Isolation and Characterization of a Novel Porcine Teschovirus 2 Strain: Incomplete PERK-Mediated Unfolded Protein Response Supports Viral Replication
Abstract
1. Introduction
2. Material and Methods
2.1. Virus Detection and Isolation
2.2. In Vitro Growth Characteristics and Virus Morphology Observation
2.3. Whole Genome Sequencing and Sequence Analysis
2.4. Phylogenetic and Recombination Analyses
2.5. Generation of Polyclonal Antibodies Against the PTV VP1 Protein
2.6. Western Blot and Indirect Immunofluorescence Assays (IFA)
2.7. Development of a qPCR Assay for PTV Viral Quantification
2.8. Analyzing the Activation of UPR Signaling Pathway Induced by PTV Infection
2.9. siRNA-Mediated PERK Knockdown
2.10. Statistical Analysis
3. Results
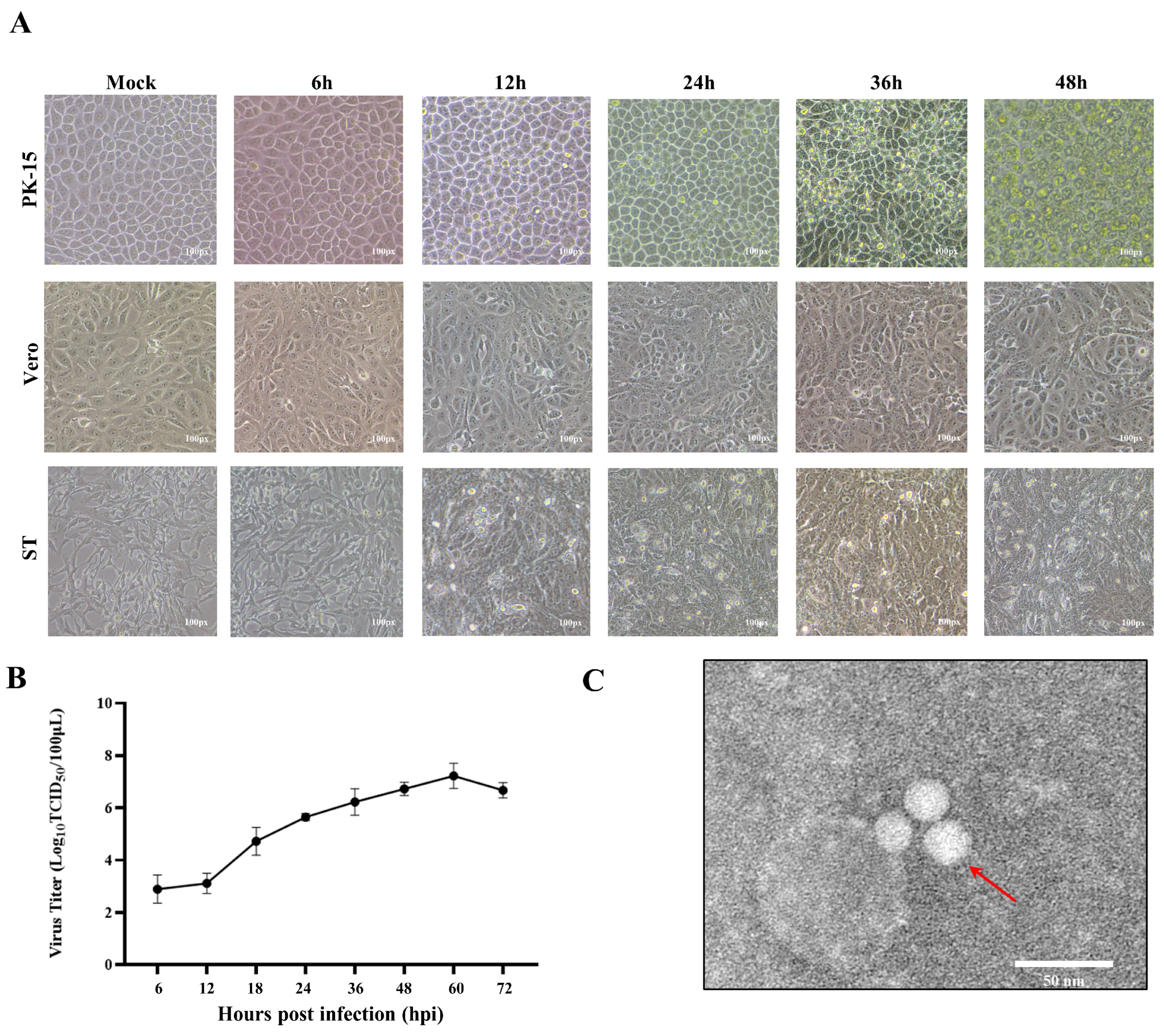
3.1. Isolation, Identification, and Growth Characteristics of PTV-GXLZ2024
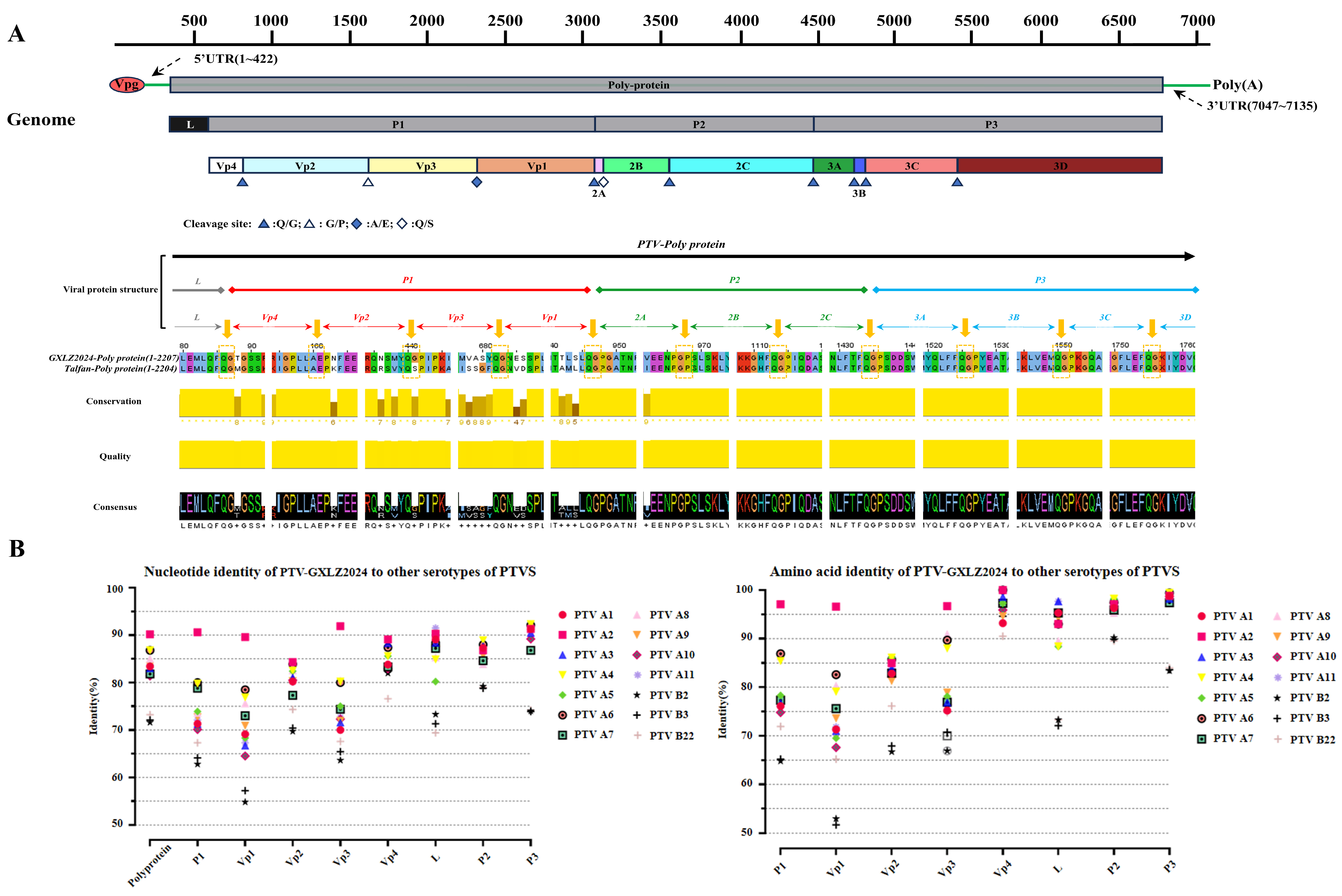
3.2. Comprehensive Genome Sequencing and Sequence Analysis of PTV-GXLZ2024
3.3. Phylogenetic and Recombination Analysis of PTV-GXLZ2024
3.4. Preparation of Polyclonal Antibody and Establishment of RT-qPCR Method for PTV-GXLZ2024
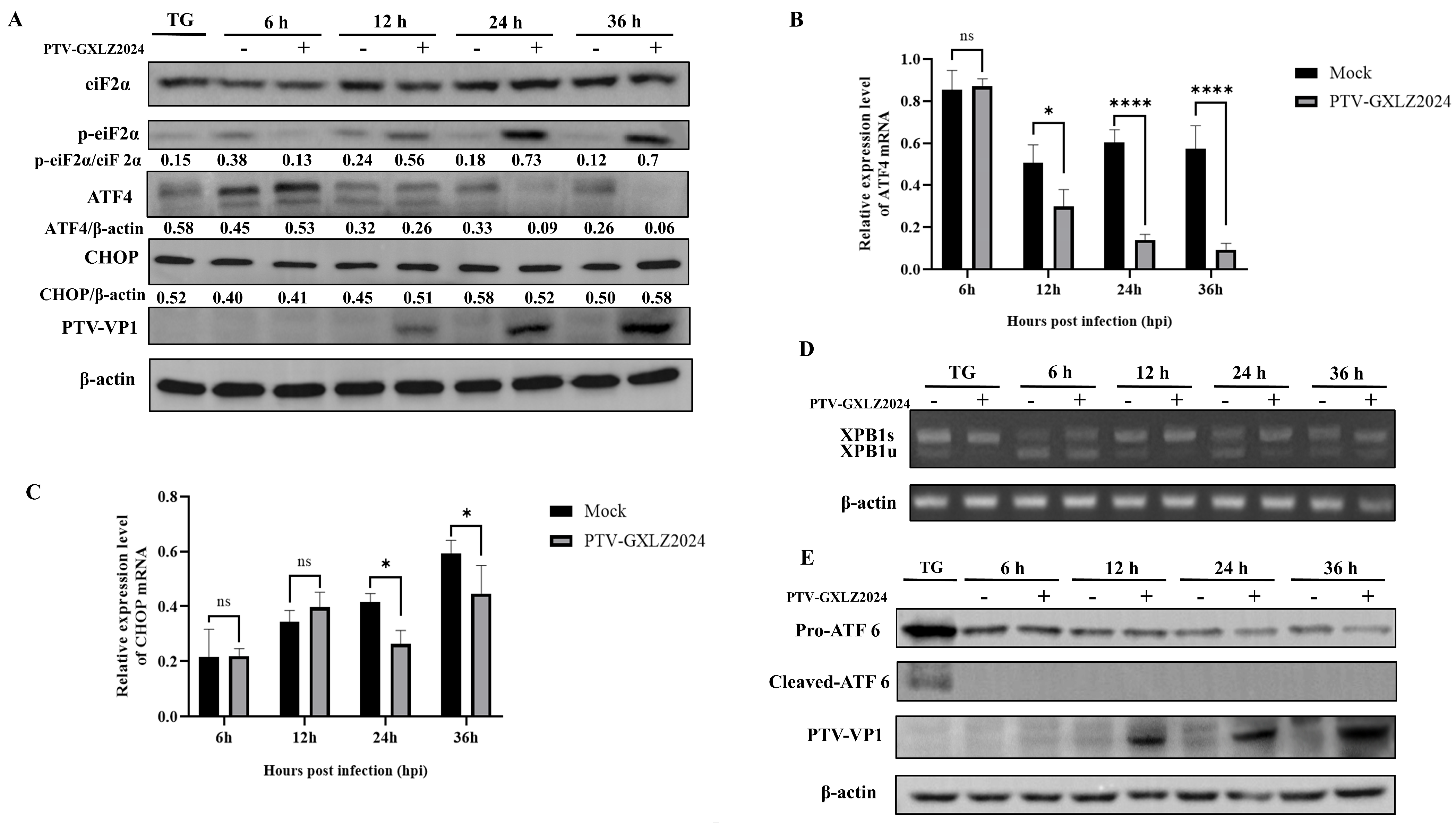
3.5. Activation of the PERK Pathway by PTV-GXLZ2024 in the UPR
3.6. Si-PERK Facilitates PTV-GXLZ2024 Replication
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bangari, D.S.; Pogranichniy, R.M.; Gillespie, T.; Stevenson, G.W. Genotyping of Porcine teschovirus from nervous tissue of pigs with and without polioencephalomyelitis in Indiana. J. Vet. Diagn. Investig. 2010, 22, 594–597. [Google Scholar] [CrossRef]
- Zhang, C.F.; Cui, S.J.; Hu, S.; Zhang, Z.; Guo, Q.; Zell, R. Isolation and characterization of the first Chinese strain of porcine Teschovirus-8. J. Virol. Methods 2010, 167, 208–213. [Google Scholar] [CrossRef]
- Souza, F.G.; Gularte, J.S.; Demoliner, M.; Lima, A.F.; Siebert, J.C.; Rigotto, C.; Henzel, A.; Eisen, A.K.A.; Spilki, F.R. Teschovirus and other swine and human enteric viruses in Brazilian watersheds impacted by swine husbandry. Braz. J. Microbiol. 2020, 51, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Villanova, F.; Cui, S.; Ai, X.; Leal, É. Analysis of full-length genomes of porcine teschovirus (PTV) and the effect of purifying selection on phylogenetic trees. Arch. Virol. 2016, 161, 1199–1208. [Google Scholar] [CrossRef]
- Liang, W.; Wu, X.; Ding, Z.; Zhong, S.; Qian, X.; Ye, P.; Liu, H.; Chen, Z.; Zhang, J.; Cao, H.; et al. Identification of a novel porcine Teschovirus 2 strain as causative agent of encephalomyelitis in suckling piglets with high mortality in China. BMC Vet. Res. 2023, 19, 2. [Google Scholar] [CrossRef]
- Vreman, S.; Caliskan, N.; Harders, F.; Boonstra, J.; Peperkamp, K.; Ho, C.K.Y.; Kuller, W.; Kortekaas, J. Two novel porcine teschovirus strains as the causative agents of encephalomyelitis in the Netherlands. BMC Vet. Res. 2020, 16, 51. [Google Scholar] [CrossRef]
- Chiu, S.C.; Hu, S.C.; Chang, C.C.; Chang, C.Y.; Huang, C.C.; Pang, V.F.; Wang, F.I. The role of porcine teschovirus in causing diseases in endemically infected pigs. Vet. Microbiol. 2012, 161, 88–95. [Google Scholar] [CrossRef]
- Carnero, J.; Prieto, C.; Polledo, L.; Martínez-Lobo, F.J. Detection of Teschovirus type 13 from two swine herds exhibiting nervous clinical signs in growing pigs. Transbound. Emerg. Dis. 2018, 65, e489–e493. [Google Scholar] [CrossRef]
- Pogranichniy, R.M.; Janke, B.H.; Gillespie, T.G.; Yoon, K.J. A prolonged outbreak of polioencephalomyelitis due to infection with a group I porcine enterovirus. J. Vet. Diagn. Investig. 2003, 15, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.T.; Kuo, C.C.; Kuo, Y.C.; Yang, J.L.; Chang, C.Y.; Wang, F.I. The urinary shedding of porcine teschovirus in endemic field situations. Vet. Microbiol. 2016, 182, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Lu, Y.; Zhang, L.; Li, X.; Chang, Y. Novel species of Teschovirus B comprises at least three distinct evolutionary genotypes. Transbound. Emerg. Dis. 2020, 67, 1015–1018. [Google Scholar] [CrossRef]
- Manuelidis, E.E.; Sprinz, H.; Horstmann, D.M. Pathology of Teschen disease; virus encephalomyelitis of swine. Am. J. Pathol. 1954, 30, 567–597. [Google Scholar]
- Kaku, Y.; Murakami, Y.; Sarai, A.; Wang, Y.; Ohashi, S.; Sakamoto, K. Antigenic properties of porcine teschovirus 1 (PTV-1) Talfan strain and molecular strategy for serotyping of PTVs. Arch. Virol. 2007, 152, 929–940. [Google Scholar] [CrossRef]
- Possatti, F.; Headley, S.A.; Leme, R.A.; Dall Agnol, A.M.; Zotti, E.; de Oliveira, T.E.S.; Alfieri, A.F.; Alfieri, A.A. Viruses associated with congenital tremor and high lethality in piglets. Transbound. Emerg. Dis. 2018, 65, 331–337. [Google Scholar] [CrossRef]
- Takahashi, M.; Seimiya, Y.M.; Seki, Y.; Yamada, M. A piglet with concurrent polioencephalomyelitis due to porcine teschovirus and postweaning multisystemic wasting syndrome. J. Vet. Med. Sci. 2008, 70, 497–500. [Google Scholar] [CrossRef]
- Hwang, J.; Qi, L. Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018, 43, 593–605. [Google Scholar] [CrossRef]
- Wiseman, R.L.; Mesgarzadeh, J.S.; Hendershot, L.M. Reshaping endoplasmic reticulum quality control through the unfolded protein response. Mol. Cell 2022, 82, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.C.; Meier, S.E.; Ingram, A.L.; Abisambra, J.F. PERK-opathies: An endoplasmic reticulum stress mechanism underlying neurodegeneration. Curr. Alzheimer Res. 2016, 13, 150–163. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Nieto-Torres, J.L.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Alvarez, E.; Oliveros, J.C.; Zhao, J.; Fett, C.; Perlman, S.; Enjuanes, L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011, 7, e1002315. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Men, Y.; Wang, D.; Xu, D.; Liu, S.; Xiao, S.; Fang, L. Porcine reproductive and respiratory syndrome virus infection induces endoplasmic reticulum stress, facilitates virus replication, and contributes to autophagy and apoptosis. Sci. Rep. 2020, 10, 13131. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Yamada, M.; Miyazaki, A.; Yamamoto, Y.; Nakamura, K.; Ito, M.; Tsunemitsu, H.; Narita, M. Experimental teschovirus encephalomyelitis in gnotobiotic pigs. J. Comp. Pathol. 2014, 150, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Fu, F.; Ma, Y.; Zhang, X.; Li, L.; Feng, L.; Liu, P. The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production. J. Virol. 2018, 92, e00431-18. [Google Scholar] [CrossRef] [PubMed]
- Groenendyk, J.; Agellon, L.B.; Michalak, M. Calcium signaling and endoplasmic reticulum stress. Int. Rev. Cell Mol. Biol. 2021, 363, 1–20. [Google Scholar]
- He, B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006, 13, 393–403. [Google Scholar] [CrossRef]
- Wang, L.; Byrum, B.; Zhang, Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230. [Google Scholar] [CrossRef]
- Wang, E.; Guo, D.; Li, C.; Wei, S.; Wang, Z.; Liu, Q.; Zhang, B.; Kong, F.; Feng, L.; Sun, D. Molecular Characterization of the ORF3 and S1 Genes of Porcine Epidemic Diarrhea Virus Non S-INDEL Strains in Seven Regions of China, 2015. PLoS ONE 2016, 11, e0160561. [Google Scholar] [CrossRef]
- Palmquist, J.M.; Munir, S.; Taku, A.; Kapur, V.; Goyal, S.M. Detection of porcine teschovirus and enterovirus type II by reverse transcription-polymerase chain reaction. J. Vet. Diagn. Investig. 2002, 14, 476–480. [Google Scholar] [CrossRef]
- Chu, D.K.; Poon, L.L.; Guan, Y.; Peiris, J.S. Novel astroviruses in insectivorous bats. J. Virol. 2008, 82, 9107–9114. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yu, X.; Luo, B.; Yan, M.; Li, R.; Qu, T.; Ren, X. Epidemiology and molecular characterization of Porcine teschovirus in Hunan, China. Transbound. Emerg. Dis. 2018, 65, 480–490. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Han, S.; Mao, L.; Liao, Y.; Sun, S.; Zhang, Z.; Mo, Y.; Liu, H.; Zhi, X.; Lin, S.; Seo, H.S.; et al. Sec62 Suppresses Foot-and-Mouth Disease Virus Proliferation by Promotion of IRE1α-RIG-I Antiviral Signaling. J. Immunol. 2019, 203, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.; Kattoor, J.J.; Sircar, S.; VinodhKumar, O.R.; Thomas, P.; Ghosh, S.; Malik, Y.S. Detection and Molecular Characterization of Porcine Teschoviruses in India: Identification of New Genotypes. Indian J. Microbiol. 2024, 64, 963–972. [Google Scholar] [CrossRef]
- Boros, Á.; Nemes, C.; Pankovics, P.; Kapusinszky, B.; Delwart, E.; Reuter, G. Porcine teschovirus in wild boars in Hungary. Arch. Virol. 2012, 157, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, K. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J. Virol. 1990, 64, 195–206. [Google Scholar] [CrossRef]
- Deng, M.Y.; Millien, M.; Jacques-Simon, R.; Flanagan, J.K.; Bracht, A.J.; Carrillo, C.; Barrette, R.W.; Fabian, A.; Mohamed, F.; Moran, K.; et al. Diagnosis of Porcine teschovirus encephalomyelitis in the Republic of Haiti. J. Vet. Diagn. Invest. 2012, 24, 671–678. [Google Scholar] [CrossRef]
- Yang, T.; Li, R.; Yao, Q.; Zhou, X.; Liao, H.; Ge, M.; Yu, X. Prevalence of Porcine teschovirus genotypes in Hunan, China: Identification of novel viral species and genotypes. J. Gen. Virol. 2018, 99, 1261–1267. [Google Scholar] [CrossRef]
- Kuiken, T.; Holmes, E.C.; McCauley, J.; Rimmelzwaan, G.F.; Williams, C.S.; Grenfell, B.T. Host species barriers to influenza virus infections. Science 2006, 312, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Pankovics, P.; Knowles, N.J.; Reuter, G. Natural interspecies recombinant bovine/porcine enterovirus in sheep. J. Gen. Virol. 2012, 93 Pt 9, 1941–1951. [Google Scholar] [CrossRef]
- Lukashev, A.N. Recombination among picornaviruses. Rev. Med. Virol. 2010, 20, 327–337. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, A. Virus-induced ER stress and the unfolded protein response. Front. Plant Sci. 2012, 3, 293. [Google Scholar] [CrossRef] [PubMed]
- Peña, J.; Harris, E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J. Biol. Chem. 2011, 286, 14226–14236. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xin, X.; Wang, T.; Wan, J.; Ou, Y.; Yang, Z.; Yu, Q.; Zhu, L.; Guo, Y.; Wu, Y. Japanese encephalitis virus induces apoptosis and encephalitis by activating the PERK pathway. J. Virol. 2019, 93, e00887-19. [Google Scholar] [CrossRef]
- Gladwyn-Ng, I.; Cordón-Barris, L.; Alfano, C.; Creppe, C.; Couderc, T.; Morelli, G.; Thelen, N.; America, M.; Bessières, B.; Encha-Razavi, F.; et al. Stress-induced unfolded protein response contributes to Zika virus-associated microcephaly. Nat. Neurosci. 2018, 21, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Turpin, J.; El-Safadi, D.; Lebeau, G.; Frumence, E.; Desprès, P.; Viranaïcken, W.; Krejbich-Trotot, P. CHOP Pro-Apoptotic Transcriptional Program in Response to ER Stress Is Hacked by Zika Virus. Int. J. Mol. Sci. 2021, 22, 3750. [Google Scholar] [CrossRef]
- Ambrose, R.L.; Mackenzie, J.M. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 2011, 85, 2723–2732. [Google Scholar] [CrossRef]



| Primer. | Sequence of Primer (5′-3′) | Cite by |
|---|---|---|
| PDcoV-F | ATCCTCCAAGGAGGCTATGC | [26] |
| PDcoV-R | GCGAATTCTGGATCGTTGTT | |
| PEDV-F | TTTTCTAATCATTTGGTCAACG | [27] |
| PEDV-R | AATACTCATACTAAAGTTGGTGG | |
| PSV-F | GTGGCGACAGGGTACAGAAGAG | [28] |
| PSV-R | GTGGCGACAGGGTACAGAAGAG | |
| PAstV-F | GARTTYGATTGGRCKCGKTAYGA | [29] |
| PAstV-R | GGYTTKACCCACATNCCRAA | |
| PTV-F | TGTTGTGTTTAAACACAGAAAT | [30] |
| PTV-R | TTCAACTGACTATACAAAGTAC |
| Primer | Sequence of Primer (5′-3′) | Product Size (bp) | Consultation |
|---|---|---|---|
| PTV-VP1-F | TGGCTGACCTGCCTGATAAA | 181 | This study |
| PTV-VP1-R | AGGTCAAGTGTCCCATCAGG | ||
| ATF4-F | CCCTTTACGTTCTTGCAAACTC | 165 | [33] |
| ATF4-R | GCTTCCTATCTCCTTCCGAGA | ||
| CHOP-F | CTCAGGAGGAAGAGGAGGAAG | 133 | [33] |
| CHOP-R | GCTAGCTGTGCCACTTTCCTT | ||
| GAPDH-F | CCTTCCGTGTCCCTACTGCCAAC | 103 | [33] |
| GAPDH-R | GACGCCTGCTTCACCACCTTCT |
| Identity% | Structure Protein | Non-Structure Protein | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference Strain | Country | GeneBank Accession No. | Complete Genome | P1 | Vp1 | Vp2 | Vp3 | Vp4 | L | P2 | P3 | |||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |||
| PTV A1/CHN-NP1-2016 | China | MF967586 | 83.4 | 76.1 | 71.3 | 76.1 | 69.1 | 70.2 | 80.2 | 82.8 | 70.0 | 75.2 | 83.8 | 93.2 | 89.1 | 95.3 | 87.3 | 96.3 | 91.3 | 98.8 |
| PTVA2/GX2020 | China | OM281048 | 90.2 | 93.6 | 90.6 | 97.1 | 89.6 | 96.6 | 84.3 | 85.0 | 91.9 | 96.7 | 89.1 | 100 | 90.3 | 93 | 86.8 | 97.1 | 91.3 | 98.8 |
| PTV A3/HuN4 | China | MF170908 | 83.0 | 79.2 | 71.3 | 76.8 | 66.7 | 71.0 | 81.1 | 83.9 | 71.6 | 76.9 | 88.3 | 98.6 | 88.4 | 97.7 | 87.7 | 97.3 | 90.5 | 98.2 |
| PTV A4/HuN23 | China | MF170927 | 86.8 | 85.5 | 79.8 | 85.4 | 76.9 | 79.1 | 82.5 | 86.1 | 80.2 | 88.0 | 85.6 | 100 | 84.9 | 88.4 | 88.9 | 98.2 | 92.3 | 99.4 |
| PTV A5/HuN33 | China | MF170937 | 83.5 | 80.2 | 73.9 | 78.3 | 68.3 | 69.5 | 82.4 | 86.1 | 75.0 | 78.1 | 85.6 | 97.3 | 80.2 | 88.4 | 87 | 98.2 | 91.3 | 98.4 |
| PTV A6/HuN25 | China | MF170929 | 86.8 | 85.7 | 80 | 86.9 | 78.5 | 82.6 | 83.9 | 85.7 | 80.0 | 89.7 | 87.4 | 100 | 88.4 | 93 | 88 | 97.5 | 92.1 | 99.4 |
| PTV A7/F_43 | USA | AF296092 | 81.8 | 76.3 | 78.8 | 77.3 | 73.0 | 75.6 | 77.3 | 82.9 | 74.7 | 76.9 | 83.3 | 97.3 | 87.2 | 95.3 | 84.6 | 95.9 | 86.8 | 97.4 |
| PTV A8/Fuyu2009-2 | China | KC757344 | 85.0 | 82.5 | 73.6 | 86.4 | 75.6 | 80.3 | 82.6 | 86.1 | 80.4 | 90.9 | 87.4 | 100 | 85.3 | 89.5 | 83.9 | 95.3 | 90.6 | 98.8 |
| PTV A9/HuN1 | China | MF170905 | 83.5 | 77.6 | 72.7 | 76.8 | 70.9 | 73.6 | 80.3 | 81.3 | 71.9 | 78.9 | 83.3 | 94.6 | 90.3 | 94.2 | 85.7 | 96.7 | 91.7 | 99 |
| PTV A10/Vir_46188 | Germany | AF296119 | 81.4 | 79.1 | 70.1 | 74.8 | 64.5 | 67.6 | 80.5 | 82.1 | 72.3 | 76.4 | 82.9 | 95.9 | 87.6 | 93 | 84.4 | 97.3 | 89.2 | 98.7 |
| PTV A11/HuN19 | China | MF170923 | 83.7 | 79.3 | 72.1 | 76.9 | 67.3 | 71.8 | 80.1 | 83.9 | 72.7 | 77.3 | 87.4 | 97.3 | 91.5 | 97.7 | 88 | 97.1 | 91.8 | 98.8 |
| PTV B1/HuN41 | China | MG875515 | 71.6 | 51.3 | 62.8 | 64.8 | 54.8 | 52.9 | 69.7 | 66.7 | 63.6 | 66.9 | 82.0 | 97.3 | 73.3 | 73.3 | 79.2 | 90.2 | 74 | 83.4 |
| PTV B3/HuN42 | China | MG875516 | 72.1 | 54.7 | 64.1 | 65.2 | 57.2 | 51.7 | 70.4 | 67.9 | 65.4 | 70.7 | 82.4 | 94.6 | 71.3 | 72.1 | 78.8 | 89.9 | 73.9 | 83.6 |
| PTV B22/JiangX1 | China | MN094632 | 73.2 | 55.9 | 67.3 | 71.9 | 64.4 | 65.2 | 74.3 | 76.1 | 67.6 | 74.8 | 76.6 | 90.5 | 69.4 | 73.3 | 79 | 89.5 | 74.2 | 84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Du, Y.; Lv, Y.; Wei, X.; Cui, C.; Qin, Y.; Lu, B.; Chen, Z.; Ouyang, K.; Chen, Y.; et al. Isolation and Characterization of a Novel Porcine Teschovirus 2 Strain: Incomplete PERK-Mediated Unfolded Protein Response Supports Viral Replication. Viruses 2025, 17, 1200. https://doi.org/10.3390/v17091200
Feng X, Du Y, Lv Y, Wei X, Cui C, Qin Y, Lu B, Chen Z, Ouyang K, Chen Y, et al. Isolation and Characterization of a Novel Porcine Teschovirus 2 Strain: Incomplete PERK-Mediated Unfolded Protein Response Supports Viral Replication. Viruses. 2025; 17(9):1200. https://doi.org/10.3390/v17091200
Chicago/Turabian StyleFeng, Xiaoying, Yiyang Du, Yueqing Lv, Xiaofang Wei, Chang Cui, Yibin Qin, Bingxia Lu, Zhongwei Chen, Kang Ouyang, Ying Chen, and et al. 2025. "Isolation and Characterization of a Novel Porcine Teschovirus 2 Strain: Incomplete PERK-Mediated Unfolded Protein Response Supports Viral Replication" Viruses 17, no. 9: 1200. https://doi.org/10.3390/v17091200
APA StyleFeng, X., Du, Y., Lv, Y., Wei, X., Cui, C., Qin, Y., Lu, B., Chen, Z., Ouyang, K., Chen, Y., Wei, Z., Huang, W., He, Y., & Qin, Y. (2025). Isolation and Characterization of a Novel Porcine Teschovirus 2 Strain: Incomplete PERK-Mediated Unfolded Protein Response Supports Viral Replication. Viruses, 17(9), 1200. https://doi.org/10.3390/v17091200









