Genomic Analysis of Rotavirus G8P[8] Strains Detected in the United States Through Active Surveillance, 2016–2017
Abstract
1. Introduction
2. Materials and Methods
2.1. The Study Enrollment
2.2. Sample Selection for Whole Genome Analysis
2.3. Viral dsRNA Extraction for NGS
2.4. Whole-Genome Sequence Analysis
2.5. Phylogenetic, Sequence, and Structural Analyses
2.6. Clinical Severity Score
2.7. Bayesian Phylodynamic Analyses
3. Results
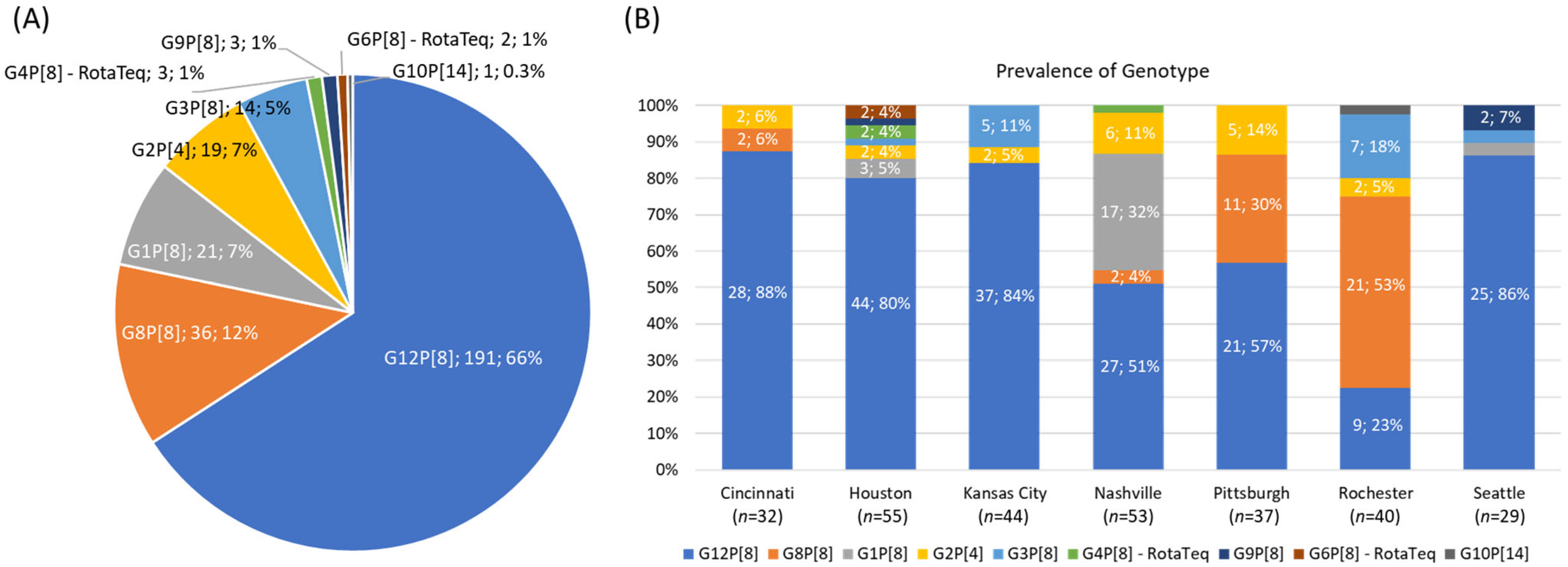
3.1. VP7 and VP4 Genotypes
3.2. Clinical Observations
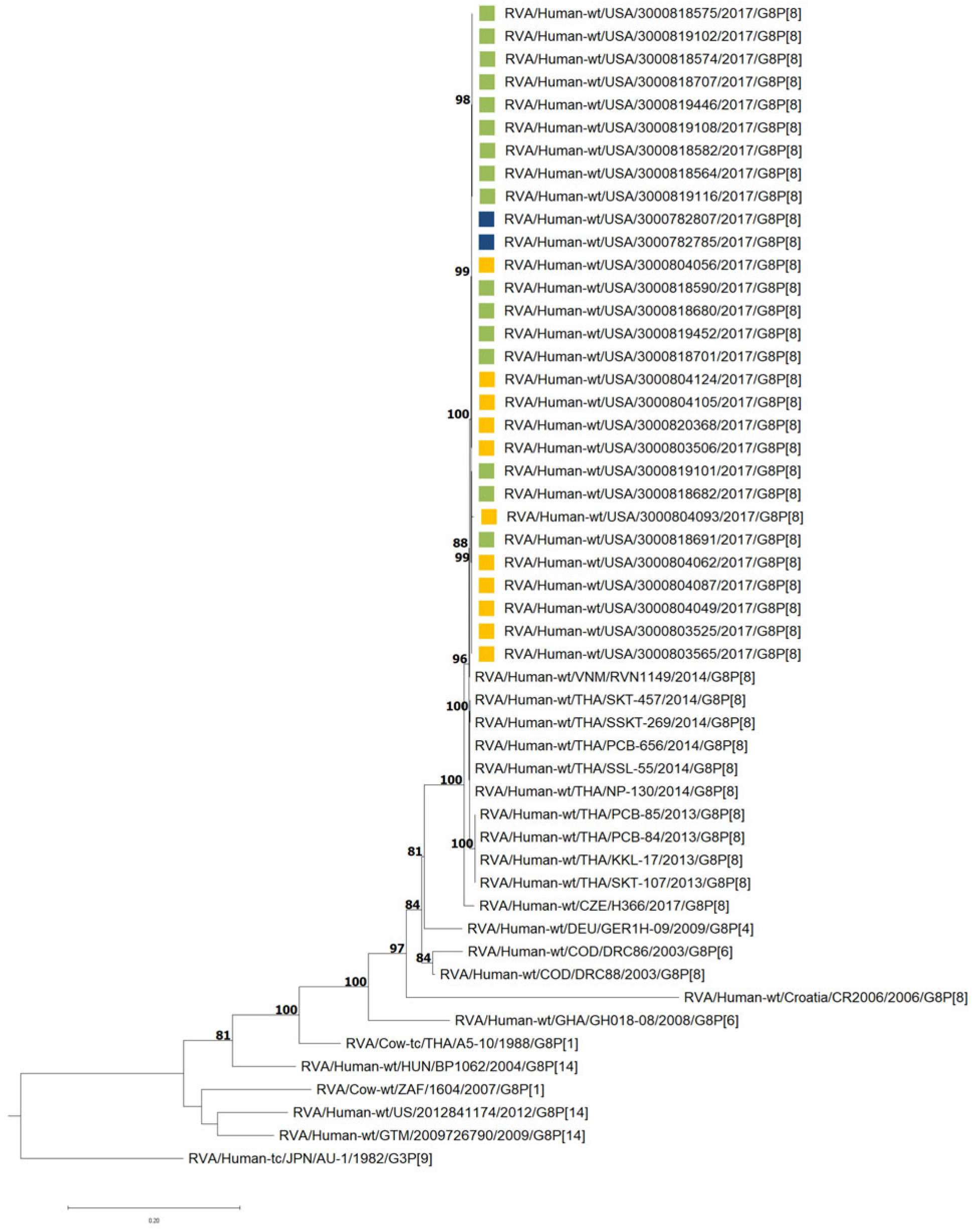
3.3. G8P[8] Whole-Genome Analysis
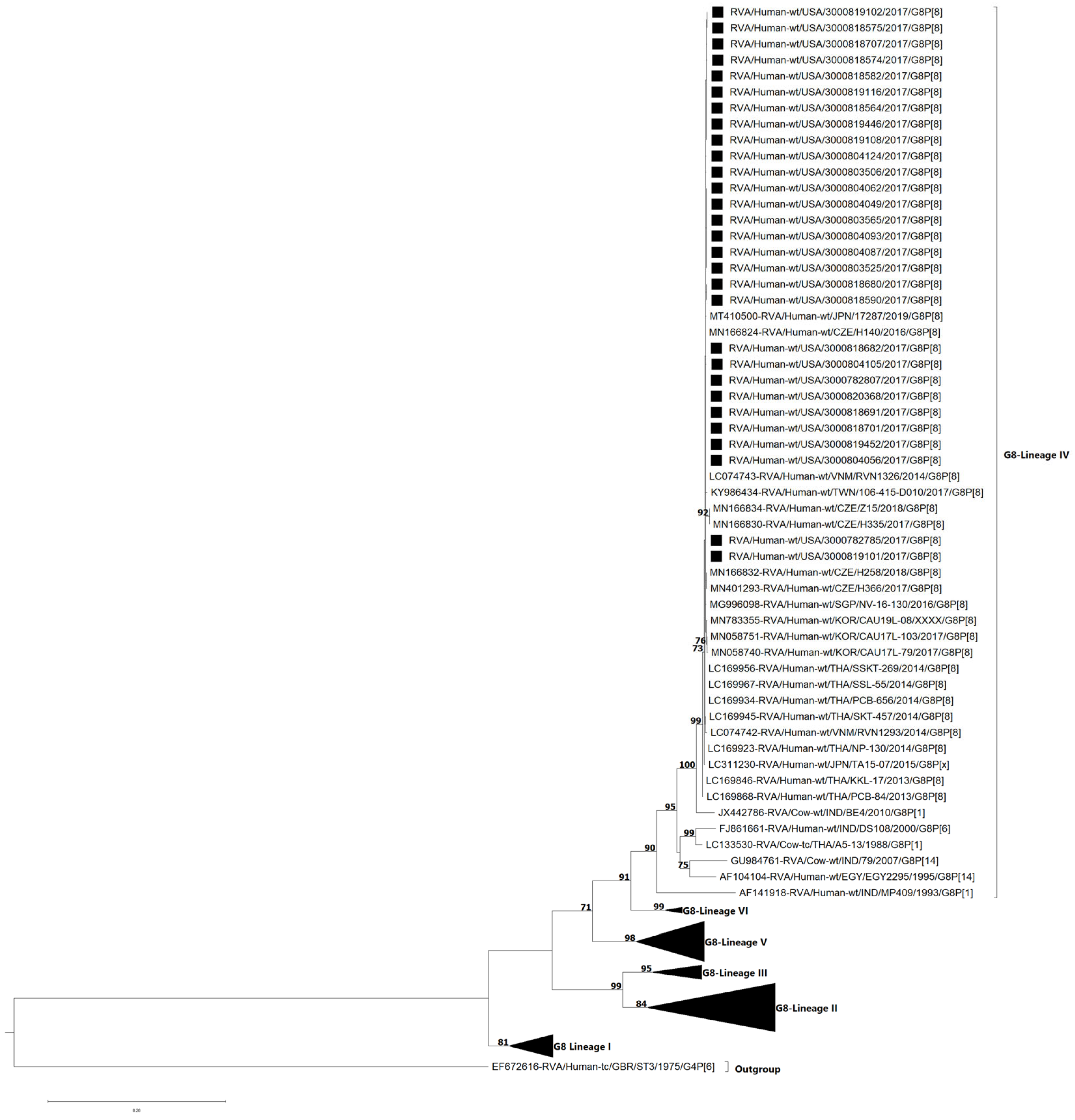
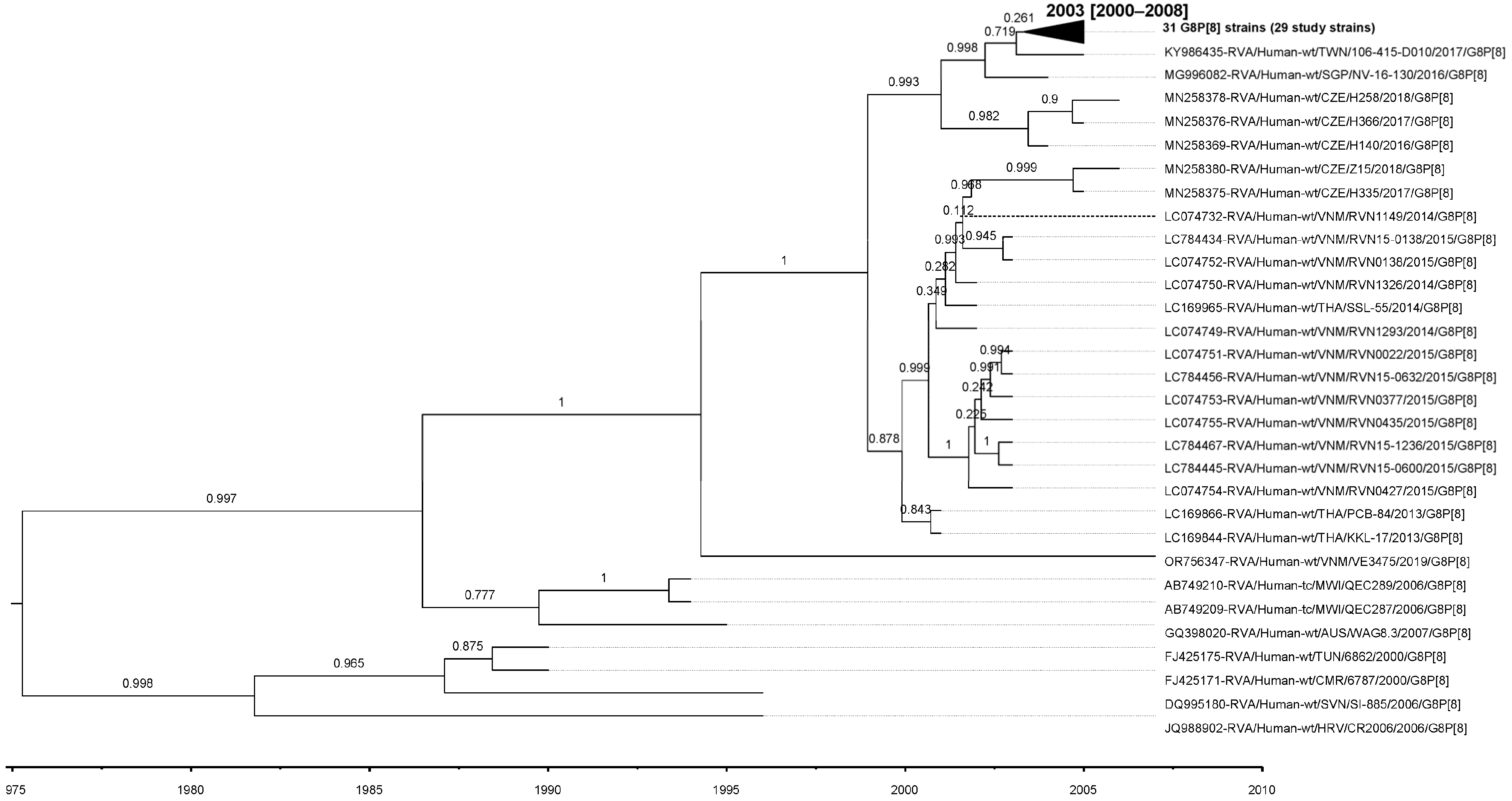
3.4. VP7 and VP4 Phylogenetic Analyses and Sequence Identity
3.5. Phylogenetic Analyses and Sequence Identity of Internal Genes
3.6. Comparative Analysis of VP7 Antigenic Epitopes with Reference Strains
3.7. Comparative Analysis of VP4 Antigenic Epitopes with Vaccine Strains
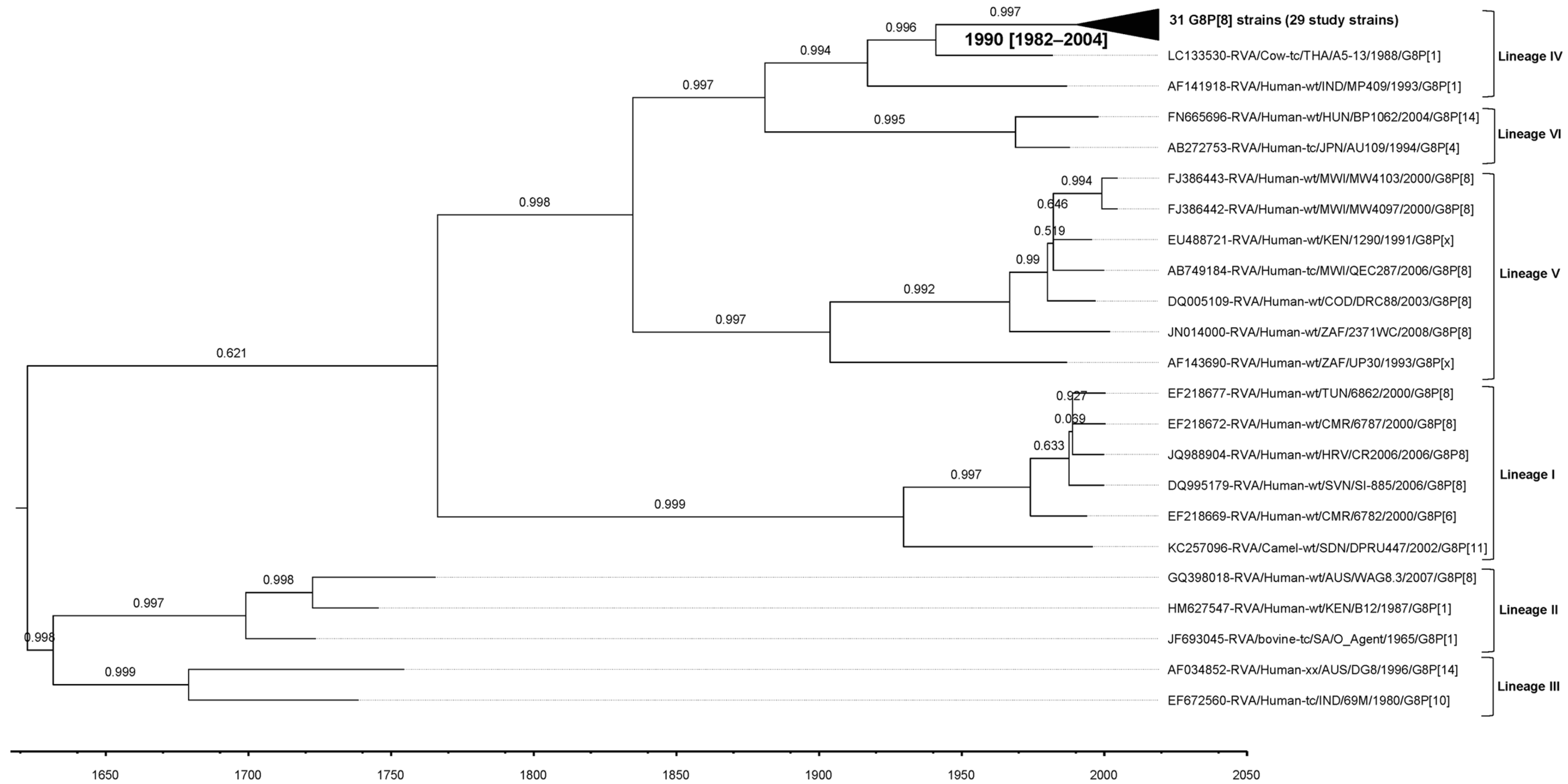
3.8. VP7 and VP4 Bayesian Evolutionary Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Disclaimer
Abbreviations
| AGE | Acute gastroenteritis |
| aa | Amino acid |
| AICc | Corrected Akaike Information Criterion |
| BLASTN | Basic Local Alignment Search Tool for Nucleotides |
| BV-BRC | Bacterial and Viral Bioinformatics Resource Center |
| CDC | Centers for Disease Control and Prevention |
| cDNA | Complementary DNA |
| dsRNA | Double-stranded RNA |
| ED | Emergency department |
| ESS | Effective Sample Size |
| HC | Healthy control |
| MCCT | Maximum Clade Credibility Tree |
| MCMC | Markov chain Monte-Carlo |
| ML | Maximum likelihood |
| MVSS | Modified Vesikari Severity Score |
| NCBI | National Center for Biotechnology Information |
| NGS | Next-generation sequencing |
| NIH | National Institutes of Health |
| NSP | Non-structural protein |
| nt | Nucleotide |
| NVSN | New Vaccine Surveillance Network |
| ORF | Open reading frame |
| qRT-PCR | Quantitative real-time reverse transcription polymerase chain reaction |
| RCWG | Rotavirus Classification Working Group |
| RNA | Ribonucleic acid |
| RVA | Group A rotavirus |
| tMRCA | Time to Most Recent Common Ancestor |
| VE | Vaccine effectiveness |
| VP | Viral protein |
References
- Black, R.E.; Perin, J.; Yeung, D.; Rajeev, T.; Miller, J.; Elwood, S.E.; Platts-Mills, J.A. Estimated global and regional causes of deaths from diarrhoea in children younger than 5 years during 2000-21: A systematic review and Bayesian multinomial analysis. Lancet Glob. Health 2024, 12, e919–e928. [Google Scholar] [CrossRef]
- Rotavirus vaccines WHO position paper: January 2013—Recommendations. Vaccine 2013, 31, 6170–6171. [CrossRef] [PubMed]
- International Vaccine Access Center (IVAC), Johns Hopkins Bloomberg School of Public Health. Available online: www.view-hub.org (accessed on 8 August 2025).
- Pindyck, T.; Tate, J.E.; Parashar, U.D. A decade of experience with rotavirus vaccination in the United States—Vaccine uptake, effectiveness, and impact. Expert. Rev. Vaccines 2018, 17, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Estes, M.; Kapikian, A.; Knipe, D.; Howley, P. Fields Virology; Lippencott, Williams and Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Bányai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef]
- Rotavirus Classification Working Group. List of Accepted Genotypes. Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/newgenotypes (accessed on 8 August 2025).
- Dóró, R.; László, B.; Martella, V.; Leshem, E.; Gentsch, J.; Parashar, U.; Bányai, K. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: Is there evidence of strain selection from vaccine pressure? Infect. Genet. Evol. 2014, 28, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Bowen, M.D.; Mijatovic-Rustempasic, S.; Esona, M.D.; Teel, E.N.; Gautam, R.; Sturgeon, M.; Azimi, P.H.; Baker, C.J.; Bernstein, D.I.; Boom, J.A.; et al. Rotavirus Strain Trends During the Postlicensure Vaccine Era: United States, 2008–2013. J. Infect. Dis. 2016, 214, 732–738. [Google Scholar] [CrossRef]
- Esona, M.D.; Ward, M.L.; Wikswo, M.E.; Rustempasic, S.M.; Gautam, R.; Perkins, C.; Selvarangan, R.; Harrison, C.J.; Boom, J.A.; Englund, J.A.; et al. Rotavirus Genotype Trends and Gastrointestinal Pathogen Detection in the United States, 2014–2016: Results From the New Vaccine Surveillance Network. J. Infect. Dis. 2021, 224, 1539–1549. [Google Scholar] [CrossRef]
- Hasegawa, A.; Inouye, S.; Matsuno, S.; Yamaoka, K.; Eko, R.; Suharyono, W. Isolation of human rotaviruses with a distinct RNA electrophoretic pattern from Indonesia. Microbiol. Immunol. 1984, 28, 719–722. [Google Scholar] [CrossRef]
- Nakagomi, T.; Doan, Y.H.; Dove, W.; Ngwira, B.; Iturriza-Gómara, M.; Nakagomi, O.; Cunliffe, N.A. G8 rotaviruses with conserved genotype constellations detected in Malawi over 10 years (1997–2007) display frequent gene reassortment among strains co-circulating in humans. J. Gen. Virol. 2013, 94, 1273–1295. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Rahman, M.; Yang, X.; Delbeke, T.; Arijs, I.; Kabue, J.P.; Muyembe, J.J.; Van Ranst, M. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 2006, 44, 1801–1809. [Google Scholar] [CrossRef]
- Hoque, S.A.; Kobayashi, M.; Takanashi, S.; Anwar, K.S.; Watanabe, T.; Khamrin, P.; Okitsu, S.; Hayakawa, S.; Ushijima, H. Role of rotavirus vaccination on an emerging G8P [8] rotavirus strain causing an outbreak in central Japan. Vaccine 2018, 36, 43–49. [Google Scholar] [CrossRef]
- Kondo, K.; Tsugawa, T.; Ono, M.; Ohara, T.; Fujibayashi, S.; Tahara, Y.; Kubo, N.; Nakata, S.; Higashidate, Y.; Fujii, Y.; et al. Clinical and Molecular Characteristics of Human Rotavirus G8P [8] Outbreak Strain, Japan, 2014. Emerg. Infect. Dis. 2017, 23, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Tacharoenmuang, R.; Komoto, S.; Guntapong, R.; Ide, T.; Sinchai, P.; Upachai, S.; Yoshikawa, T.; Tharmaphornpilas, P.; Sangkitporn, S.; Taniguchi, K. Full Genome Characterization of Novel DS-1-Like G8P [8] Rotavirus Strains that Have Emerged in Thailand: Reassortment of Bovine and Human Rotavirus Gene Segments in Emerging DS-1-Like Intergenogroup Reassortant Strains. PLoS ONE 2016, 11, e0165826. [Google Scholar] [CrossRef] [PubMed]
- Hoa-Tran, T.N.; Nakagomi, T.; Vu, H.M.; Do, L.P.; Gauchan, P.; Agbemabiese, C.A.; Nguyen, T.T.; Nakagomi, O.; Thanh, N.T. Abrupt emergence and predominance in Vietnam of rotavirus A strains possessing a bovine-like G8 on a DS-1-like background. Arch. Virol. 2016, 161, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yi, D.Y.; Lim, I.; Ward, A.C.; Kim, W. Detection of an unusual G8P [8] rotavirus in a Rotarix-vaccinated child with acute gastroenteritis using Nanopore MinION sequencing: A case report. Medicine 2020, 99, e22641. [Google Scholar] [CrossRef]
- Wang, S.-J.; Chen, L.-N.; Wang, S.-M.; Zhou, H.-L.; Qiu, C.; Jiang, B.; Qiu, T.-Y.; Chen, S.-L.; Von Seidlein, L.; Wang, X.-Y. Genetic characterization of two G8P [8] rotavirus strains isolated in Guangzhou, China, in 2020/21: Evidence of genome reassortment. BMC Infect. Dis. 2022, 22, 579. [Google Scholar] [CrossRef]
- Degiuseppe, J.I.; Stupka, J.A. Emergence of unusual rotavirus G9P [4] and G8P [8] strains during post vaccination surveillance in Argentina, 2017–2018. Infect. Genet. Evol. 2021, 93, 104940. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Y.; Chen, N.; Pang, B.; Liu, M.; Cai, K.; Kobayashi, N. Surveillance of Human Rotaviruses in Wuhan, China (2019–2022): Whole-Genome Analysis of Emerging DS-1-like G8P [8] Rotavirus. Int. J. Mol. Sci. 2023, 24, 12189. [Google Scholar] [CrossRef]
- Le, L.K.T.; Chu, M.N.T.; Tate, J.E.; Jiang, B.; Bowen, M.D.; Esona, M.D.; Gautam, R.; Jaimes, J.; Pham, T.P.T.; Huong, N.T.; et al. Genetic diversity of G9, G3, G8 and G1 rotavirus group A strains circulating among children with acute gastroenteritis in Vietnam from 2016 to 2021. Infect. Genet. Evol. 2024, 118, 105566. [Google Scholar] [CrossRef]
- Moutelikova, R.; Sauer, P.; Dvorakova Heroldova, M.; Hola, V.; Prodelalova, J. Emergence of Rare Bovine-Human Reassortant DS-1-Like Rotavirus A Strains with G8P [8] Genotype in Human Patients in the Czech Republic. Viruses 2019, 11, 1015. [Google Scholar] [CrossRef]
- Kim, K.G.; Kee, H.Y.; Park, H.J.; Chung, J.K.; Kim, T.S.; Kim, M.J. The Long-Term Impact of Rotavirus Vaccines in Korea, 2008–2020; Emergence of G8P [8] Strain. Vaccines 2021, 9, 406. [Google Scholar] [CrossRef]
- Staat, M.A.; Payne, D.C.; Halasa, N.; Weinberg, G.A.; Donauer, S.; Wikswo, M.; McNeal, M.; Edwards, K.M.; Szilagyi, P.G.; Bernstein, D.I.; et al. Continued Evidence of the Impact of Rotavirus Vaccine in Children Less Than 3 Years of Age From the United States New Vaccine Surveillance Network: A Multisite Active Surveillance Program, 2006–2016. Clin. Infect. Dis. 2020, 71, e421–e429. [Google Scholar] [CrossRef] [PubMed]
- Mijatovic-Rustempasic, S.; Jaimes, J.; Perkins, C.; Ward, M.L.; Esona, M.D.; Gautam, R.; Lewis, J.; Sturgeon, M.; Panjwani, J.; Bloom, G.A.; et al. Rotavirus Strain Trends in United States, 2009–2016: Results from the National Rotavirus Strain Surveillance System (NRSSS). Viruses 2022, 14, 1775. [Google Scholar] [CrossRef] [PubMed]
- Esona, M.D.; McDonald, S.; Kamili, S.; Kerin, T.; Gautam, R.; Bowen, M.D. Comparative evaluation of commercially available manual and automated nucleic acid extraction methods for rotavirus RNA detection in stools. J. Virol. Methods 2013, 194, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.D.; Martella, V.; et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef]
- NCBI. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Esona, M.D.; Roy, S.; Rungsrisuriyachai, K.; Gautam, R.; Hermelijn, S.; Rey-Benito, G.; Bowen, M.D. Molecular characterization of a human G20P [28] rotavirus a strain with multiple genes related to bat rotaviruses. Infect. Genet. Evol. 2018, 57, 166–170. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- QIAGEN. QIAGEN CLC Genomics Workbench: NGS Data Analysis for any Species, any Platform, any Workflow. Available online: https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/ (accessed on 2 September 2022).
- Wikswo, M.E.; Weinberg, G.A.; Szilagyi, P.G.; Selvarangan, R.; Harrison, C.J.; Klein, E.J.; Englund, J.A.; Sahni, L.C.; Boom, J.A.; Halasa, N.B.; et al. Evaluation of a Modified Vesikari Severity Score as a Research Tool for Assessing Pediatric Acute Gastroenteritis. J. Pediatr. Infect. Dis. Soc. 2024, 13, 547–550. [Google Scholar] [CrossRef]
- Baele, G.; Ji, X.; Hassler, G.W.; McCrone, J.T.; Shao, Y.; Zhang, Z.; Holbrook, A.J.; Lemey, P.; Drummond, A.J.; Rambaut, A.; et al. BEAST X for Bayesian phylogenetic, phylogeographic and phylodynamic inference. Nat. Methods 2025, 22, 1653–1656. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.R.; Suchard, M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 2008, 36, 355–368. [Google Scholar] [CrossRef]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.; Kobayashi, M.; Nagasawa, K.; Hatazawa, R.; Thi Kim Pham, N.; Miyashita, H.; Komoto, S.; Tajima, T.; Baba, T.; Okitsu, S.; et al. Whole genome sequencing and evolutionary analysis of G8P [8] rotaviruses emerging in Japan. Virusdisease 2022, 33, 215–218. [Google Scholar] [CrossRef]
- Chia, G.; Ho, H.J.; Ng, C.G.; Neo, F.J.; Win, M.K.; Cui, L.; Leo, Y.S.; Chow, A. An unusual outbreak of rotavirus G8P [8] gastroenteritis in adults in an urban community, Singapore, 2016. J. Clin. Virol. 2018, 105, 57–63. [Google Scholar] [CrossRef]
- Aoki, S.T.; Settembre, E.C.; Trask, S.D.; Greenberg, H.B.; Harrison, S.C.; Dormitzer, P.R. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 2009, 324, 1444–1447. [Google Scholar] [CrossRef]
- McDonald, S.M.; Matthijnssens, J.; McAllen, J.K.; Hine, E.; Overton, L.; Wang, S.; Lemey, P.; Zeller, M.; Van Ranst, M.; Spiro, D.J.; et al. Evolutionary dynamics of human rotaviruses: Balancing reassortment with preferred genome constellations. PLoS Pathog. 2009, 5, e1000634. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Sun, Z.Y.; Wagner, G.; Harrison, S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. Embo J. 2002, 21, 885–897. [Google Scholar] [CrossRef]
- Ludert, J.E.; Ruiz, M.C.; Hidalgo, C.; Liprandi, F. Antibodies to rotavirus outer capsid glycoprotein VP7 neutralize infectivity by inhibiting virion decapsidation. J. Virol. 2002, 76, 6643–6651. [Google Scholar] [CrossRef]
- Monnier, N.; Higo-Moriguchi, K.; Sun, Z.Y.; Prasad, B.V.; Taniguchi, K.; Dormitzer, P.R. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J. Virol. 2006, 80, 1513–1523. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Burns, J.W.; Morita, Y.; Tanaka, T.; Estes, M.K. Localization of rotavirus VP4 neutralization epitopes involved in antibody-induced conformational changes of virus structure. J. Virol. 1994, 68, 3955–3964. [Google Scholar] [CrossRef]
- McKinney, B.A.; Kallewaard, N.L.; Crowe, J.E., Jr.; Meiler, J. Using the natural evolution of a rotavirus-specific human monoclonal antibody to predict the complex topography of a viral antigenic site. Immunome Res. 2007, 3, 8. [Google Scholar] [CrossRef][Green Version]
- Dormitzer, P.R.; Nason, E.B.; Prasad, B.V.; Harrison, S.C. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 2004, 430, 1053–1058. [Google Scholar] [CrossRef]
- Mijatovic-Rustempasic, S.; Roy, S.; Sturgeon, M.; Rungsrisuriyachai, K.; Reisdorf, E.; Cortese, M.M.; Bowen, M.D. Full-Genome Sequence of the First G8P [14] Rotavirus Strain Detected in the United States. Genome Announc. 2015, 3, e00677-15. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, G.A.; Payne, D.C.; Teel, E.N.; Mijatovic-Rustempasic, S.; Bowen, M.D.; Wikswo, M.; Gentsch, J.R.; Parashar, U.D. First reports of human rotavirus G8P [4] gastroenteritis in the United States. J. Clin. Microbiol. 2012, 50, 1118–1121. [Google Scholar] [CrossRef] [PubMed]
- Mijatovic-Rustempasic, S.; Williams, F.; Bonenfant, G.; Jaimes, J.; Wikswo, M.; Tate, J.; Mirza, S.; Mattison, C.; Selvarangan, R.; Halasa, N.; et al. Prevalence and Phylogenetic Analysis of Rotavirus Genotypes Detected in the United States, 2017–2024: Results from the New Vaccine Surveillance Network. Centers for Disease Control and Prevention: Atlanta, GA, USA, 2026; manuscript in preparation; to be submitted. [Google Scholar]
- Zeller, M.; Patton, J.T.; Heylen, E.; De Coster, S.; Ciarlet, M.; Van Ranst, M.; Matthijnssens, J. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J. Clin. Microbiol. 2012, 50, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Motayo, B.O.; Oluwasemowo, O.O.; Olusola, B.A.; Opayele, A.V.; Faneye, A.O. Phylogeography and evolutionary analysis of African Rotavirus a genotype G12 reveals district genetic diversification within lineage III. Heliyon 2019, 5, e02680. [Google Scholar] [CrossRef]
- Degiuseppe, J.I.; Torres, C.; Mbayed, V.A.; Stupka, J.A. Phylogeography of Rotavirus G8P [8] Detected in Argentina: Evidence of Transpacific Dissemination. Viruses 2022, 14, 2223. [Google Scholar] [CrossRef]
- Zeller, M.; Heylen, E.; Damanka, S.; Pietsch, C.; Donato, C.; Tamura, T.; Kulkarni, R.; Arora, R.; Cunliffe, N.; Maunula, L.; et al. Emerging OP354-Like P [8] Rotaviruses Have Rapidly Dispersed from Asia to Other Continents. Mol. Biol. Evol. 2015, 32, 2060–2071. [Google Scholar] [CrossRef]
- Liu, X.; Wang, M.; Li, S.; Li, J.; Xiao, J.; Li, H.; Zhang, Q.; Kong, X.; Wang, H.; Li, D.; et al. Genomic and evolutionary characteristics of G9P [8], the dominant group a rotavirus in China (2016–2018). Front. Microbiol. 2022, 13, 997957. [Google Scholar] [CrossRef]



| RVA Positive (n = 253) | G8P[8] Positive (n = 35) | Non-G8P[8] Positive (n = 218) | p-Value a | |
|---|---|---|---|---|
| Age Group | No. (%) | |||
| 0–11 months | 54 (21%) | 3 (9%) | 51 (23%) | |
| 12–23 months | 73 (29%) | 11 (31%) | 62 (28%) | |
| 24–35 months | 55 (22%) | 9 (26%) | 46 (21%) | |
| 36–47 months | 32 (13%) | 5 (14%) | 27 (12%) | |
| 48–59 months | 10 (4%) | 3 (9%) | 7 (3%) | |
| 60+ months | 29 (11%) | 4 (11%) | 25 (11%) | |
| Age in months, median (IQR) | 23 (14–38) | 23 (13–38) | 25 (16–41) | 0.2934 |
| Vaccination Characteristics | No. (%) | |||
| Eligible patients b | 243 (96%) | 35 (100%) | 208 (96.4%) | |
| Product | ||||
| Unvaccinated | 74 (30%) | 7 (20%) | 67 (32%) | |
| Rotarix (partial or complete series) | 42 (17%) | 8 (23%) | 34 (16%) | |
| RotaTeq (partial or complete series) | 102 (42%) | 17 (49%) | 85 (41%) | |
| Mixed | 12 (5%) | 2 (6%) | 10 (5%) | |
| Unknown series | 13 (5%) | 1 (3%) | 12 (6%) | |
| Hospitalization Status | No. (%) | |||
| ED/outpatient | 127 (50%) | 17 (49%) | 110 (50%) | |
| Hospitalized/inpatient | 126 (50%) | 18 (51%) | 108 (50%) | |
| Length of stay, median (IQR) | 2 (1–2) | 1 (1–2) | 2 (1–3) | 0.1398 |
| Severity Score | No. (%) | |||
| MVSS, median (IQR) | 13 (10–15) | 13 (10–15) | 13 (10–15) | 0.9695 |
| MVSS | ||||
| 0–10 mild | 75 (30%) | 9 (26%) | 66 (30%) | |
| 11–15 moderately severe | 116 (46%) | 19 (54%) | 97 (44%) | |
| 16–20 very severe | 47 (19%) | 5 (14%) | 42 (19%) | |
| Unable to calculate | 15 (6%) | 2 (6%) | 13 (6%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casey-Moore, M.C.; Esona, M.D.; Mijatovic-Rustempasic, S.; Jaimes, J.; Gautam, R.; Wikswo, M.E.; Williams, J.V.; Halasa, N.; Chappell, J.D.; Payne, D.C.; et al. Genomic Analysis of Rotavirus G8P[8] Strains Detected in the United States Through Active Surveillance, 2016–2017. Viruses 2025, 17, 1230. https://doi.org/10.3390/v17091230
Casey-Moore MC, Esona MD, Mijatovic-Rustempasic S, Jaimes J, Gautam R, Wikswo ME, Williams JV, Halasa N, Chappell JD, Payne DC, et al. Genomic Analysis of Rotavirus G8P[8] Strains Detected in the United States Through Active Surveillance, 2016–2017. Viruses. 2025; 17(9):1230. https://doi.org/10.3390/v17091230
Chicago/Turabian StyleCasey-Moore, Mary C., Mathew D. Esona, Slavica Mijatovic-Rustempasic, Jose Jaimes, Rashi Gautam, Mary E. Wikswo, John V. Williams, Natasha Halasa, James D. Chappell, Daniel C. Payne, and et al. 2025. "Genomic Analysis of Rotavirus G8P[8] Strains Detected in the United States Through Active Surveillance, 2016–2017" Viruses 17, no. 9: 1230. https://doi.org/10.3390/v17091230
APA StyleCasey-Moore, M. C., Esona, M. D., Mijatovic-Rustempasic, S., Jaimes, J., Gautam, R., Wikswo, M. E., Williams, J. V., Halasa, N., Chappell, J. D., Payne, D. C., Allen Staat, M., Weinberg, G. A., & Bowen, M. D. (2025). Genomic Analysis of Rotavirus G8P[8] Strains Detected in the United States Through Active Surveillance, 2016–2017. Viruses, 17(9), 1230. https://doi.org/10.3390/v17091230






