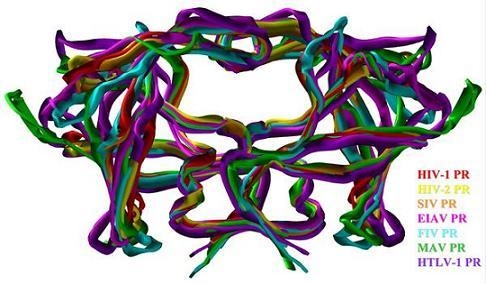
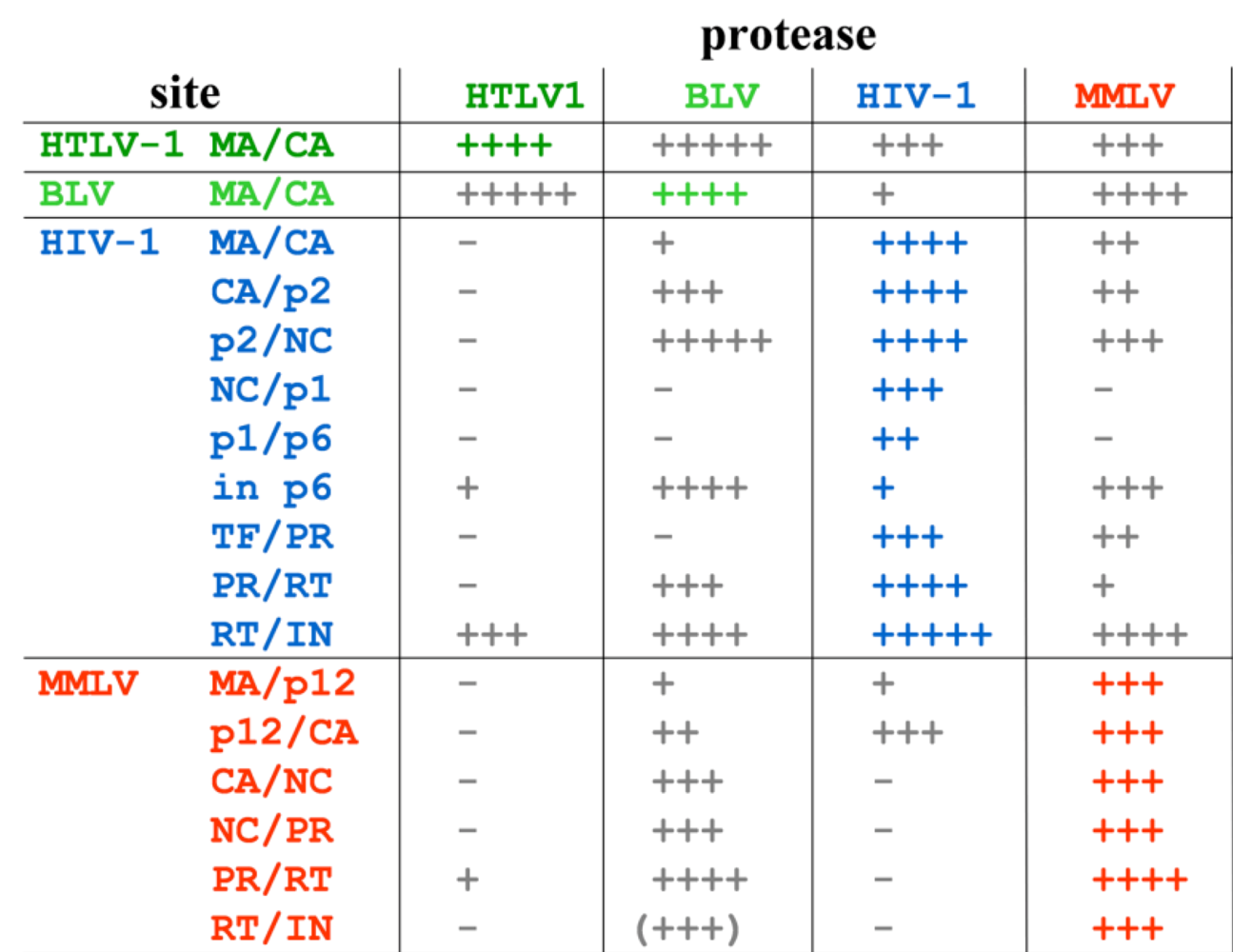
Comparative Studies on Retroviral Proteases: Substrate Specificity
Abstract
:1. Introduction
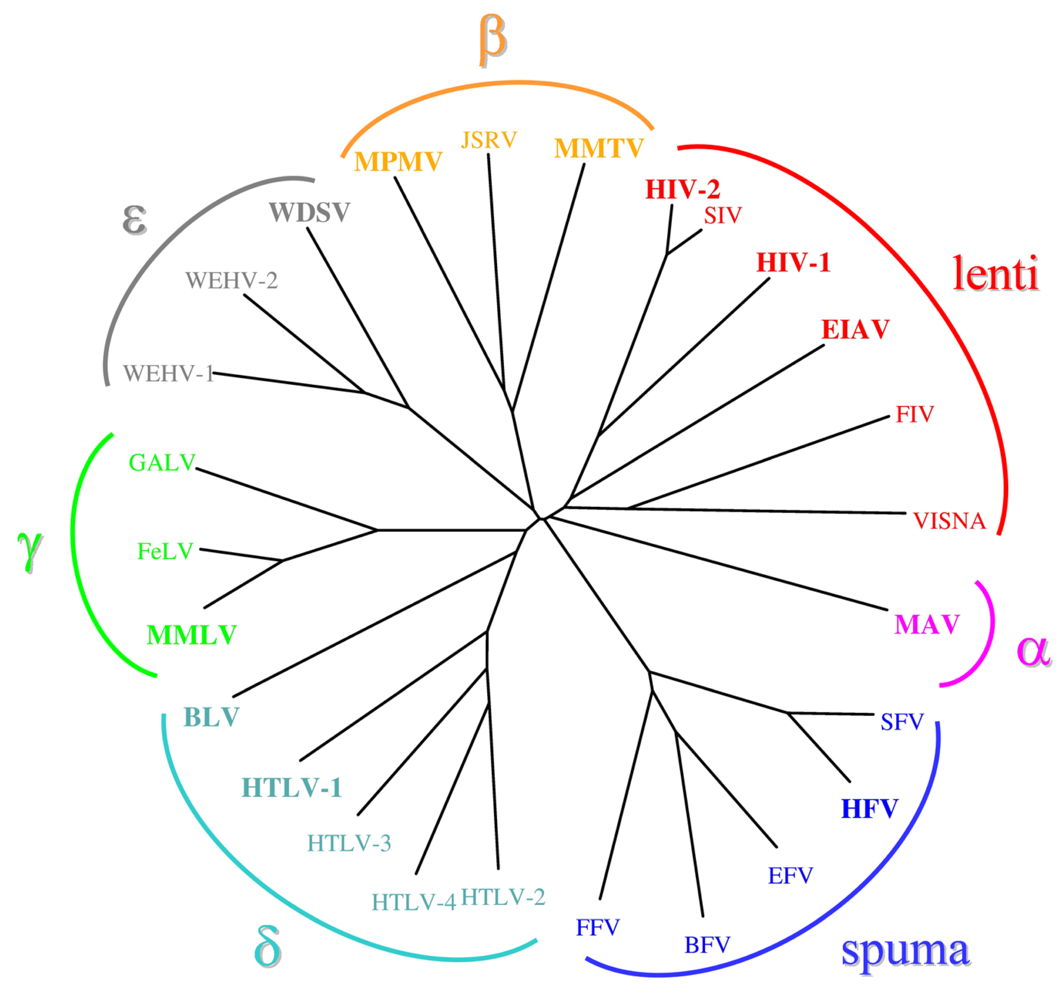
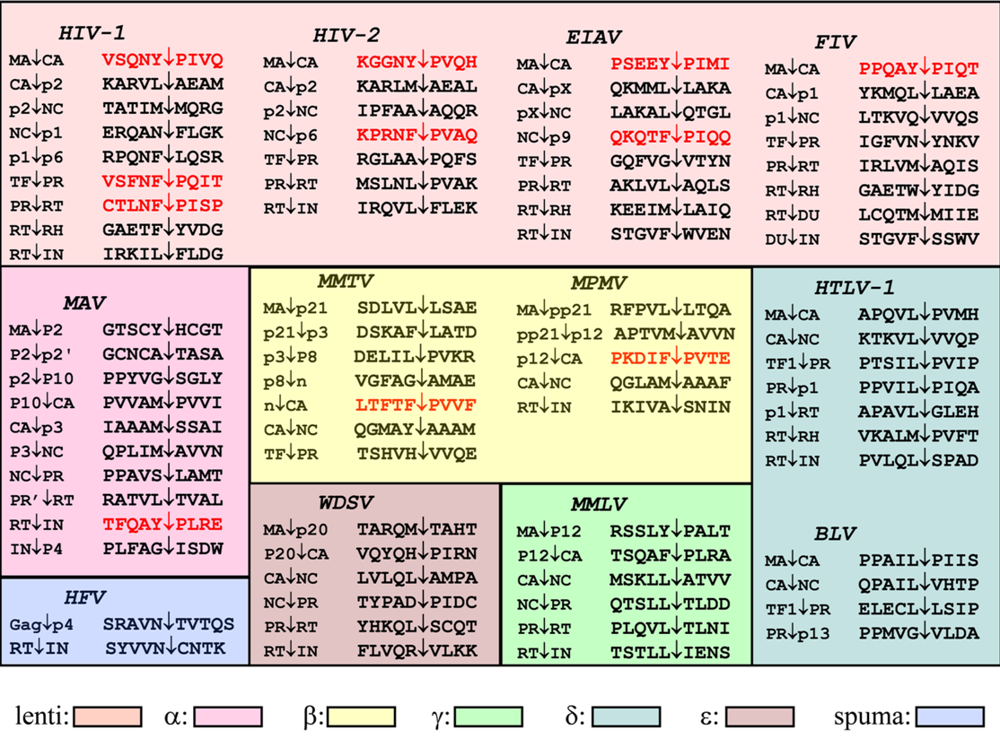
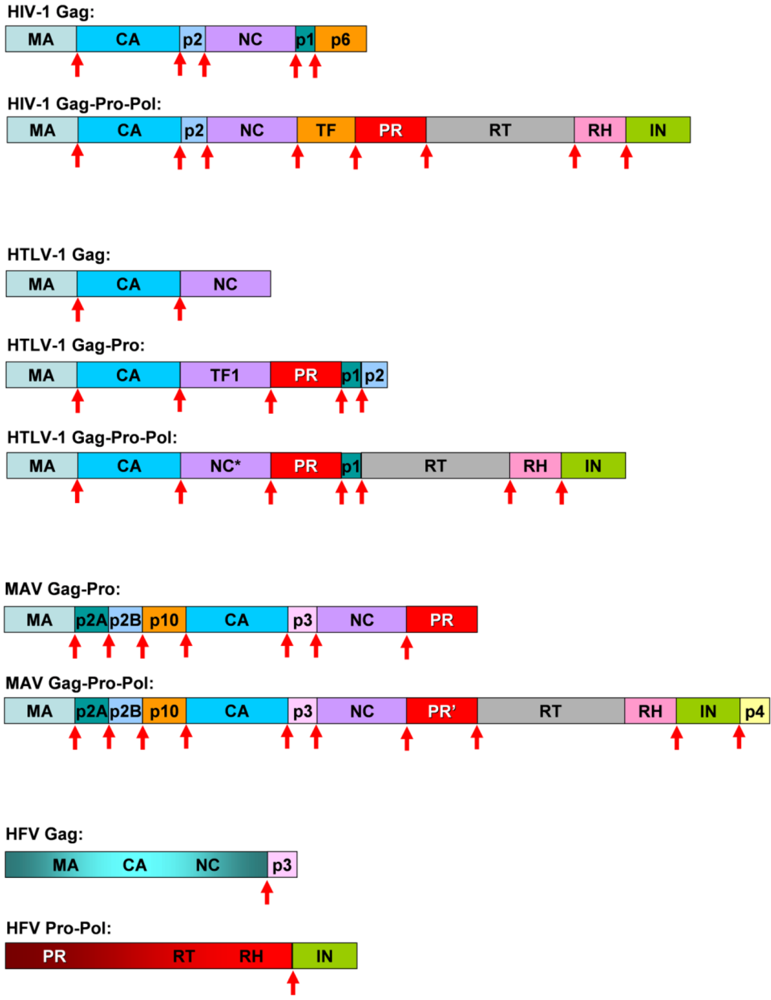

2. HIV-1 protease
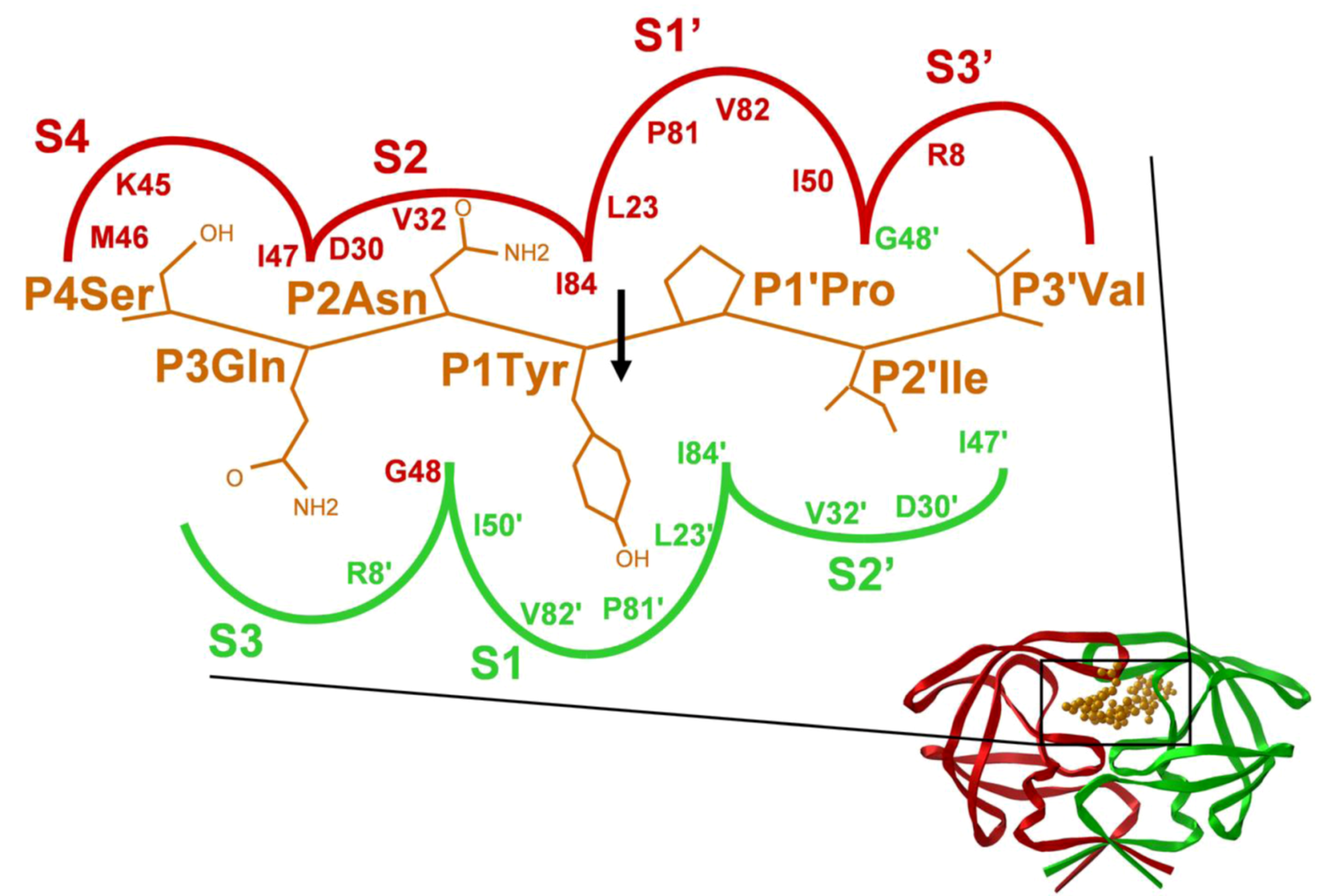
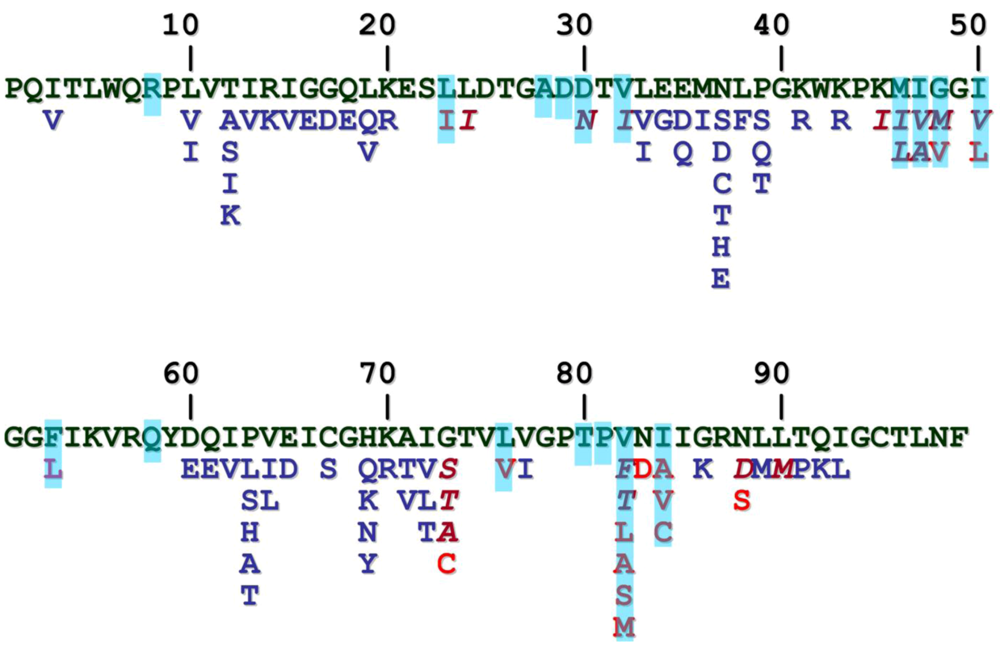
3. HIV-2 protease
Acknowledgments
Referneces
- Weiss, R.A. The discovery of endogenous retroviruses. Retrovirology 2006, 3, 67. [Google Scholar] [CrossRef] [PubMed]
- Oroszlan, S.; Luftig, R.B. Retroviral proteinases. Curr. Top. Microbiol. Immunol. 1990, 157, 153–185. [Google Scholar] [PubMed]
- Henderson, L.E.; Benveniste, R.E.; Sowder, R.; Copeland, T.D.; Schultz, A.M.; Oroszlan, S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne). J. Virol. 1988, 62, 2587–2595. [Google Scholar] [PubMed]
- Pettit, S.C.; Simsic, J.; Loeb, D.D.; Everitt, L.; Hutchison 3rd, C.A.; Swanstrom, R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 1991, 266, 14539–14547. [Google Scholar] [PubMed]
- Griffiths, J.T.; Phylip, L.H.; Konvalinka, J.; Strop, P.; Gustchina, A.; Wlodawer, A.; Davenport, R.J.; Briggs, R.; Dunn, B.M.; Kay, J. Different requirements for productive interaction between the active site of HIV-1 proteinase and substrates containing -hydrophobic*hydrophobic- or -aromatic*pro- cleavage sites. Biochemistry 1992, 31, 5193–51200. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J.; Bagossi, P.; Weber, I.T.; Louis, J.M.; Copeland, T.D.; Oroszlan, S. Studies on the symmetry and sequence context dependence of the HIV-1 proteinase specificity. J. Biol. Chem. 1997, 272, 16807–16814. [Google Scholar] [CrossRef] [PubMed]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Amino- and carboxyl-terminal amino acid sequences of proteins coded by gag gene of murine leukemia virus. Proc. Natl. Acad. Sci. USA 1978, 75, 1404–1408. [CrossRef]
- Weber, I.T.; Agniswamy, J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses 2009, 1, 1110–1136. [Google Scholar] [CrossRef]
- Dunn, B.M.; Goodenow, M.M.; Gustchina, A.; Wlodawer, A. Retroviral proteases. Genome Biol. 2002, 3, reviews3006.1–reviews3006.7. [Google Scholar] [CrossRef]
- Tomasselli, A.G.; Heinrikson, R.L. Specificity of retroviral proteases: an analysis of viral and nonviral protein substrates. Methods Enzymol. 1994, 241, 279–301. [Google Scholar] [PubMed]
- Dunn, B.M.; Gustchina, A.; Wlodawer, A.; Kay, J. Subsite preferences of retroviral proteinases. Methods Enzymol. 1994, 241, 254–278. [Google Scholar] [PubMed]
- Louis, J.M.; Weber, I.T.; Tözsér, J.; Clore, G.M.; Gronenborn, A.M. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 2000, 49, 111–146. [Google Scholar] [PubMed]
- Tözsér, J.; Oroszlan, S. Proteolytic events of HIV-1 replication as targets for therapeutic intervention. Curr. Pharm. Des. 2003, 9, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Moody, M.D.; Wehbie, R.S.; Kaplan, A.H.; Nantermet, P.V.; Klein, C.A.; Swanstrom, R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994, 68, 8017–8027. [Google Scholar] [PubMed]
- Boross, P.; Bagossi, P.; Copeland, T.D.; Oroszlan, S.; Louis, J.M.; Tözsér, J. Effect of substrate residues on the P2' preference of retroviral proteinases. Eur. J. Biochem. 1999, 264, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Prabu-Jeyabalan, M.; Nalivaika, E.; Schiffer, C.A. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 2002, 10, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Tie, Y.; Boross, P.I.; Wang, Y.F.; Gaddis, L.; Liu, F.; Chen, X.; Tözsér, J.; Harrison, R.W.; Weber, I.T. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2005, 272, 5265–5277. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J.; Weber, I.T.; Gustchina, A.; Bláha, I.; Copeland, T.D.; Louis, J.M.; Oroszlan, S. Kinetic and modeling studies of S3-S3' subsites of HIV proteinases. Biochemistry 1992, 31, 4793–4800. [Google Scholar] [CrossRef] [PubMed]
- Shafer, R.W.; Schapiro, J.M. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008, 10, 67–84. [Google Scholar] [PubMed]
- Weber, I.T.; Zhang, Y.; Tözsér, J. HIV-1 Protease and AIDS Therapy. Lendeckel U.; Hooper, M.N., Ed.; Springer: Berlin, Germany, 2009; pp. 25–45. [Google Scholar]
- Gulnik, S.V.; Suvorov, L.I.; Liu, B.; Yu, B.; Anderson, B.; Mitsuya, H.; Erickson, J.W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 1995, 34, 9282–9287. [Google Scholar] [CrossRef] [PubMed]
- King, N.M.; Prabu-Jeyabalan, M.; Nalivaika, E.A.; Schiffer, C.A. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 2004, 11, 1333–1338. [Google Scholar] [PubMed]
- Bock, P.J.; Markovitz, D.M. Infection with HIV-2. AIDS 2001, 15, S35–S45. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J.; Bláha, I.; Copeland, T.D.; Wondrak, E.M.; Oroszlan, S. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 1991, 281, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.C.; Carr, S.F.; Jarnagin, K.; Kirsher, S.; Barnett, J.; Chow, J.; Chan, H.W.; Chen, M.S.; Medzihradszky, D.; Yamashiro, D.; et al. Synthetic HIV-2 protease cleaves the GAG precursor of HIV-1 with the same specificity as HIV-1 protease. Arch. Biochem. Biophys. 1990, 277, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Pichuantes, S.; Babé, L.M.; Barr, P.J.; DeCamp, D.L.; Craik, C.S. Recombinant HIV2 protease processes HIV1 Pr53gag and analogous junction peptides in vitro. J. Biol. Chem. 1990, 265, 13890–13898. [Google Scholar] [PubMed]
- Tomasselli, A.G.; Hui, J.O.; Sawyer, T.K.; Staples, D.J.; Bannow, C.; Reardon, I.M.; Howe, W.J.; DeCamp, D.L.; Craik, C.S.; Heinrikson, R.L. Specificity and inhibition of proteases from human immunodeficiency viruses 1 and 2. J. Biol. Chem. 1990, 265, 14675–14683. [Google Scholar] [PubMed]
- Poorman, R.A.; Tomasselli, A.G.; Heinrikson, R.L.; Kézdy, F.J. A cumulative specificity model for proteases from human immunodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate database. J. Biol. Chem. 1991, 266, 14554–14561. [Google Scholar] [PubMed]
- Tözsér, J.; Gustchina, A.; Weber, I.T.; Blaha, I.; Wondrak, E.M.; Oroszlan S. Studies on the role of the S4 substrate binding site of HIV proteinases. FEBS Lett. 1991, 279, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Bagossi, P.; Cheng, Y.S.; Oroszlan, S.; Tözsér, J. Comparison of the specificity of homo- and heterodimeric linked HIV-1 and HIV-2 proteinase dimers. Protein Eng. 1998, 11, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Brower, E.T.; Bacha, U.M.; Kawasaki, Y.; Freire, E. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem Biol Drug Des. 2008, 71, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L.; Tözsér, J. HIV-1 protease inhibitors: effects on HIV-2 replication and resistance. Trends Pharmacol Sci. 2008, 29, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Tomasselli, A.G.; Bannow, C.A.; Deibel Jr., M.R.; Hui, J.O.; Zurcher-Neely, H.A.; Reardon, I.M.; Smith, C.W.; Heinrikson, R.L. Chemical synthesis of a biotinylated derivative of the simian immunodeficiency virus protease. Purification by avidin affinity chromatography and autocatalytic activation. J. Biol. Chem. 1992, 267, 10232–10237. [Google Scholar] [PubMed]
- Rushlow, K.; Peng, X.X.; Montelaro, R.C.; Shih, D.S. Expression of the protease gene of equine infectious anemia virus in Escherichia coli: formation of the mature processed enzyme and specific cleavage of the gag precursor. Virology 1992, 188, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Luukkonen, B.G.; Tan, W.; Fenyö, E.M.; Schwartz, S. Analysis of cross reactivity of retrovirus proteases using a vaccinia virus-T7 RNA polymerase-based expression system. J. Gen. Virol. 1995, 76, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.J.; Bur, D.; Wlodawer, A.; Gustchina, A.; Payne, S.L.; Dunn, B.M.; Kay, J. Expression, characterisation and mutagenesis of the aspartic proteinase from equine infectious anaemia virus. Eur. J. Biochem. 1996, 241, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Weber, I.T.; Tözsér, J.; Wu, J.; Friedman, D.; Oroszlan S. Molecular model of equine infectious anemia virus proteinase and kinetic measurements for peptide substrates with single amino acid substitutions. Biochemistry 1993, 32, 3354–3362. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J.; Friedman, D.; Weber, I.T.; Bláha, I.; Oroszlan, S. Studies on the substrate specificity of the proteinase of equine infectious anemia virus using oligopeptide substrates. Biochemistry 1993, 32, 3347–3353. [Google Scholar] [CrossRef] [PubMed]
- Wlodawer, A.; Gustchina, A.; Reshetnikova, L.; Lubkowski, J.; Zdanov, A.; Hui, K.Y.; Angleton, E.L.; Farmerie, W.G.; Goodenow, M.M.; Bhatt, D.; et al. Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus. Nat. Struct. Biol. 1995, 2, 480–488. [Google Scholar] [CrossRef]
- Lin, Y.C.; Beck, Z.; Lee, T.; Le, V.D.; Morris, G.M.; Olson, A.J.; Wong, C.H.; Elder, J.H. Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease. J. Virol. 2000, 74, 4710–4720. [Google Scholar] [CrossRef] [PubMed]
- Beck, Z.Q.; Lin, Y.C.; Elder, J.H. Molecular basis for the relative substrate specificity of human immunodeficiency virus type 1 and feline immunodeficiency virus proteases. J. Virol. 2001, 75, 9458–9469. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Beck, Z.; Morris, G.M.; Olson, A.J.; Elder, J.H. Structural basis for distinctions between substrate and inhibitor specificities for feline immunodeficiency virus and human immunodeficiency virus proteases. J. Virol. 2003, 77, 6589–6600. [Google Scholar] [CrossRef] [PubMed]
- Schnölzer, M.; Rackwitz, H.R.; Gustchina, A.; Laco, G.S.; Wlodawer, A.; Elder, J.H.; Kent, S.B. Comparative properties of feline immunodeficiency virus (FIV) and human immunodeficiency virus type 1 (HIV-1) proteinases prepared by total chemical synthesis. Virology 1996, 224, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Bagossi, P.; Sperka, T.; Fehér, A.; Kádas, J.; Zahuczky, G.; Miklóssy, G.; Boross, P.; Tözsér, J. Amino acid preferences for a critical substrate binding subsite of retroviral proteases in type 1 cleavage sites. J. Virol. 2005, 79, 4213–4218. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Jaskólski, M.; Rao, J.K.; Leis, J.; Wlodawer, A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature 1989, 337, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Konvalinka, J.; Horejsí, M.; Andreánsky, M.; Novek, P.; Pichová, I.; Bláha, I.; Fábry, M.; Sedlácek, J.; Foundling, S.; Strop, P. An engineered retroviral proteinase from myeloblastosis associated virus acquires pH dependence and substrate specificity of the HIV-1 proteinase. EMBO J. 1992, 11, 1141–1144. [Google Scholar] [PubMed]
- Grinde, B.; Cameron, C.E.; Leis, J.; Weber, I.T.; Wlodawer, A.; Burstein, H.; Bizub, D.; Skalka, A.M. Mutations that alter the activity of the Rous sarcoma virus protease. J. Biol. Chem. 1992, 267, 9481–9490. [Google Scholar] [PubMed]
- Grinde, B.; Cameron, C.E.; Leis, J.; Weber, I.T.; Wlodawer, A.; Burstein, H.; Skalka AM. Analysis of substrate interactions of the Rous sarcoma virus wild type and mutant proteases and human immunodeficiency virus-1 protease using a set of systematically altered peptide substrates. J. Biol. Chem. 1992, 267, 9491–9498. [Google Scholar] [PubMed]
- Cameron, C.E.; Grinde, B.; Jacques, P.; Jentoft, J.; Leis, J.; Wlodawer, A.; Weber, I.T. Comparison of the substrate-binding pockets of the Rous sarcoma virus and human immunodeficiency virus type 1 proteases. J. Biol. Chem. 1993, 268, 11711–11720. [Google Scholar] [PubMed]
- Cameron, C.E.; Ridky, T.W.; Shulenin, S.; Leis, J.; Weber, I.T.; Copeland, T.; Wlodawer, A.; Burstein, H.; Bizub-Bender, D.; Skalka, A.M. Mutational analysis of the substrate binding pockets of the Rous sarcoma virus and human immunodeficiency virus-1 proteases. J. Biol. Chem. 1994, 269, 11170–11177. [Google Scholar] [PubMed]
- Strop, P.; Konvalinka, J.; Stys, D.; Pavlickova, L.; Blaha, I.; Velek, J.; Travnicek, M.; Kostka, V.; Sedlacek, J. Specificity studies on retroviral proteinase from myeloblastosis-associated virus. Biochemistry 1991, 30, 3437–3443. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J.; Bagossi, P.; Weber, I.T; Copeland, T.D.; Oroszlan, S. Comparative studies on the substrate specificity of avian myeloblastosis virus proteinase and lentiviral proteinases. J. Biol. Chem. 1996, 271, 6781–6788. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L.; Young, M.; Oroszlan, S. Purification and characterization of the mouse mammary tumor virus protease expressed in Escherichia coli. J. Biol. Chem. 1992, 267, 24134–24139. [Google Scholar] [PubMed]
- Hrusková-Heidingsfeldová, O.; Andreansky, M.; Fábry, M.; Bláha, I.; Strop, P.; Hunter, E. Cloning, bacterial expression, and characterization of the Mason-Pfizer monkey virus proteinase. J. Biol. Chem. 1995, 270, 15053–15058. [Google Scholar] [CrossRef] [PubMed]
- Rumlová, M.; Ruml, T.; Pohl, J.; Pichová, I. Specific in vitro cleavage of Mason-Pfizer monkey virus capsid protein: evidence for a potential role of retroviral protease in early stages of infection. Virology 2003, 310, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J.; Shulenin, S.; Kádas, J.; Boross, P.; Bagossi, P.; Copeland, T.D.; Nair, B.C.; Sarngadharan, M.G.; Oroszlan, S. Human immunodeficiency virus type 1 capsid protein is a substrate of the retroviral proteinase while integrase is resistant toward proteolysis. Virology 2003, 310, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaka, Y.; Katoh, I.; Copeland, T.D.; Oroszlan, S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. USA 1985, 82, 1618–1622. [Google Scholar] [CrossRef]
- Menendez-Arias, L.; Tözsér, J.; Oroszlan, S. Moloney murine leukemia virus retropepsin, 2ndBarrett, A.J., Rawlings, N.D., Woessner, J.F., Eds.; Elsevier: London, UK, 2004; pp. 176–178. [Google Scholar]
- Menendez-Arias, L.; Gotte, D.; Oroszlan, S. Moloney murine leukemia virus protease: bacterial expression and characterization of the purified enzyme. Virology 1993, 196, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A.; Boross, P.; Sperka, T.; Miklóssy, G.; Kádas, J.; Bagossi, P.; Oroszlan, S.; Weber, I.T.; Tözsér, J. Characterization of the murine leukemia virus protease and its comparison with the human immunodeficiency virus type 1 protease. J. Gen. Virol. 2006, 87, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L.; Weber, I.T.; Soss, J.; Harrison, R.W.; Gotte, D.; Oroszlan, S. Kinetic and modeling studies of subsites S4-S3' of Moloney murine leukemia virus protease. J. Biol. Chem. 1994, 269, 16795–16801. [Google Scholar] [PubMed]
- Cannon, K.; Qin, L.; Schumann, G.; Boeke, J.D. Moloney murine leukemia virus protease expressed in bacteria is enzymatically active. Arch. Virol. 1998, 143, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Harrod, R.; Franchini, G. Molecular biology and pathogenesis of the human T-cell leukaemia/lymphotropic virus Type-1 (HTLV-1). Int. J. Exp. Pathol. 2001, 82, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Hrusková-Heidingsfeldová, O.; Bláha, I.; Urban, J.; Strop, P.; Pichová, I. Substrates and inhibitors of human T-cell leukemia virus type 1 (HTLV-1) proteinase. Leukemia 1997, 11, 45–46. [Google Scholar] [PubMed]
- Sperka, T.; Miklóssy, G.; Tie, Y.; Bagossi, P.; Zahuczky, G.; Boross, P.; Matúz, K.; Harrison, R.W.; Weber, I.T.; Tözsér, J. Bovine leukemia virus protease: comparison with human T-lymphotropic virus and human immunodeficiency virus proteases. J. Gen. Virol. 2007, 88, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Kádas, J.; Weber, I.T.; Bagossi, P.; Miklóssy, G.; Boross, P.; Oroszlan, S.; Tözsér, J. Narrow substrate specificity and sensitivity toward ligand-binding site mutations of human T-cell Leukemia virus type 1 protease. J. Biol. Chem. 2004, 279, 27148–27157. [Google Scholar] [CrossRef] [PubMed]
- Zahuczky, G.; Boross, P.; Bagossi, P.; Emri, G.; Copeland, T.D.; Oroszlan, S.; Louis, J.M.; Tözsér, J. Cloning of the bovine leukemia virus proteinase in Escherichia coli and comparison of its specificity to that of human T-cell leukemia virus proteinase. Biochim. Biophys. Acta. 2000, 1478, 1–8. [Google Scholar] [PubMed]
- Flügel, R.M.; Pfrepper, K.I. Proteolytic processing of foamy virus Gag and Pol proteins. Curr. Top. Microbiol. Immunol. 2003, 277, 63–88. [Google Scholar] [PubMed]
- Khan, A.S. Simian foamy virus infection in humans: prevalence and management. Expert Rev. Anti Infect. Ther. 2009, 7, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Pfrepper, K.I.; Flügel, R.M. Molecular characterization of proteolytic processing of the Gag proteins of human spumaretrovirus. Methods Mol. Biol. 2005, 304, 435–444. [Google Scholar] [PubMed]
- Roy, J.; Linial, M.L. Role of the foamy virus Pol cleavage site in viral replication. J. Virol. 2007, 81, 4956–4962. [Google Scholar] [CrossRef] [PubMed]
- Konvalinka, J.; Löchelt, M.; Zentgraf, H.; Flügel, R.M.; Kräusslich, H.G. Active foamy virus proteinase is essential for virus infectivity but not for formation of a Pol polyprotein. J. Virol. 1995, 69, 7264–7268. [Google Scholar] [PubMed]
- Fenyöfalvi, G.; Bagossi, P.; Copeland, T.D.; Oroszlan, S.; Boross, P.; Tözsér, J. Expression and characterization of human foamy virus proteinase. FEBS Lett. 1999, 462, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Eizert, H.; Bander, P.; Bagossi, P.; Sperka, T.; Miklóssy, G.; Boross, P.; Weber, I.T.; Tözsér, J. Amino acid preferences of retroviral proteases for amino-terminal positions in a type 1 cleavage site. J. Virol. 2008, 82, 10111–10117. [Google Scholar] [CrossRef] [PubMed]
- Fodor, S.K.; Vogt, V.M. Characterization of the protease of a fish retrovirus, walleye dermal sarcoma virus. J. Virol. 2002, 76, 4341–4349. [Google Scholar] [CrossRef] [PubMed]
- Holzschu, D.L.; Martineau, D.; Fodor, S.K.; Vogt, V.M.; Bowser, P.R.; Casey, J.W. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J. Virol. 1995, 69, 5320–5331. [Google Scholar] [PubMed]
- Konvalinka, J.; Heuser, A.M.; Hruskova-Heidingsfeldova, O.; Vogt, V.M.; Sedlacek, J.; Strop, P.; Kräusslich, H.G. Proteolytic processing of particle-associated retroviral polyproteins by homologous and heterologous viral proteinases. Eur. J. Biochem. 1995, 228, 191–188. [Google Scholar] [CrossRef] [PubMed]
- Snásel, J.; Shoeman, R.; Horejsí, M.; Hrusková-Heidingsfeldová, O.; Sedlácek, J.; Ruml, T.; Pichová, I. Cleavage of vimentin by different retroviral proteases Arch. Biochem. Biophys. 2000, 377, 241–245. [Google Scholar] [CrossRef]
- Alvarez, E.; Castelló, A.; Menéndez-Arias, L.; Carrasco, L. HIV protease cleaves poly(A)-binding protein. Biochem. J. 2006, 396, 219–226. [Google Scholar] [CrossRef] [PubMed]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Tözsér, J. Comparative Studies on Retroviral Proteases: Substrate Specificity. Viruses 2010, 2, 147-165. https://doi.org/10.3390/v2010147
Tözsér J. Comparative Studies on Retroviral Proteases: Substrate Specificity. Viruses. 2010; 2(1):147-165. https://doi.org/10.3390/v2010147
Chicago/Turabian StyleTözsér, József. 2010. "Comparative Studies on Retroviral Proteases: Substrate Specificity" Viruses 2, no. 1: 147-165. https://doi.org/10.3390/v2010147
APA StyleTözsér, J. (2010). Comparative Studies on Retroviral Proteases: Substrate Specificity. Viruses, 2(1), 147-165. https://doi.org/10.3390/v2010147