Evaluation of Porcine Gastric Mucin-Based Method for Extraction of Noroviruses from Seaweed Salad
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Seaweed Samples
2.3. Artificial Inoculation
2.4. PGM-MB Preparation
2.5. Extraction Methodologies
2.5.1. ISO 15216–1
2.5.2. Magnetic Silica Beads
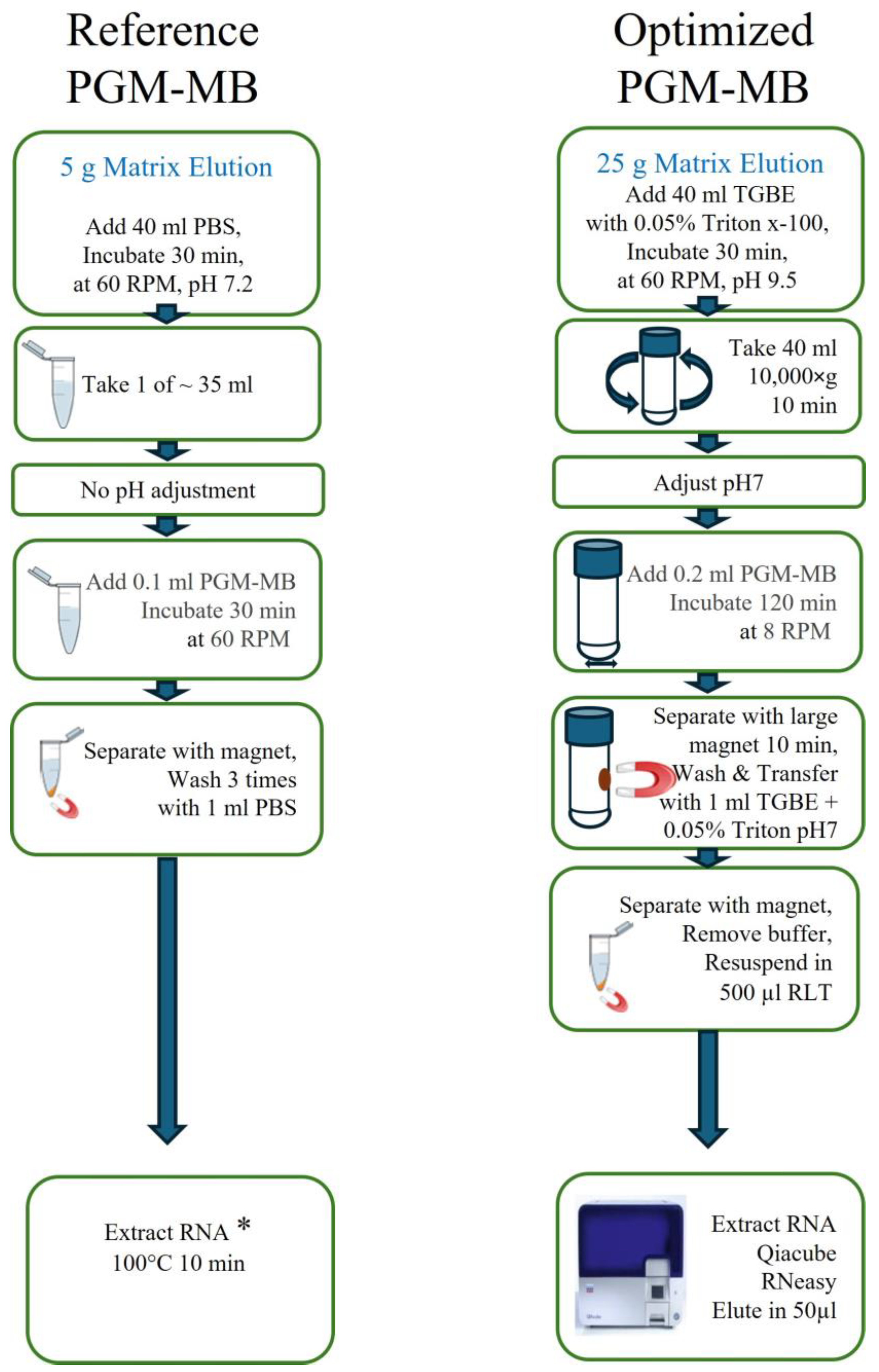
2.5.3. Reference PGM-MB Protocol
2.5.4. PGM-MB Protocol Optimization
2.5.5. The Optimized PGM-MB Protocol
2.6. RT-qPCR Detection
2.7. Recovery Rates
2.8. Sensitivity
2.9. Inhibition Measurement
2.10. Robustness
2.10.1. Analyst
2.10.2. PGM-MB Preparation
2.10.3. Competitor
2.10.4. Genotype
2.10.5. Seaweed Salad
2.11. Matrix Extension
2.12. Statistical Analysis
3. Results
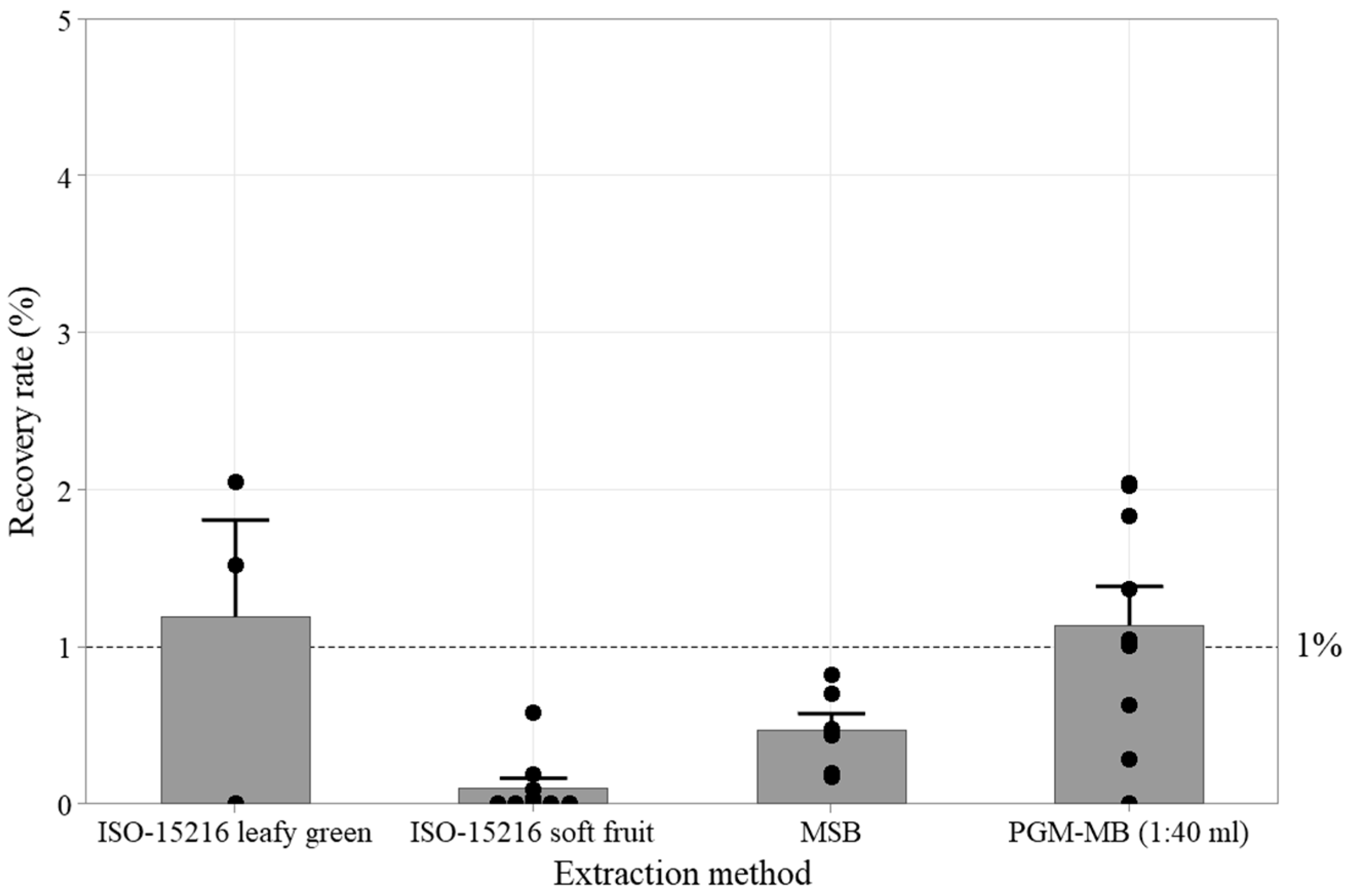
3.1. Reference Protocol Recovery Rates
3.2. PGM-MB Optimization
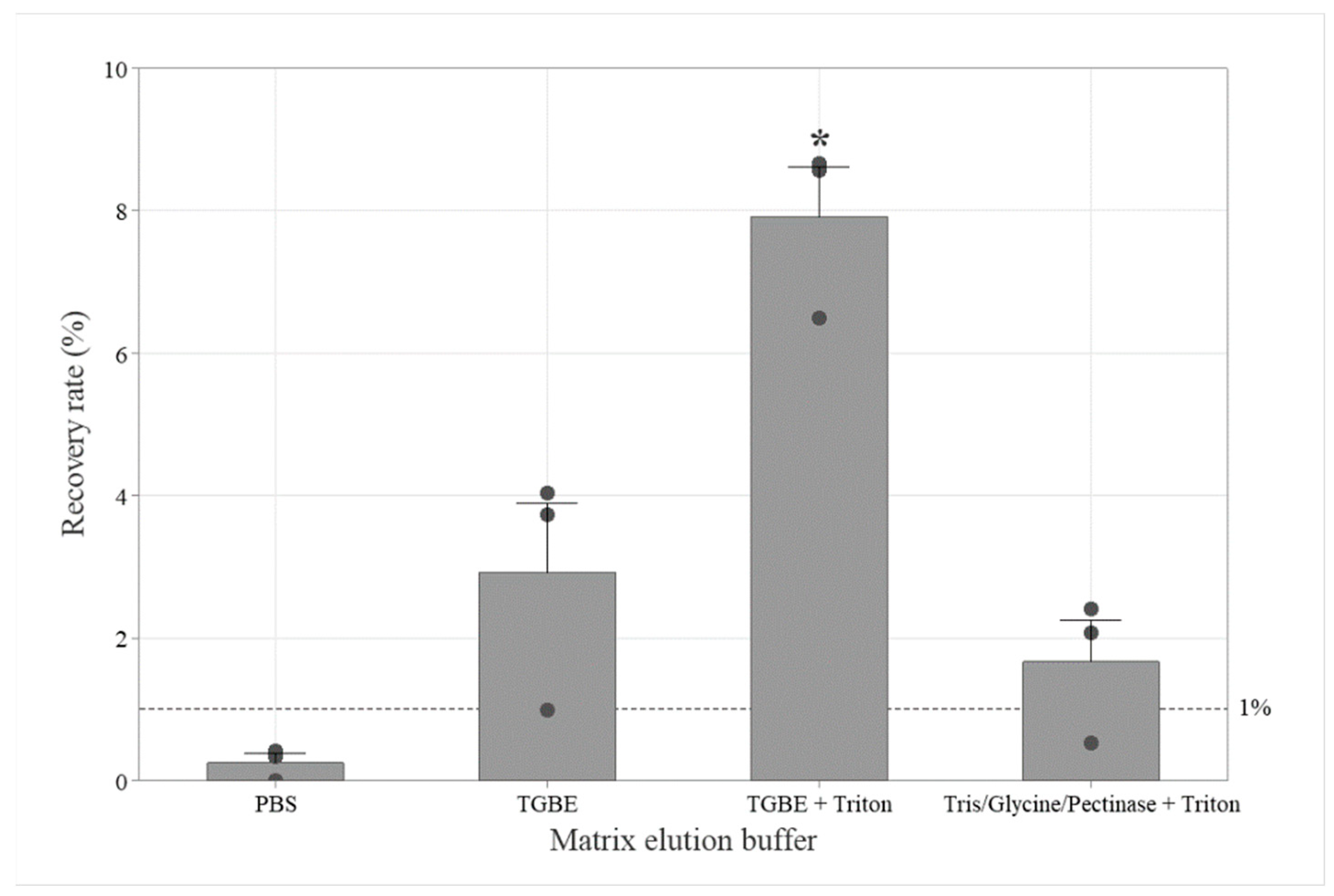
3.2.1. Buffer Selection
3.2.2. The PGM-MB Ratio
3.2.3. The pH Adjustment
3.2.4. Optimization of the Centrifugation Time
3.2.5. Virus Incubation Time
3.2.6. PGM-MB Aging
3.2.7. PGM Sources
3.3. Validation
3.3.1. LOD
3.3.2. Inhibition
3.3.3. Robustness
Inter-Analyst Repeatability
Competition
Genotype
Seaweed Salad Brands
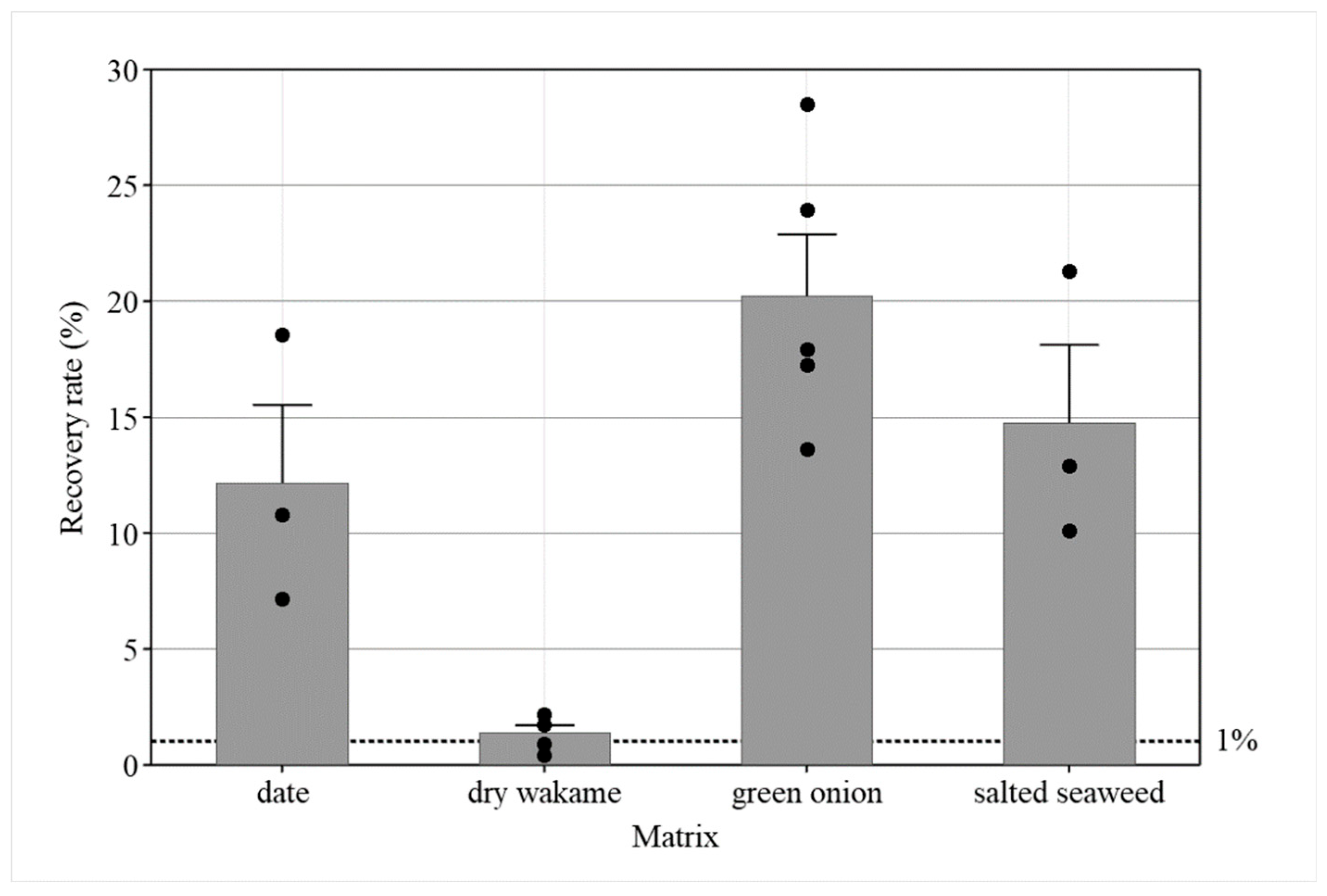
3.4. Extension
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Ludwig-Begall, L.F.; Mauroy, A.; Thiry, E. Noroviruses—The State of the Art, Nearly Fifty Years after Their Initial Discovery. Viruses 2021, 13, 1541. [Google Scholar] [CrossRef]
- Anonymous. National Enteric Surveillance Program Annual Summary 2017. 2018, ISBN: 2292-8561. Available online: https://canadacommons.ca/artifacts/3604696/annual-summary/4409630/ (accessed on 21 November 2023).
- CDC. Surveillance for Foodborne Disease Outbreaks, United States, 2017, Annual Report; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA; U.S. Department of Health and Human Services: Washington, DC, USA, 2019. Available online: https://stacks.cdc.gov/view/cdc/152191/cdc_152191_DS1.pdf (accessed on 21 November 2023).
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.-W.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Olaimat, A.N.; Taybeh, A.O.; Al-Nabulsi, A.; Al-Holy, M.; Hatmal, M.M.; Alzyoud, J.; Aolymat, I.; Abughoush, M.H.; Shahbaz, H.; Alzyoud, A.; et al. Common and Potential Emerging Foodborne Viruses: A Comprehensive Review. Life 2024, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.; Knight, A.; Richards, G.P. Persistence and Elimination of Human Norovirus in Food and on Food Contact Surfaces: A Critical Review. J. Food Prot. 2016, 79, 1273–1294. [Google Scholar] [CrossRef] [PubMed]
- Nasheri, N.; Vester, A.; Petronella, N. Foodborne viral outbreaks associated with frozen produce. Epidemiology Infect. 2019, 147, e291. [Google Scholar] [CrossRef]
- Cook, N.; Bertrand, I.; Gantzer, C.; Pinto, R.M.; Bosch, A. Persistence of Hepatitis A Virus in Fresh Produce and Production Environments, and the Effect of Disinfection Procedures: A Review. Food Environ. Virol. 2018, 10, 253–262. [Google Scholar] [CrossRef]
- Sakon, N.; Sadamasu, K.; Shinkai, T.; Hamajima, Y.; Yoshitomi, H.; Matsushima, Y.; Takada, R.; Terasoma, F.; Nakamura, A.; Komano, J.; et al. Foodborne Outbreaks Caused by Human Norovirus GII.P17-GII.17–Contaminated Nori, Japan, 2017. Emerg. Infect. Dis. 2018, 24, 920–923. [Google Scholar] [CrossRef]
- Donnan, E.J.; Fielding, J.E.; Gregory, J.E.; Lalor, K.; Rowe, S.; Goldsmith, P.; Antoniou, M.; Fullerton, K.E.; Knope, K.; Copland, J.G.; et al. A Multistate Outbreak of Hepatitis A Associated With Semidried Tomatoes in Australia, 2009. Clin. Infect. Dis. 2012, 54, 775–781. [Google Scholar] [CrossRef]
- Nasheri, N.; Harlow, J.; Chen, A.; Corneau, N.; Bidawid, S. Survival and Inactivation by Advanced Oxidative Process of Foodborne Viruses in Model Low-Moisture Foods. Food Environ. Virol. 2021, 13, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Raymond, P.; Blain, R.; Nasheri, N. Detection of Foodborne Viruses in Dates Using ISO 15216 Methodology. Viruses 2025, 17, 174. [Google Scholar] [CrossRef]
- Park, J.H.; Jeong, H.S.; Lee, J.S.; Lee, S.W.; Choi, Y.H.; Choi, S.J.; Joo, I.S.; Kim, Y.R.; Park, Y.K.; Youn, S.K. First norovirus outbreaks associated with consumption of green seaweed (Enteromorpha spp.) in South Korea. Epidemiology Infect. 2014, 143, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, J. Norway Norovirus Outbreaks Linked to Seaweed Salad from China. 2019. Available online: https://www.foodsafetynews.com/2019/09/norway-norovirus-outbreaks-linked-to-seaweed-salad-from-china/ (accessed on 21 November 2023).
- Anonymous. Modello di Richiamo Asiantrade Srl-Antipasto Giapponese di Alghe-Congelato-Goma Wakame; c17; Ministero della Saute: Comunicazione Istituzionale. 2024. Available online: https://www.salute.gov.it/new/sites/default/files/external_data/avvisi_sicurezza_alimentare/cartello%20di%20richiamo%20Goma%20Wakame_1743371339.pdf. (accessed on 4 April 2025).
- FAO and WHO. Report of the Expert Meeting on Food Safety for Seaweed–Current Status and Future Perspectives; Rome, 2–29 October 2021. 2022. Available online: https://www.who.int/publications/i/item/9789240058538 (accessed on 21 November 2023).
- Løvdal, T.; Lunestad, B.T.; Myrmel, M.; Rosnes, J.T.; Skipnes, D. Microbiological Food Safety of Seaweeds. Foods 2021, 10, 2719. [Google Scholar] [CrossRef]
- ISO 15216-1:2017; Microbiology of the Food Chain—Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part 1: Method for Quantification. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/65681.html (accessed on 21 November 2023).
- Oliveira, J.M.; Pardal, M.A.; Pereira, L.; Matos, A.M.; Rodrigues, E.T. Portuguese macroalgae and halophytes for human consumption: Minimal risk of norovirus and Salmonella infection. Food Control 2024, 164, 110600. [Google Scholar] [CrossRef]
- Tian, P.; Engelbrektson, A.; Mandrell, R. Two-Log Increase in Sensitivity for Detection of Norovirus in Complex Samples by Concentration with Porcine Gastric Mucin Conjugated to Magnetic Beads. Appl. Environ. Microbiol. 2008, 74, 4271–4276. [Google Scholar] [CrossRef]
- Dancho, B.A.; Chen, H.; Kingsley, D.H. Discrimination between infectious and non-infectious human norovirus using porcine gastric mucin. Int. J. Food Microbiol. 2012, 155, 222–226. [Google Scholar] [CrossRef]
- Cannon, J.L.; Vinjé, J. Histo-Blood Group Antigen Assay for Detecting Noroviruses in Water. Appl. Environ. Microbiol. 2008, 74, 6818–6819. [Google Scholar] [CrossRef][Green Version]
- Tian, P.; Brandl, M.; Mandrell, R. Porcine gastric mucin binds to recombinant norovirus particles and competitively inhibits their binding to histo-blood group antigens and Caco-2 cells. Lett. Appl. Microbiol. 2005, 41, 315–320. [Google Scholar] [CrossRef]
- Esseili, M.A.; Wang, Q.; Saif, L.J. Binding of Human GII.4 Norovirus Virus-Like Particles to Carbohydrates of Romaine Lettuce Leaf Cell Wall Materials. Appl. Environ. Microbiol. 2012, 78, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Suresh, M.; Harlow, J.; Nasheri, N. Evaluation of porcine gastric mucin assay for detection and quantification of human norovirus in fresh herbs and leafy vegetables. Food Microbiol. 2019, 84, 103254. [Google Scholar] [CrossRef]
- Raymond, P.; Paul, S.; Perron, A.; Deschênes, L. Norovirus Extraction from Frozen Raspberries Using Magnetic Silica Beads. Food Environ. Virol. 2021, 13, 248–258. [Google Scholar] [CrossRef]
- Wilrich, C.; Wilrich, P.-T. Estimation of the POD Function and the LOD of a Qualitative Microbiological Measurement Method. J. AOAC Int. 2009, 92, 1763–1772. [Google Scholar] [CrossRef] [PubMed]
- Raymond, P.; Paul, S.; Perron, A.; Deschênes, L.; Hara, K. Extraction of human noroviruses from leafy greens and fresh herbs using magnetic silica beads. Food Microbiol. 2021, 99, 103827. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Pu, C.; Suo, Y.; Zhang, X.; Qu, Y.; Feng, B.; Huang, L.; Shao, Y.; Dai, Y. Exploring magnetic capture to improve the detection of human norovirus in strawberries. Food Front. 2023, 4, 1819–1830. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Dillen, P.M.W.-V.; van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Plante, D.; Barrera, J.A.B.; Lord, M.; Harlow, J.; Iugovaz, I.; Nasheri, N. Examining the efficiency of porcine gastric mucin-coated magnetic beads in extraction of noroviruses from frozen berries. Food Microbiol. 2023, 120, 104461. [Google Scholar] [CrossRef]
- Sim, M.-C.; Ho, C.-L.; Phang, S.-M. A simple and effective method for RNA isolation and cDNA library construction from the brown seaweed Sargassum polycystum (Fucales, Phaeophyceae). J. Appl. Phycol. 2013, 25, 1277–1285. [Google Scholar] [CrossRef]
- Tian, P.; Yang, D.; Jiang, X.; Zhong, W.; Cannon, J.; Iii, W.B.; Woods, J.; Hartman, G.; Lindesmith, L.; Baric, R.; et al. Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J. Appl. Microbiol. 2010, 109, 1753–1762. [Google Scholar] [CrossRef]
- Murakami, K.; Tenge, V.R.; Karandikar, U.C.; Lin, S.-C.; Ramani, S.; Ettayebi, K.; Crawford, S.E.; Zeng, X.-L.; Neill, F.H.; Ayyar, B.V.; et al. Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. Proc. Natl. Acad. Sci. USA 2020, 117, 1700–1710. [Google Scholar] [CrossRef]
- Feigelstock, D.; Thompson, P.; Mattoo, P.; Zhang, Y.; Kaplan, G.G. The Human Homolog of HAVcr-1 Codes for a Hepatitis A Virus Cellular Receptor. J. Virol. 1998, 72, 6621–6628. [Google Scholar] [CrossRef] [PubMed]
- Lowther, J.; Bosch, A.; Butot, S.; Ollivier, J.; Mäde, D.; Rutjes, S.; Hardouin, G.; Lombard, B.; Veld, P.I.; Leclercq, A. Validation of EN ISO method 15216—Part 1—Quantification of hepatitis A virus and norovirus in food matrices. Int. J. Food Microbiol. 2019, 288, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Williams-Woods, J.; Rodriguez, R.; Marchant, J.; Swinford, A.G.; Burkhardt, W.I. Chapter 26—Concentration, Extraction and Detection of Enteric Viruses from Food. In Bacteriological Analytical Manual; Food and Drug Administration: Silver Spring, MD, USA, 2022. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-26-and-appendices-concentration-extraction-and-detection-enteric-viruses-food (accessed on 21 November 2023).
- Zhang, Q.; Pan, L.; Li, X.; Fang, Y.; Tian, P. Comparative Detection of Human Noroviruses in Green Onion and Grape Using Porcine Gastric Mucin-Conjugated Magnetic Beads and Polyethylene Glycol Enrichment. Food Sci. 2012, 33, 241–245. Available online: https://www.spkx.net.cn/EN/abstract/abstract28296.shtml (accessed on 21 November 2023).
| Beads Volume (µL) | HuNoV GII | |
|---|---|---|
| Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) | |
| 100 | 3.1 × 103 | 31 ± 3 |
| 200 | 3.1 × 103 | 37 ± 3 |
| Binding pH | HuNoV GI | HuNoV GII | ||
|---|---|---|---|---|
| Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) | Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) | |
| 7.2 | 4.2 × 102 | 16 ± 4 | 4.4 × 103 | 15 ± 3 |
| 3.5 | 4.2 × 102 | 11 ± 2 | 4.5 × 103 | 4 ± 2 |
| Centrifugation 10,000× g (min) | HuNoV GII | |
|---|---|---|
| Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) | |
| 10 | 1.6 × 103 | 16 ± 3 |
| 30 | 1.6 × 103 | 11 ± 4 |
| Incubation Time and Temperature | HuNoV GI | HuNoV GII | ||
|---|---|---|---|---|
| Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) | Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) | |
| 30 min @ RT | 1.5 × 103 | 19 ± 3 | 1.9 × 103 | 17 ± 4 |
| 48 h @ 4 °C | 1.0 × 103 | 9 ± 2 | 1.5 × 103 | 19 ± 7 |
| PGM Types and Lots | Brands | Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) |
|---|---|---|---|
| Type III mucin lot A | MilliporeSigma | 9.0 × 102 | 13 ± 4 |
| Type III mucin lot A | MilliporeSigma | 9.0 × 102 | 23 ± 3 |
| Type III mucin lot B | MilliporeSigma | 7.0 × 102 | 5 ± 3 |
| Native porcine stomach mucin | Biovenic | 7.0 × 102 | 5 ± 2 |
| Analyst | HuNoV GI | HuNoV GII | ||
|---|---|---|---|---|
| Inoculum (gEq per 25 g) | Recovery Rates Average ± sd | Inoculum (gEq per 25 g) | Recovery Rates Average ± sd | |
| 1 | 1.5 × 103 | 19 ± 3 (n = 5) | 1.91 × 103 | 17 ± 4 (n = 5) |
| 2 | 1.5 × 103 | 13 ± 2 (n = 4) | 2.3 × 103 | 19 ± 2 (n = 5) |
| 3 | nt | 9.0 × 102 | 23 ± 3 (n = 4) | |
| Inoculum Description | HuNoV GI | HuNoV GII | ||
|---|---|---|---|---|
| Inoculum (gEq per 25 g) | Recovery Rates Average ± sd | Inoculum (gEq per 25 g) | Recovery Rates Average ± sd | |
| High GI/MedGII | 11,183 | 12 ± 1 | 1364 | 12 ± 3 |
| Med GI/High GII | 1099 | 13 ± 5 | 15,624 | 14 ± 6 |
| Med GI | 1508 | 19 ± 3 | na | |
| Med GII | na | 1906 | 17 ± 4 | |
| Genotype | Virus Sample Number | GenBank Locus | Inoculum (gEq per 25 g) | Recovery Rates (Average ± sd) |
|---|---|---|---|---|
| GI.2 | CFIA-FVR-001 | MT745876 | 7 × 102 | 13 ± 1 (n = 3) |
| GI.3 | CFIA-FVR-002 | MW558948 | 7 × 102 | 21 ± 3 (n = 3) |
| GI.4 | CFIA-FVR-003 | MT750326 | 7 × 102 | 12 ± 2 (n = 3) |
| GI.5 | CFIA-FVR-022 | OL345567 | 3.8 × 102 | 19 ± 3 (n = 5) |
| GII.3 | CFIA-FVR-014 | MT808055 | 1.5 × 103 | 1.1 ± 1 (n = 3) |
| GII.4 | CFIA-FVR-018 | na | 1.4 × 103 | 14 ± 3 (n = 3) |
| GII.4 | CFIA-FVR-020 | MT754281 | 1.9 × 103 | 17 ± 4 (n = 5) |
| GII.7 | CFIA-FVR-030 | na | 1.6 × 103 | 7 ± 1 (n = 3) |
| Brands * | HuNoV GI | HuNoV GII | ||
|---|---|---|---|---|
| Inoculum gEq | Recovery Rates (Average ± sd) | Inoculum gEq | Recovery Rates (Average ± sd) | |
| A | 3.8 × 102 | 19 ± 3 (n = 5) | 1.9 × 103 | 17 ± 4 (n = 5) |
| B | 7 × 102 | 11 ± 2 (n = 3) | 1 × 103 | 9 ± 2 (n = 3) |
| C | 7 × 102 | 28 ± 6 (n = 3) | 1 × 103 | 21 ± 7 (n = 3) |
| D ** | 7 × 102 | 11 ± 2 (n = 3) | 1 × 103 | 9 ± 2 (n = 3) |
| E | 7 × 102 | 10 ± 2 (n = 3) | 1 × 103 | 26 ± 8 (n = 3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raymond, P.; Paul, S.; Blain, R.; Nasheri, N. Evaluation of Porcine Gastric Mucin-Based Method for Extraction of Noroviruses from Seaweed Salad. Viruses 2025, 17, 1245. https://doi.org/10.3390/v17091245
Raymond P, Paul S, Blain R, Nasheri N. Evaluation of Porcine Gastric Mucin-Based Method for Extraction of Noroviruses from Seaweed Salad. Viruses. 2025; 17(9):1245. https://doi.org/10.3390/v17091245
Chicago/Turabian StyleRaymond, Philippe, Sylvianne Paul, Roxanne Blain, and Neda Nasheri. 2025. "Evaluation of Porcine Gastric Mucin-Based Method for Extraction of Noroviruses from Seaweed Salad" Viruses 17, no. 9: 1245. https://doi.org/10.3390/v17091245
APA StyleRaymond, P., Paul, S., Blain, R., & Nasheri, N. (2025). Evaluation of Porcine Gastric Mucin-Based Method for Extraction of Noroviruses from Seaweed Salad. Viruses, 17(9), 1245. https://doi.org/10.3390/v17091245





