C. Histone H1e interacts with small hepatitis delta antigen and affects hepatitis delta virus replication. Virology 2008, 375, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D.; Cao, D.; Huang, Y.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat. Struct. Mol. Biol. 2008, 15, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Jeong, S.H.; Hwang, S.B. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology 2007, 132, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Park, C.Y.; Oh, S.H.; Kang, S.M.; Lim, Y.S.; Hwang, S.B. Hepatitis delta virus large antigen sensitizes to TNF-alpha-induced NF-kappaB signaling. Mol. Cells 2009, 28, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.F.; Baker, S.C.; Soe, L.H.; Kamahora, T.; Keck, J.G.; Makino, S.; Govindarajan, S.; Lai, M.M. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 1988, 62, 2403–2410. [Google Scholar] [PubMed]
- Mu, J.J.; Wu, H.L.; Chiang, B.L.; Chang, R.P.; Chen, D.S.; Chen, P.J. Characterization of the phosphorylated forms and the phosphorylated residues of hepatitis delta virus delta antigens. J. Virol. 1999, 73, 10540–10545. [Google Scholar] [PubMed]
- Mu, J.J.; Chen, D.S.; Chen, P.J. The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 2001, 75, 9087–9095. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Jeng, K.S.; Lai, M.M. Transcription of subgenomic mRNA of hepatitis delta virus requires a modified hepatitis delta antigen that is distinct from antigenomic RNA synthesis. J. Virol. 2008, 82, 9409–9416. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.S.; Lee, Y.H. Assembly of hepatitis delta virus particles: package of multimeric hepatitis delta virus genomic RNA and role of phosphorylation. Virology 1998, 249, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Bernier-Villamor, V.; Sampson, D.A.; Matunis, M.J.; Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 2002, 108, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Boggio, R.; Chiocca, S. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 2006, 9, 430–436. [Google Scholar] [CrossRef]
- Zhao, J. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 2007, 64, 3017–3033. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.; Freitas, N.; Cunha, C. Characterization of the nuclear localization signal of the hepatitis delta virus antigen. Virology 2008, 370, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.P.; Yeh, C.T.; Ou, J.H.; Lai, M.M. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol. 1992, 66, 914–921. [Google Scholar] [PubMed]
- Chou, H.C.; Hsieh, T.Y.; Sheu, G.T.; Lai, M.M. Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J. Virol. 1998, 72, 3684–3690. [Google Scholar] [PubMed]
- Lee, C.H.; Chang, S.C.; Wu, C.H.; Chang, M.F. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 2001, 276, 8142–8148. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.S.; Chang, S.C.; Wang, Y.H.; Sun, C.Y.; Chang, M.F. Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: an enhancement on ribozyme catalysis. Virology 2000, 271, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Sikora, D.; Greco-Stewart, V.S.; Miron, P.; Pelchat, M. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology 2009, 390, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Circle, D.A.; Neel, O.D.; Robertson, H.D.; Clarke, P.A.; Mathews, M.B. Surprising specificity of PKR binding to delta agent genomic RNA. RNA 1997, 3, 438–448. [Google Scholar] [PubMed]
- Robertson, H.D.; Manche, L.; Mathews, M.B. Paradoxical interactions between human delta hepatitis agent RNA and the cellular protein kinase PKR. J. Virol. 1996, 70, 5611–5617. [Google Scholar] [PubMed]
- Greco-Stewart, V.S.; Schissel, E.; Pelchat, M. The hepatitis delta virus RNA genome interacts with the human RNA polymerases I and III. Virology 2009, 386, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Greco-Stewart, V.S.; Miron, P.; Abrahem, A.; Pelchat, M. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virology 2007, 357, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Nie, X.; Chang, H.E.; Han, Z.; Taylor, J. Transcription of hepatitis delta virus RNA by RNA polymerase II. J. Virol. 2008, 82, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Abrahem, A.; Pelchat, M. Formation of an RNA polymerase II preinitiation complex on an RNA promoter derived from the hepatitis delta virus RNA genome. Nucleic Acids Res. 2008, 36, 5201–5211. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, E.; Brueckner, F.; Cramer, P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 2007, 450, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Greco-Stewart, V.S.; Thibault, C.S.; Pelchat, M. Binding of the polypyrimidine tract-binding protein-associated splicing factor (PSF) to the hepatitis delta virus RNA. Virology 2006, 356, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, R.; Tuteja, N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 1998, 33, 407–436. [Google Scholar] [CrossRef] [PubMed]
- Ghisolfi, L.; Joseph, G.; Amalric, F.; Erard, M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 1992, 267, 2955–2959. [Google Scholar] [PubMed]
- Han, Z.; Alves, C.; Gudima, S.; Taylor, J. Intracellular localization of hepatitis delta virus proteins in the presence and absence of viral RNA accumulation. J. Virol. 2009, 83, 6457–6463. [Google Scholar] [CrossRef] [PubMed]
- Hiscox, J.A. The nucleolus--a gateway to viral infection? Arch. Virol. 2002, 147, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Okuwaki, M. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J. Biochem. 2008, 143, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, A.; Herrera, J.E.; Olson, M.O. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry 1995, 34, 8037–8042. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, A.; Mehrotra, B.; Baumann, A.; Adam, S.A.; Wingfield, P.T.; Olson, M.O. Nucleolar protein B23 stimulates nuclear import of the HIV-1 Rev protein and NLS-conjugated albumin. Biochemistry 1997, 36, 3941–3949. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Chromatin structure: a repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [PubMed]
- Kornberg, R.D.; Thomas, J.O. Chromatin structure; oligomers of the histones. Science 1974, 184, 865–868. [Google Scholar] [PubMed]
- Happel, N.; Doenecke, D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 2009, 431, 1–12. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Bezy, O.; Elabd, C.; Cochet, O.; Petersen, R.K.; Kristiansen, K.; Dani, C.; Ailhaud, G.; Amri, E.Z. Delta-interacting protein A, a new inhibitory partner of CCAAT/enhancer-binding protein beta, implicated in adipocyte differentiation. J. Biol. Chem. 2005, 280, 11432–11438. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, Q.; Hirohashi, Y.; Greene, M.I. DIPA, which can localize to the centrosome, associates with p78/MCRS1/MSP58 and acts as a repressor of gene transcription. Exp. Mol. Pathol. 2006, 81, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Veretnik, S.; Gribskov, M. RNA binding domain of HDV antigen is homologous to the HMG box of SRY. Arch. Virol. 1999, 144, 1139–1158. [Google Scholar] [CrossRef] [PubMed]
- Mota, S.; Mendes, M.; Penque, D.; Coelho, A.V.; Cunha, C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens. J. Proteomics 2008, 71, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.; Sheu, G.T.; Lai, M.M. Inhibition of Cellular RNA polymerase II transcription by delta antigen of hepatitis delta virus. Virology 1998, 247, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Ganem, D. Activation of heterologous gene expression by the large isoform of hepatitis delta antigen. J. Virol. 1998, 72, 2089–2096. [Google Scholar] [PubMed]
- Goto, T.; Kato, N.; Ono-Nita, S.K.; Yoshida, H.; Otsuka, M.; Shiratori, Y.; Omata, M. Large isoform of hepatitis delta antigen activates serum response factor-associated transcription. J. Biol. Chem. 2000, 275, 37311–37316. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Kato, N.; Yoshida, H.; Otsuka, M.; Moriyama, M.; Shiratori, Y.; Koike, K.; Matsumura, M.; Omata, M. Synergistic activation of the serum response element-dependent pathway by hepatitis B virus x protein and large-isoform hepatitis delta antigen. J. Infect. Dis. 2003, 187, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Williams, V.; Brichler, S.; Radjef, N.; Lebon, P.; Goffard, A.; Hober, D.; Fagard, R.; Kremsdorf, D.; Deny, P.; Gordien, E. Hepatitis delta virus proteins repress hepatitis B virus enhancers and activate the alpha/beta interferon-inducible MxA gene. J. Gen. Virol. 2009, 90, 2759–2767. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.C.; Chen, P.J.; Kuo, M.Y.; Lee, S.D.; Chen, D.S.; Ting, L.P. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J. Virol. 1991, 65, 1099–1104. [Google Scholar] [PubMed]
- Derynck, R.; Zhang, Y.; Feng, X.H. Smads: transcriptional activators of TGF-beta responses. Cell 1998, 95, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Gallagher, E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 2005, 57, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Trougakos, I.P.; Gonos, E.S. Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in ageing and age-related diseases. Free Radic. Res. 2006, 40, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.T.; Lee, Y.J.; Ko, J.L.; Tsai, C.C.; Tseng, C.J.; Sheu, G.T. Hepatitis delta virus epigenetically enhances clusterin expression via histone acetylation in human hepatocellular carcinoma cells. J. Gen. Virol. 2009, 90, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.B.; Taylor, J. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 1993, 67, 6965–6972. [Google Scholar] [PubMed]
- MacNaughton, T.B.; Gowans, E.J.; McNamara, S.P.; Burrell, C.J. Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology 1991, 184, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Chang, J.; Taylor, J.M. Alternative processing of hepatitis delta virus antigenomic RNA transcripts. J. Virol. 2004, 78, 4517–4524. [Google Scholar] [CrossRef] [PubMed]
- Gudima, S.; Wu, S.Y.; Chiang, C.M.; Moraleda, G.; Taylor, J. Origin of hepatitis delta virus mRNA. J. Virol. 2000, 74, 7204–7210. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.Y.; Chao, M.; Coates, L.; Taylor, J. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J. Virol. 1990, 64, 3192–3198. [Google Scholar] [PubMed]
- Bichko, V.V.; Taylor, J.M. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J. Virol. 1996, 70, 8064–8070. [Google Scholar] [PubMed]
- Beard, M.R.; MacNaughton, T.B.; Gowans, E.J. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J. Virol. 1996, 70, 4986–4995. [Google Scholar] [PubMed]
- Filipovska, J.; Konarska, M.M. Specific HDV RNA-templated transcription by pol II in vitro. RNA 2000, 6, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.Y.; Chen, P.J. Phosphorylation of Serine 177 of the Small Hepatitis Delta Antigen Regulates Viral Antigenomic RNA Replication by Interacting with the Processive RNA Polymerase II. J. Virol. 2009. [Google Scholar] [CrossRef]
- Nedialkov, Y.A.; Gong, X.Q.; Hovde, S.L.; Yamaguchi, Y.; Handa, H.; Geiger, J.H.; Yan, H.; Burton, Z.F. NTP-driven translocation by human RNA polymerase II. J. Biol. Chem. 2003, 278, 18303–18312. [Google Scholar] [CrossRef] [PubMed]
- Modahl, L.E.; Macnaughton, T.B.; Zhu, N.; Johnson, D.L.; Lai, M.M. RNA-Dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 2000, 20, 6030–6039. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Macnaughton, T.; Gao, L.; Lai, M.M. RNA-templated replication of hepatitis delta virus: genomic and antigenomic RNAs associate with different nuclear bodies. J. Virol. 2006, 80, 6478–6486. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Chen, Y.S.; Chen, P.J. Nucleolar targeting of hepatitis delta antigen abolishes its ability to initiate viral antigenomic RNA replication. J. Virol. 2008, 82, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Niranjanakumari, S.; Lasda, E.; Brazas, R.; Garcia-Blanco, M.A. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 2002, 26, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Chang, M.F.; Baker, S.C.; Govindarajan, S.; Lai, M.M. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J. Virol. 1990, 64, 4051–4058. [Google Scholar] [PubMed]
- Lee, C.Z.; Lin, J.H.; Chao, M.; McKnight, K.; Lai, M.M. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol. 1993, 67, 2221–2227. [Google Scholar] [PubMed]
- Poisson, F.; Roingeard, P.; Baillou, A.; Dubois, F.; Bonelli, F.; Calogero, R.A.; Goudeau, A. Characterization of RNA-binding domains of hepatitis delta antigen. J. Gen. Virol. 1993, 74, 2473–2478. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Chang, T.C.; Lin, C.W.; Tsui, H.L.; Chu, P.B.; Chen, B.S.; Huang, Z.S.; Wu, H.N. Nucleic acid binding properties of the nucleic acid chaperone domain of hepatitis delta antigen. Nucleic Acids Res. 2003, 31, 6481–6492. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.S.; Netter, H.J.; Bayer, M.; Taylor, J. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 1993, 67, 3281–3287. [Google Scholar] [PubMed]
- Lin, B.C.; Defenbaugh, D.A.; Casey, J.L. Multimerization of hepatitis delta antigen is a critical determinant of RNA binding specificity. J. Virol. 2009. [Google Scholar]
- Chao, Y.C.; Chang, M.F.; Gust, I.; Lai, M.M. Sequence conservation and divergence of hepatitis delta virus RNA. Virology 1990, 178, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.; Hsieh, S.Y.; Taylor, J. The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J. Virol. 1991, 65, 4057–4062. [Google Scholar] [PubMed]
- Polson, A.G.; Bass, B.L.; Casey, J.L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 1996, 380, 454–456. [Google Scholar] [CrossRef] [PubMed]
- Polson, A.G.; Ley 3rd., H.L.; Bass, B.L.; Casey, J.L. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 1998, 18, 1919–1926. [Google Scholar] [PubMed]
- Sirover, M.A. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell. Biochem. 2005, 95, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Roeder, R.G.; Luo, Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 2003, 114, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M.; Jespersen, L. Enhancement of hammerhead ribozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. J. Mol. Biol. 1996, 257, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Manche, L.; Green, S.R.; Schmedt, C.; Mathews, M.B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992, 12, 5238–5248. [Google Scholar] [PubMed]
- Brands, J.H.; Maassen, J.A.; van Hemert, F.J.; Amons, R.; Moller, W. The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur. J. Biochem. 1986, 155, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. Translation elongation factor-1 alpha interacts with the 3' stem-loop region of West Nile virus genomic RNA. J. Virol. 1997, 71, 6433–6444. [Google Scholar] [PubMed]
- Joshi, R.L.; Ravel, J.M.; Haenni, A.L. Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. EMBO J. 1986, 5, 1143–1148. [Google Scholar] [PubMed]
- Das, T.; Mathur, M.; Gupta, A.K.; Janssen, G.M.; Banerjee, A.K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc. Natl. Acad. Sci. USA 1998, 95, 1449–1454. [Google Scholar] [CrossRef]
- Harris, K.S.; Xiang, W.; Alexander, L.; Lane, W.S.; Paul, A.V.; Wimmer, E. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994, 269, 27004–27014. [Google Scholar] [PubMed]
- Wu-Baer, F.; Lane, W.S.; Gaynor, R.B. Identification of a group of cellular cofactors that stimulate the binding of RNA polymerase II and TRP-185 to human immunodeficiency virus 1 TAR RNA. J. Biol. Chem. 1996, 271, 4201–4208. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.M. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 1998, 244, 1–12. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Zipori, D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002, 531, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Horowitz, D.S.; Kobayashi, R.; Krainer, A.R. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993, 21, 4085–4092. [Google Scholar] [CrossRef] [PubMed]
- Emili, A.; Shales, M.; McCracken, S.; Xie, W.; Tucker, P.W.; Kobayashi, R.; Blencowe, B.J.; Ingles, C.J. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 2002, 8, 1102–1111. [Google Scholar] [CrossRef]
- Greco-Stewart, V.S.; Pelchat, M. Unpublished work. University of Ottawa: Ottawa, Ontario, Canada, 2010. [Google Scholar]
- Clark, J.; Lu, Y.J.; Sidhar, S.K.; Parker, C.; Gill, S.; Smedley, D.; Hamoudi, R.; Linehan, W.M.; Shipley, J.; Cooper, C.S. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 1997, 15, 2233–2239. [Google Scholar] [PubMed]
- Benn, D.E.; Dwight, T.; Richardson, A.L.; Delbridge, L.; Bambach, C.P.; Stowasser, M.; Gordon, R.D.; Marsh, D.J.; Robinson, B.G. Sporadic and familial pheochromocytomas are associated with loss of at least two discrete intervals on chromosome 1p. Cancer Res. 2000, 60, 7048–7051. [Google Scholar] [PubMed]
- Li, L.; Feng, T.; Lian, Y.; Zhang, G.; Garen, A.; Song, X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 12956–12961. [Google Scholar] [CrossRef]
- Song, X.; Sui, A.; Garen, A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 621–626. [Google Scholar] [CrossRef]
- Song, X.; Sun, Y.; Garen, A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl. Acad. Sci. USA 2005, 102, 12189–12193. [Google Scholar] [CrossRef]
- Wang, G.; Cui, Y.; Zhang, G.; Garen, A.; Song, X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 16794–16798. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Greco-Stewart, V.; Pelchat, M. Interaction of Host Cellular Proteins with Components of the Hepatitis Delta Virus. Viruses 2010, 2, 189-212. https://doi.org/10.3390/v2010189
Greco-Stewart V, Pelchat M. Interaction of Host Cellular Proteins with Components of the Hepatitis Delta Virus. Viruses. 2010; 2(1):189-212. https://doi.org/10.3390/v2010189
Chicago/Turabian StyleGreco-Stewart, Valerie, and Martin Pelchat. 2010. "Interaction of Host Cellular Proteins with Components of the Hepatitis Delta Virus" Viruses 2, no. 1: 189-212. https://doi.org/10.3390/v2010189
APA StyleGreco-Stewart, V., & Pelchat, M. (2010). Interaction of Host Cellular Proteins with Components of the Hepatitis Delta Virus. Viruses, 2(1), 189-212. https://doi.org/10.3390/v2010189




