Diversity of RNA Viruses and Circular Viroid-like Elements in Heterobasidion spp. in Near-Natural Forests of Bosnia and Herzegovina
Abstract
1. Introduction
2. Materials and Methods
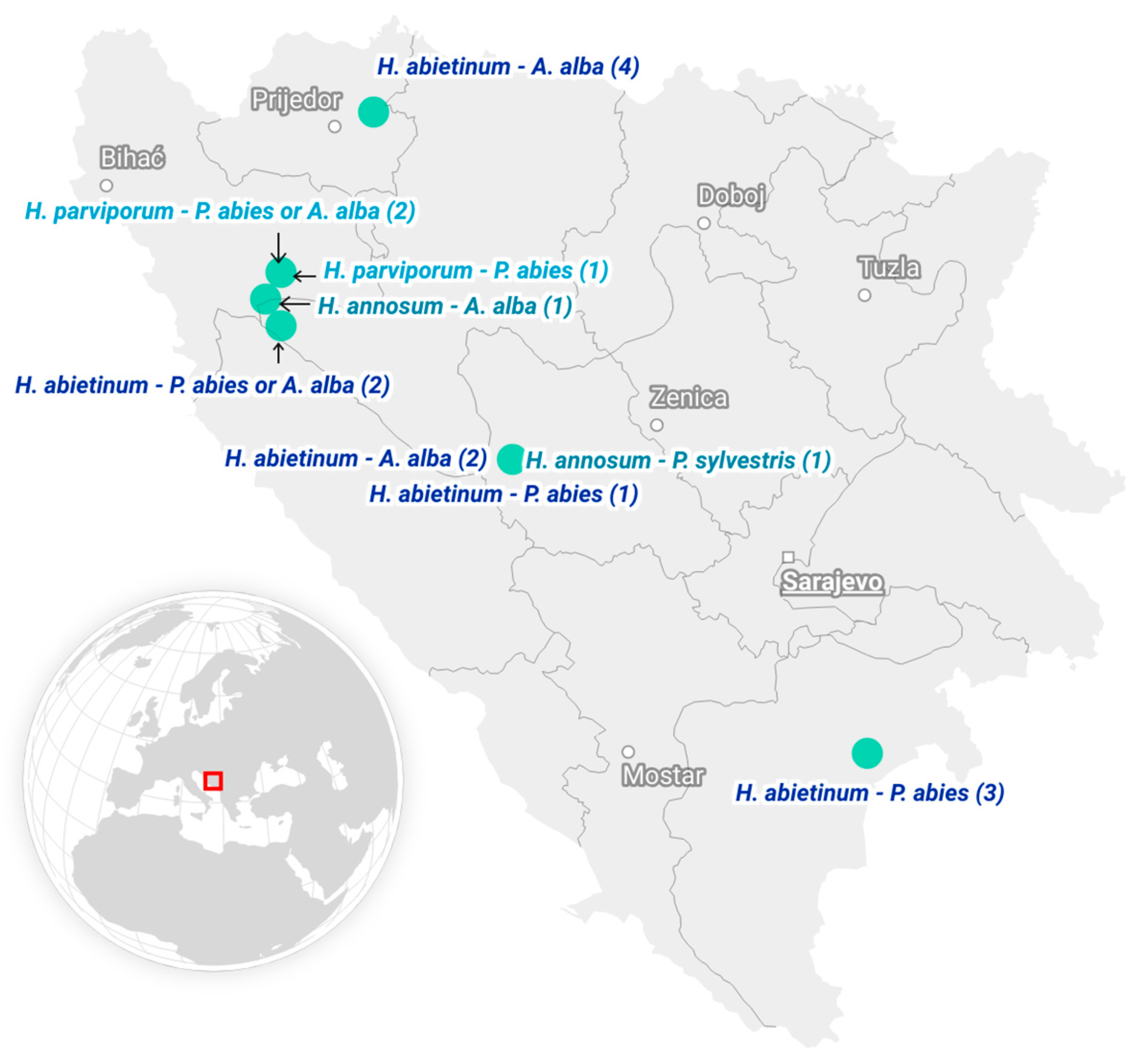
2.1. Collection of Heterobasidion Isolates
2.2. RNA Extraction
2.3. Total RNA Sequencing
2.4. Bioinformatics
2.5. Phylogenetic Analysis and Pairwise Comparison
2.6. DAPC Analysis of Ambiviruses
3. Results and Discussion
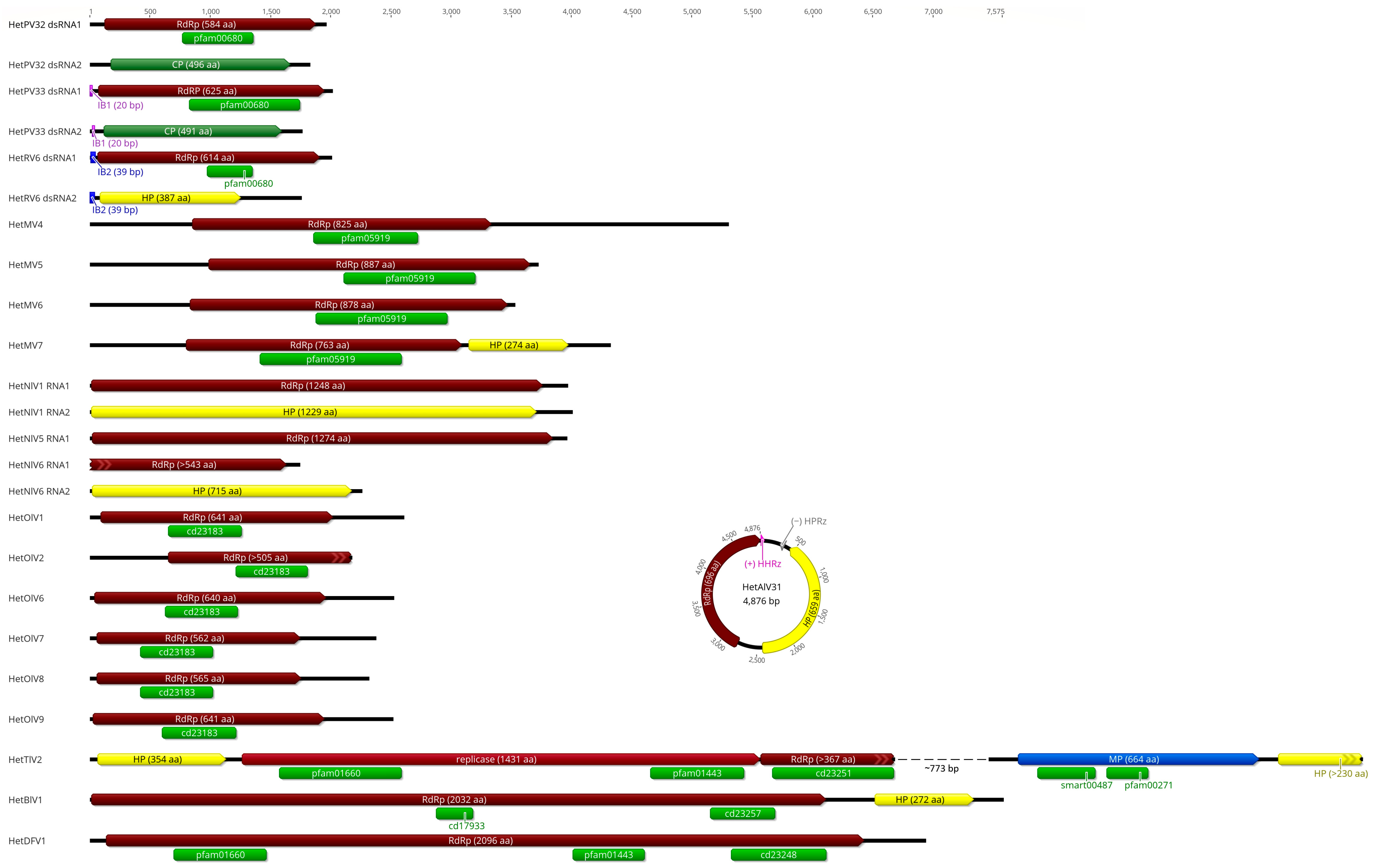
3.1. Virus and Viroid-like Agents Discovery
3.2. Alphapartitiviruses
| Mycovirus | Acronym | Segment | GenBank Accession | Host Species | Length (nt) | % GC | Mapped Reads f | Mean Depth | BLASTX First g | Identity | Query Cover | E Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heterobasidion partitivirus 32 | HetPV32 | dsRNA1 | PP626369 | H. abietinum | 1934 d | 47.7% d | 260,018 d | 18,931 d |
Sarcosphaera coronaria partitivirus (QLC36816) | 50.1% | 89% | 0 |
| dsRNA2 | PP626370 | H. abietinum | 1816 | 54.7% | 25,327 | 1939 | Carrot cryptic virus (ACL93279) | 43.0% | 70% | 3.0 × 10−92 | ||
| Heterobasidion partitivirus 33 | HetPV33 | dsRNA1 | PP626371 | H. abietinum | 1970 d | 46.5%d | 12,617 d | 916 d |
Heterobasidion partitivirus 4 (ADV15443) | 70.5% | 94% | 0 |
| dsRNA2 | PP626372 | H. abietinum | 1755 | 53.3% | 59,967 | 4803 |
Tulasnella partitivirus 3 (BDB07480) | 49.9% | 83% | 1.0 × 10−130 | ||
| Heterobasidion RNA virus 6 | HetRV6 | dsRNA1 | PP626373 | H. abietinum OR H. annosum | 1996 | 56.3% | 1172 | 78 |
Heterobasidion RNA virus 6 (ADW82833) | 93.1% | 91% | 0 |
| dsRNA2 | PP626374 | H. abietinum OR H. annosum | 1748 | 63.4% | 915 | 69 |
Heterobasidion RNA virus 6 (QED55787) | 93.7% | 62% | 0 | ||
| Heterobasidion mitovirus 4 | HetMV4 | RNA1 | PP626375 | H. abietinum a | 5292 | 42.1% | 67,004 | 1685 |
Heterobasidion mitovirus 3 (QED55406) h | 69.2% h | 46% h | 0 h |
| Heterobasidion mitovirus 5 | HetMV5 | RNA1 | PP626376 | H. abietinum | 3715 | 40.9% | 676,330 | 25,276 |
Heterobasidion mitovirus 1 (AIF33766) h | 63.6% h | 71% h | 0 h |
| Heterobasidion mitovirus 6 | HetMV6 | RNA1 | PP626377 | H. abietinum | 3521 | 41.4% | 644,309 | 26,115 |
Heterobasidion mitovirus 1 (AIF33766) h | 63.4% h | 74%h | 0 h |
| Heterobasidion mitovirus 7 | HetMV7 | RNA1 | PP626378 | H. abietinum OR H. annosum b | 4314 | 41.1% | 538,636 | 17,624 |
Lentinula edodes mitovirus 1 (QOX06058)h | 38.8% h | 51%h | 2.0 × 10−160h |
| Heterobasidion narna-like virus 1 | HetNlV1 | RNA1 | PP626379 | H. abietinum a | 3956 | 54.0% | 34,130 | 1221 |
Heterobasidion narna-like virus 1 (UHK02569) | 92.8% | 93% | 0 |
| RNA2 | PP626380 | H. abietinum a | 3998 | 55.8% | 27,091 | 960 |
Heterobasidion narna-like virus 1 (UHK02570) | 91.8% | 92% | 0 | ||
| Heterobasidion narna-like virus 5 | HetNlV5 | RNA1 | PP626381 | H. abietinum OR H. annosum | 3953 | 55.3% | 77,698 | 2714 |
Heterobasidion narna-like virus 4 (WBE16499) | 78.9% | 96% | 0 |
| Heterobasidion narna-like virus 6 | HetNlV6 | RNA1 | PP626382 | H. abietinum OR H. annosum | 1731 e | 51.2 | 4745 | 379 |
Mbeech associated narna-like virus 5 (WPR16644) | 53.0% | 93% | 0 |
| RNA2 | PP626383 | H. abietinum OR H. annosum | 2252 | 51.5 | 6832 | 394 |
Botrytis cinerea binarnavirus 2 (QLF49184) | 45.9% | 92% | 0 | ||
| Heterobasidion ourmia-like virus 1 | HetOlV1 | RNA1 | PP626384 | H. parviporum | 2601 | 49.5 | 214,903 | 11,422 |
Heterobasidion ourmia-like virus 1 (UHK02571) | 96.6% | 73% | 0 |
| Heterobasidion ourmia-like virus 2 | HetOlV2 | RNA1 | PP626385 | H. abietinum a | 2169 e | 58.0 | 10,251 | 640 |
Heterobasidion ourmia-like virus 2 (UOX39320) | 93.0% | 67% | 0 |
| Heterobasidion ourmia-like virus 6 | HetOlV6 | RNA1 | PP626386 | H. abietinum | 2516 | 51.2 | 12,337 | 658 |
Heterobasidion ourmia-like virus 1 (WLV75612) | 76.9% | 74% | 0 |
| Heterobasidion ourmia-like virus 7 | HetOlV7 | RNA1 | PP626387 | H. abietinum a | 2366 | 50.5 | 11,364 | 566 |
Heterobasidion ourmia-like virus 1 (WLV75612) | 79.4% | 72% | 0 |
| Heterobasidion ourmia-like virus 8 | HetOlV8 | RNA1 | PP626388 | H. abietinum | 2305 | 50.4 | 16,611 | 884 |
Heterobasidion ourmia-like virus 1 (WLV75612) | 77.1% | 76% | 0 |
| Heterobasidion ourmia-like virus 9 | HetOlV9 | RNA1 | PP626389 | H. abietinum OR H. annosum | 2505 | 50.3 | 118,718 | 6411 |
Heterobasidion ourmia-like virus 1 (WLV75612) | 81.4% | 74% | 0 |
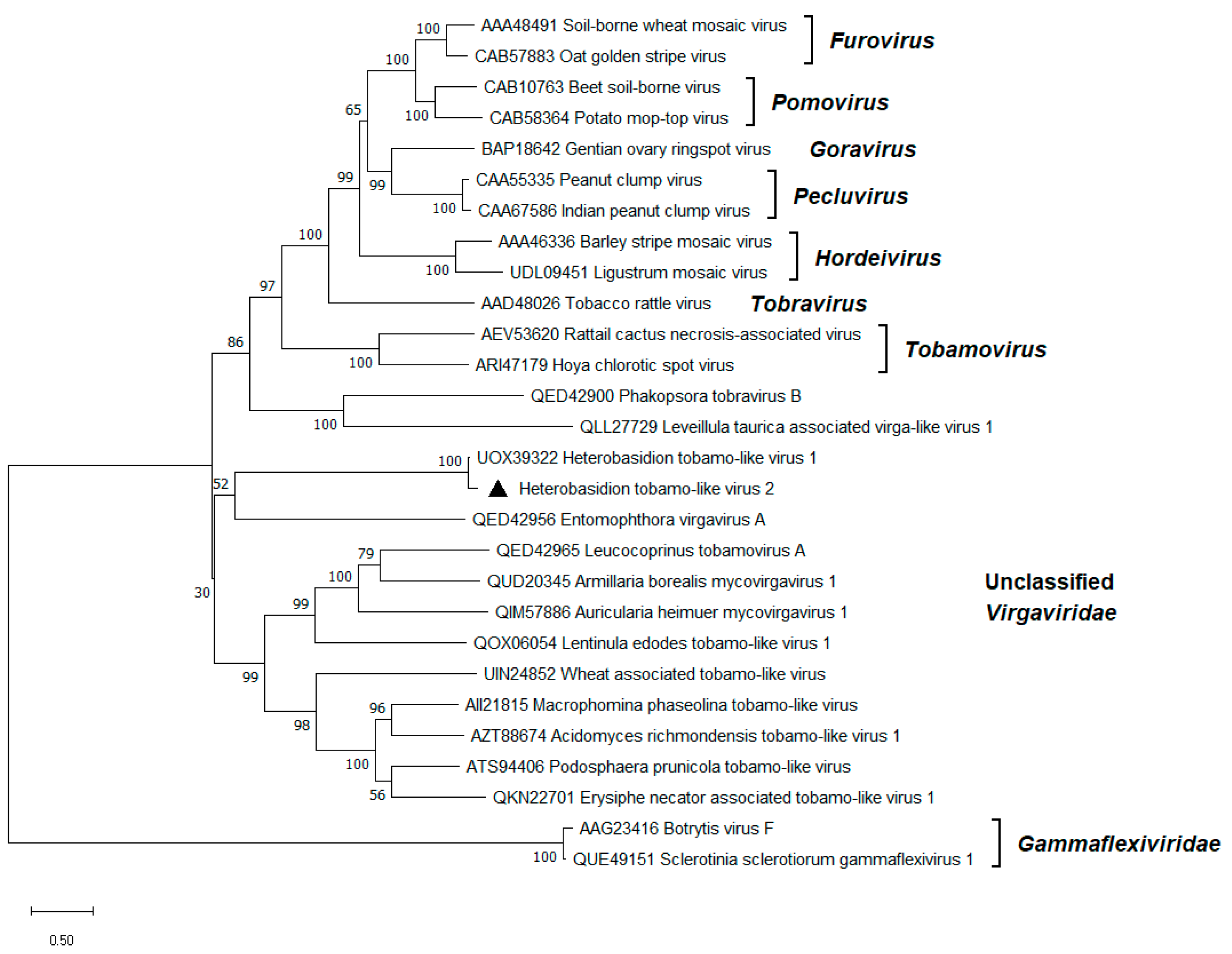
| Heterobasidion tobamo-like virus 2 | HetTlV2 | RNA1 | PP626390 | H. abietinum OR H. annosum | 6675 e | 49.3 | 1880 | 38 |
Heterobasidion tobamo-like virus 1 (UOX39322) | 87.6% | 64% | 0 |
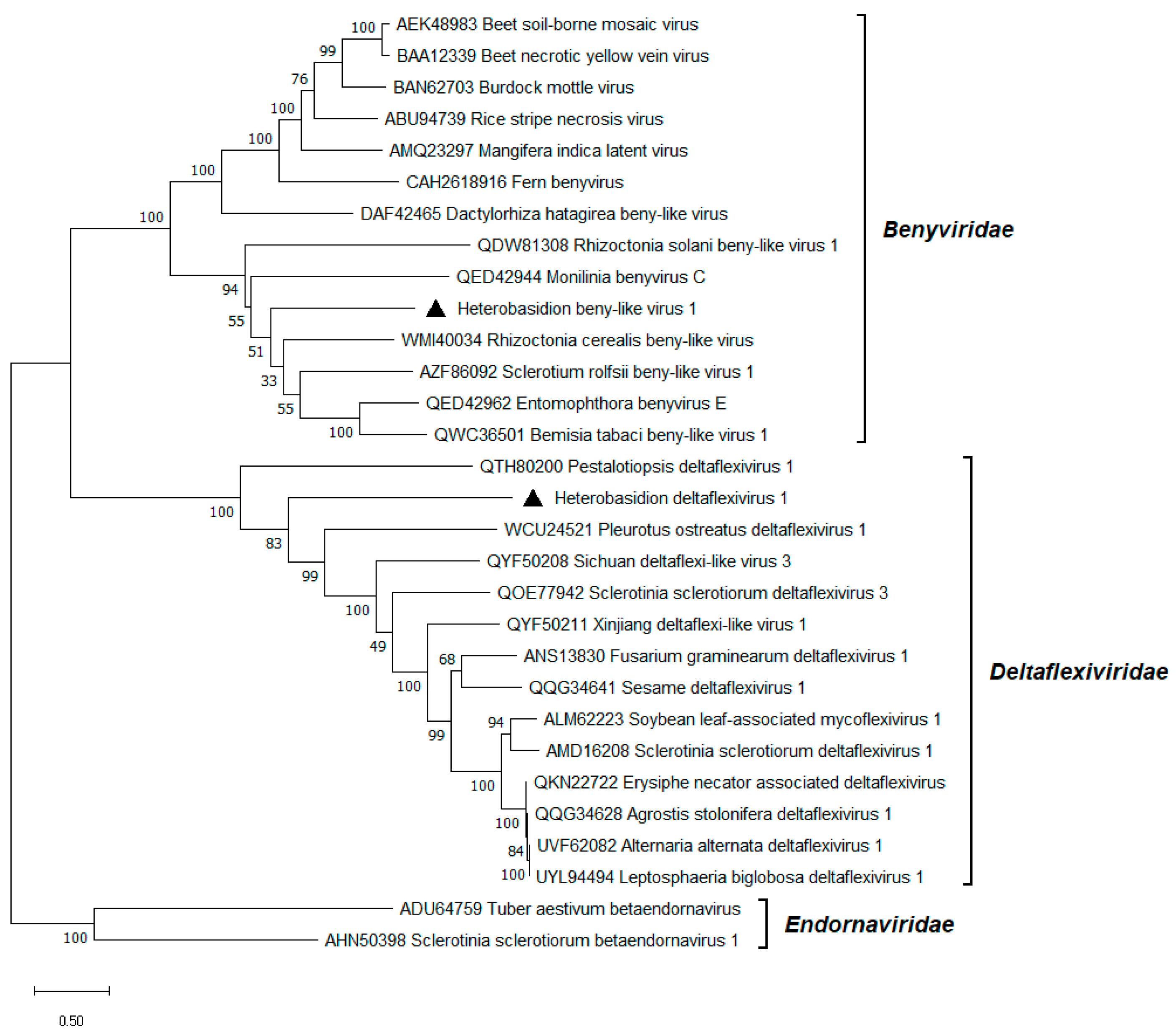
| Heterobasidion beny-like virus 1 | HetBlV1 | RNA1 | PP626391 | H. abietinum OR H. annosum a | 7563 d | 57.2 d | 268,134 d | 4956 d |
Rhizoctonia cerealis beny-like virus (WMI40034) | 31.1% | 64% | 2.0 × 10−162 |
| Heterobasidion deltaflexivirus 1 | HetDFV1 | RNA1 | PP626392 | H. abietinum OR H. annosum a | 6928 | 64.1 | 123,445 | 2322 |
Rhizoctonia solani flexi-like virus 1 (QDW81317) | 33.1% | 34% | 2.0 × 10−107 |
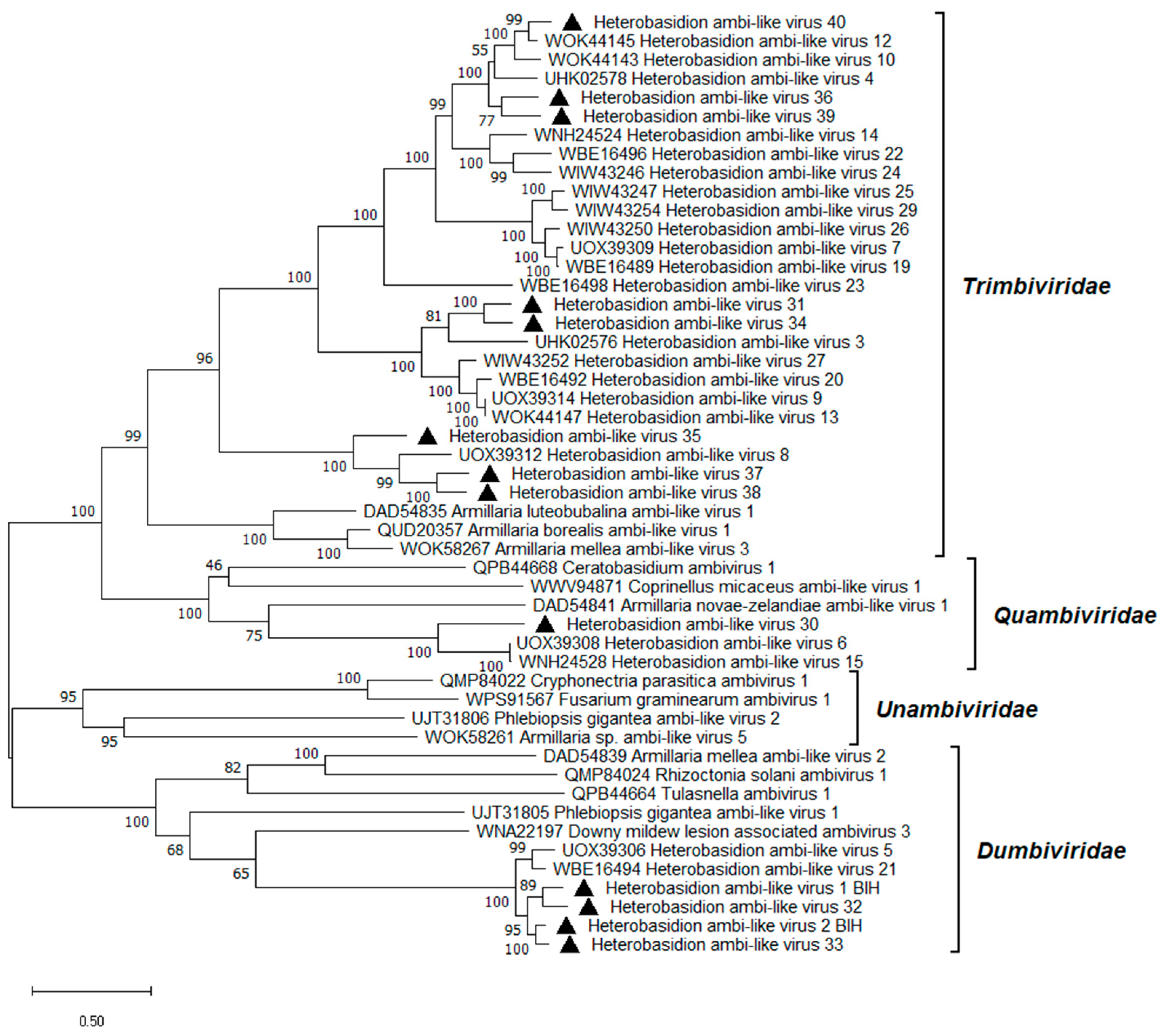
| Heterobasidion ambi-like virus 1 | HetAlV1 | RNA1 | PP744446 | H. abietinum | 4754 | 49.2 | 822,963 | 24,597 |
Heterobasidion ambi-like virus 1 (UHK02572) | 97.8% | 38% | 0 |
| Heterobasidion ambi-like virus 2 | HetAlV2 | RNA1 | PP744447 | H. abietinum OR H. annosum | 4286 | 49.2 | 938,195 | 30,286 |
Heterobasidion ambi-like virus 2 (UHK02574) | 95.6% | 41% | 0 |
| Heterobasidion ambi-like virus 30 | HetAlV30 | RNA1 | PP744448 | H. abietinum | 5085 | 49.2 | 8663 | 241 |
Heterobasidion ambi-like virus 6 (UOX39308) | 58.0% | 42% | 0 |
| Heterobasidion ambi-like virus 31 | HetAlV31 | RNA1 | PP744449 | H. abietinum c | 4876 | 49.9 | 32,263 | 922 |
Heterobasidion ambi-like virus 3 (UHK02576) | 68.3% | 42% | 0 |
| Heterobasidion ambi-like virus 32 | HetAlV32 | RNA1 | PP744450 | H. abietinum | 4450 | 49.2 | 1,172,645 | 37,056 |
Heterobasidion ambi-like virus 1 (UHK02572) | 83.3% | 40% | 0 |
| Heterobasidion ambi-like virus 33 | HetAlV33 | RNA1 | PP744451 | H. abietinum | 4368 | 49.2 | 505,254 | 16,380 |
Heterobasidion ambi-like virus 2 (UHK02574) | 92.6% | 40% | 0 |
| Heterobasidion ambi-like virus 34 | HetAlV34 | RNA1 | PP744452 | H. abietinum OR H. annosum | 4956 | 50.2 | 110,813 | 3101 |
Heterobasidion ambi-like virus 3 (UHK02576) | 67.3% | 42% | 0 |
| Heterobasidion ambi-like virus 35 | HetAlV35 | RNA1 | PP744453 | H. abietinum OR H. annosum | 4851 | 48.4 | 11,912 | 336 |
Heterobasidion ambi-like virus 8 (UOX39312) | 61.6% | 40% | 0 |
| Heterobasidion ambi-like virus 36 | HetAlV36 | RNA1 | PP744454 | H. abietinum OR H. annosum | 4660 | 49.7 | 61,156 | 1779 |
Heterobasidion ambi-like virus 12 (WOK44145) | 76.0% | 44% | 0 |
| Heterobasidion ambi-like virus 37 | HetAlV37 | RNA1 | PP744455 | H. abietinum OR H. annosum | 4767 | 48.8 | 54,368 | 1574 |
Heterobasidion ambi-like virus 8 (UOX39312) | 65.9% | 41% | 0 |
| Heterobasidion ambi-like virus 38 | HetAlV38 | RNA1 | PP744456 | H. abietinum OR H. annosum | 4762 | 48.0 | 87,791 | 2543 |
Heterobasidion ambi-like virus 8 (UOX39312) | 66.3% | 41% | 0 |
| Heterobasidion ambi-like virus 39 | HetAlV39 | RNA1 | PP744457 | H. abietinum OR H. annosum | 4632 | 49.0 | 21,324 | 602 |
Heterobasidion ambi-like virus 10 (WOK44143) | 74.1% | 44% | 0 |
| Heterobasidion ambi-like virus 40 | HetAlV40 | RNA1 | PP744458 | H. abietinum OR H. annosum | 4287 | 48.4 | 53,587 | 1644 |
Heterobasidion ambi-like virus 12 (WOK44145) | 87.7% | 45% | 0 |
| Heterobasidion circular RNA 1 | HetcRNA1 | RNA1 | PQ298351 | H. abietinum | 919 | 60.1 | 206,319 | 33,675 | n.s. | |||
| Heterobasidion circular RNA 2 | HetcRNA2 | RNA1 | PQ298352 | H. abietinum | 918 | 60.3 | 22,977 | 3754 | n.s. |
3.3. Orthocurvulavirus Annosi
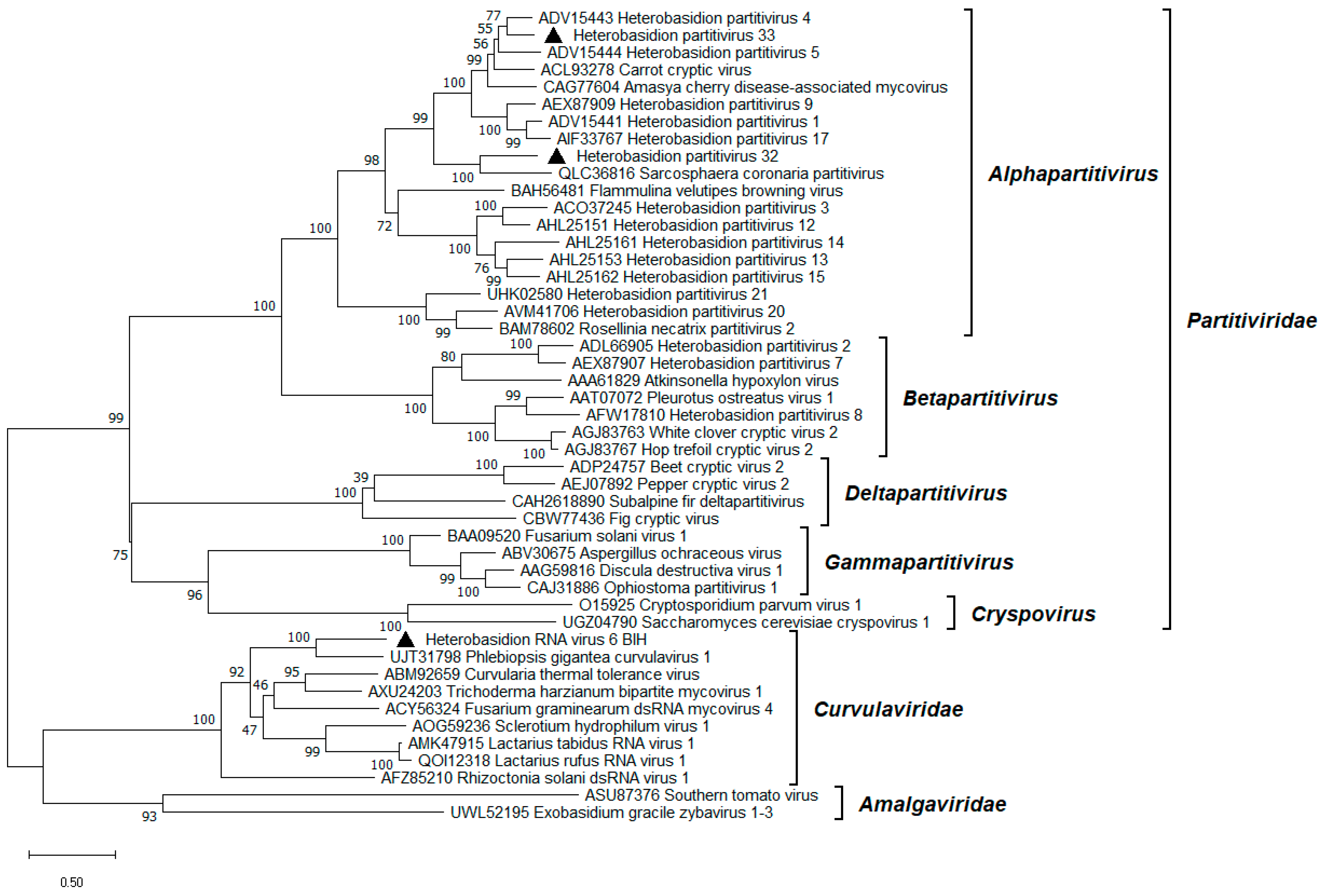
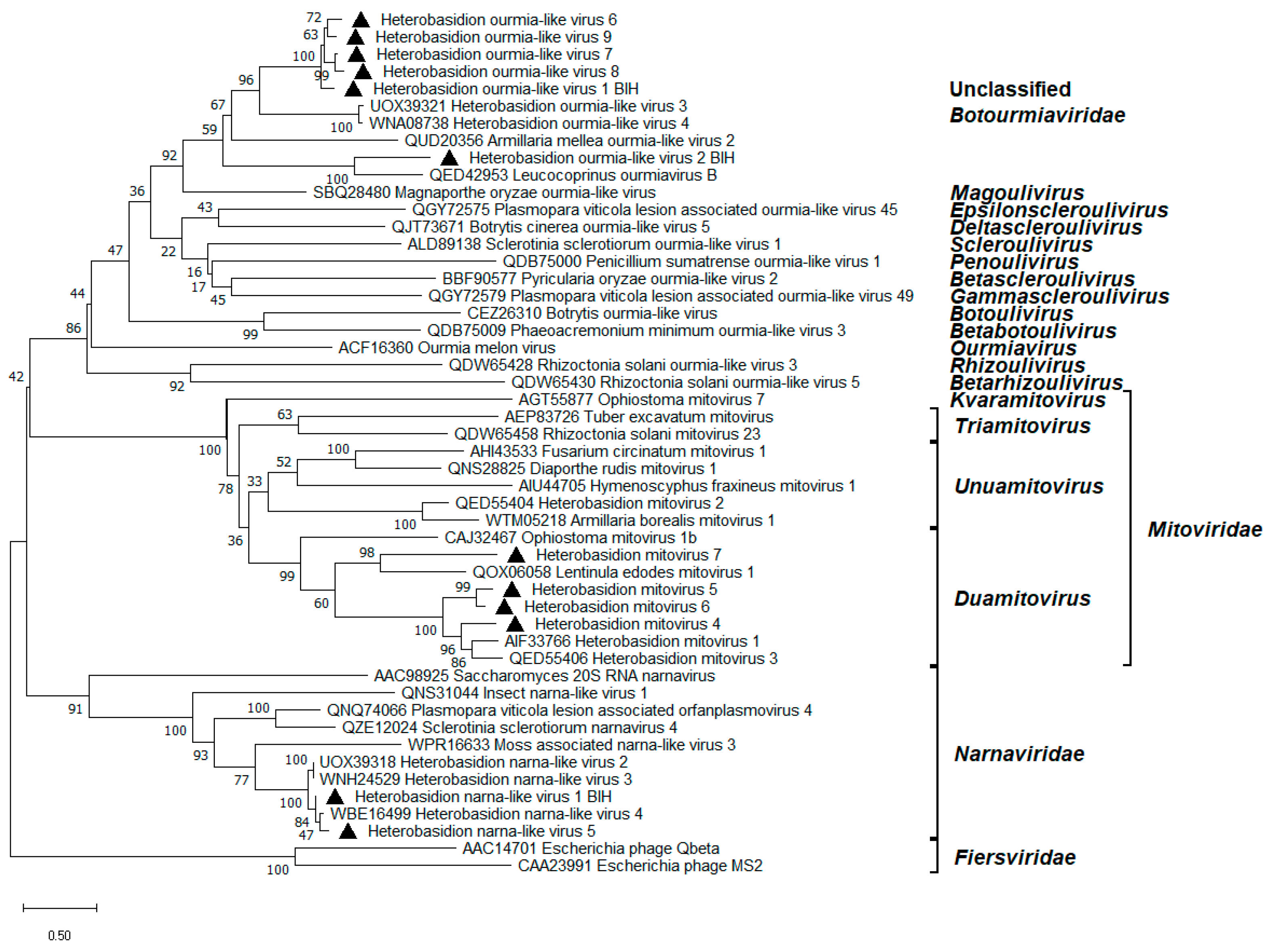
3.4. Mitoviruses
3.5. Narna-like Viruses
3.6. Ourmia-like Viruses
3.7. Tobamo-like Virus
3.8. Beny-like Virus
3.9. Deltaflexivirus
3.10. Ambi-like Viruses
3.11. Viroid-like cRNAs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mataruga, M.; Ballian, D.; Terzić, R.; Daničić, V.; Cvjetković, B. State of forests in Bosnia and Herzegovina: Ecological and vegetation distribution, management and genetic variability. In Forests of Southeast Europe under a Changing Climate; Šijačić-Nikolić, M., Milovanović, J., Nonić, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 65, pp. 3–19. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Govedar, Z.; Krstić, M.; Keren, S.; Babić, V.; Zlokapa, B.; Kanjevac, B. Actual and balanced stand structure: Examples from beech-fir-spruce old-growth forests in the area of the Dinarides in Bosnia and Herzegovina. Sustainability 2018, 10, 540. [Google Scholar] [CrossRef]
- Motta, R.; Alberti, G.; Ascoli, D.; Berretti, R.; Bilic, S.; Bono, A.; Milic, C.; Vojislav, D.; Finsinger, W.; Garbarino, M.; et al. Old-growth forests in the Dinaric Alps of Bosnia-Herzegovina and Montenegro: A continental hot-spot for research and biodiversity. Front. For. Glob. Change 2024, 7, 1371144. [Google Scholar] [CrossRef]
- Drinić, P. Taksacioni elementi sastojina jele, smrče i bukve prašumskog tipa u Bosni [Taxation elements of the fir-spruce-beech old-growth forests in Bosnia]. Rad. Šumarskog Fak. Univ. u Sarajev. 1956, 4, 107–160. [Google Scholar] [CrossRef]
- Drašković, B.; Gutalj, M.; Stjepanović, S.; Miletić, B. Estimating recent forest losses in Bosnia and Herzegovina by using the Copernicus and Corine land cover databases. Šumarski List 2021, 145, 581–589. [Google Scholar] [CrossRef]
- Vukmir, G.; Stanišljević, L.; Cero, M.; Cacan, M.; Marković, M.; Rudež, M.; Laganin, O.; Kostić, R.; Oprašić, S.; Ćatović, S.; et al. Initial National Communication (INC) of Bosnia and Herzegovina Under the United Nations Framework Convention on Climate Change; UNDP (United Nations Development Programme): Banja Luka, Bosnia and Herzegovina, 2019; Available online: https://www.undp.org/bosnia-herzegovina/publications/initial-national-communication-inc (accessed on 17 June 2025).
- Camarero, J.J.; Pizarro, M.; Gernandt, D.S.; Gazol, A. Smaller conifers are more resilient to drought. Agric. For. Meteorol. 2024, 350, 109993. [Google Scholar] [CrossRef]
- Stanivuković, Z.; Vasiljević, R. The most significant biotic harmful agents and their influence on the intensity of spruce (Picea abies Karst.) dieback on the Romanian plateau. Šumarstvo 2019, 3–4, 21–41. [Google Scholar]
- Zahirović, K.; Treštić, T.; Čabaravdić, A.; Dautbašić, M.; Mujezinović, O. Causitive agents of decay of Norway spruce/Picea abies (L.) Karst./on the mountain Zvijezda. Šumarski List 2019, 143, 155–160. [Google Scholar] [CrossRef]
- Karadžić, D.; Stanivuković, Z.; Milanović, S.; Milenković, I. Najznačajniji Prouzrokovači Infektivnih Bolesti u Šumama Republike Srpske [The Most Important Causes of Infectious Diseases in the Forests of the Republic of Srpska]; University of Banja Luka, Faculty of Forestry: Banja Luka, Bosnia and Herzegovina, 2019; 324p, /unibl/book/idKnjiga:2140, 978-99938-56-42-9; Available online: https://pub.unibl.org/s/lat/item/48960 (accessed on 11 August 2025).
- Piri, T.; Vainio, E.J.; Hantula, J. Preventing mycelial spread of Heterobasidion annosum in young Scots pine stands using fungal and viral biocontrol agents. Biol. Control 2023, 184, 105263. [Google Scholar] [CrossRef]
- Ihrmark, K.; Zheng, J.; Stenström, E.; Stenlid, J. Presence of double-stranded RNA in Heterobasidion annosum. For. Pathol. 2001, 31, 387–394. [Google Scholar] [CrossRef]
- Vainio, E.J.; Hantula, J. Taxonomy, biogeography and importance of Heterobasidion viruses. Virus Res. 2016, 219, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Dálya, L.B.; Černý, M.; de la Peña, M.; Poimala, A.; Vainio, E.J.; Hantula, J.; Botella, L. Diversity and impact of single-stranded RNA viruses in Czech Heterobasidion populations. MSystems 2024, 9, e0050624. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Hakanpää, J.; Dai, Y.C.; Hansen, E.; Korhonen, K.; Hantula, J. Species of Heterobasidion host a diverse pool of partitiviruses with global distribution and interspecies transmission. Fungal Biol. 2011, 115, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Hyder, R.; Aday Kaya, A.G.; Hansen, E.; Piri, T.; Doğmuş-Lehtijärvi, H.T.; Lehtijärvi, A.; Korhonen, K.; Hantula, J. Population structure of a novel putative mycovirus infecting the conifer root-rot fungus Heterobasidion annosum sensu lato. Virology 2012, 422, 366–376. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 January 2025).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.L.; Pham, S.; Pevzner, P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Botella, L.; Hejna, O.; Kudláček, T.; Kovačiková, K.; Rost, M.; Forgia, M.; Raco, M.; Milenković, I.; Corcobado, T.; Maia, C.; et al. The virome of the panglobal, wide host-range plant pathogen Phytophthora cinnamomi: Phylogeography and evolutionary insights. Virus Evol. 2025, 11, veaf020. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Forgia, M.; Navarro, B.; Daghino, S.; Cervera, A.; Gisel, A.; Perotto, S.; Aghayeva, D.N.; Akinyuwa, M.F.; Gobbi, E.; Zheludev, I.N.; et al. Hybrids of RNA viruses and viroid-like elements replicate in fungi. Nat. Commun. 2023, 14, 2591. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA package 2.0. Algorit. Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Sutela, S.; Piri, T.; Vainio, E.J. Discovery and community dynamics of novel ssRNA mycoviruses in the conifer pathogen Heterobasidion parviporum. Front. Microbiol. 2021, 12, 770787. [Google Scholar] [CrossRef]
- Vainio, E.J.; Capretti, P.; Motta, E.; Hantula, J. Molecular characterization of HetRV8-ir1, a partitivirus of the invasive conifer pathogenic fungus Heterobasidion irregulare. Arch. Virol. 2013, 158, 1613–1615. [Google Scholar] [CrossRef]
- Jurvansuu, J.; Kashif, M.; Vaario, L.; Vainio, E.J.; Hantula, J. Partitiviruses of a fungal forest pathogen have species-specific quantities of genome segments and transcripts. Virology 2014, 462–463, 25–33. [Google Scholar] [CrossRef]
- Márquez, L.M.; Redman, R.S.; Rodriguez, R.J.; Roossinck, M.J. A virus in a fungus in a plant: Three-way symbiosis required for thermal tolerance. Science 2007, 315, 513–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wu, J.; Zhu, X.; Chen, J. Complete nucleotide sequences of dsRNA2 and dsRNA7 detected in the phytopathogenic fungus Sclerotium hydrophilum and their close phylogenetic relationship to a group of unclassified viruses. Virus Genes 2016, 52, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, M.; Redda, E.T.; Mei, J.; Zhang, J.; Elena, S.F.; Wu, B.; Jiang, X. Complete nucleotide sequence of a novel mycovirus from Trichoderma harzianum in China. Arch. Virol. 2019, 164, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Sutela, S.; Vainio, E.J. Virus population structure in the ectomycorrhizal fungi Lactarius rufus and L. tabidus at two forest sites in Southern Finland. Virus Res. 2020, 285, 197993. [Google Scholar] [CrossRef]
- Drenkhan, T.; Sutela, S.; Veeväli, V.; Vainio, E.J. Phlebiopsis gigantea strains from Estonia show potential as native biocontrol agents against Heterobasidion root rot and contain diverse dsRNA and ssRNA viruses. Biol. Control 2022, 167, 104837. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Vainio, E.J. Mitoviruses in the conifer root rot pathogens Heterobasidion annosum and H. parviporum. Virus Res. 2019, 271, 197681. [Google Scholar] [CrossRef]
- Guo, M.; Shen, G.; Wang, J.; Liu, M.; Bian, Y.; Xu, Z. Mycoviral diversity and characteristics of a negative-stranded RNA virus LeNSRV1 in the edible mushroom Lentinula edodes. Virology 2021, 555, 89–101. [Google Scholar] [CrossRef]
- Vainio, E.J.; Jurvansuu, J.; Streng, J.; Rajamäki, M.-L.; Hantula, J.; Valkonen, J.P.T. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J. Gen. Virol. 2015, 96, 714–725. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a Maximum-Likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Li, W.; Sun, H.; Cao, S.; Zhang, A.; Zhang, H.; Shu, Y.; Chen, H. Extreme diversity of mycoviruses present in single strains of Rhizoctonia cerealis, the pathogen of wheat sharp eyespot. Microbiol Spectr. 2023, 11, e0052223. [Google Scholar] [CrossRef]
- Canuti, M.; Rodrigues, B.; Lang, A.S.; Dufour, S.C.; Verhoeven, J.T.P. Novel divergent members of the Kitrinoviricota discovered through metagenomics in the intestinal contents of red-backed voles (Clethrionomys gapperi). Int. J. Mol. Sci. 2023, 24, 131. [Google Scholar] [CrossRef]
- Forgia, M.; Isgandarli, E.; Aghayeva, D.N.; Huseynova, I.; Turina, M. Virome characterization of Cryphonectria parasitica isolates from Azerbaijan unveiled a new mymonavirus and a putative new RNA virus unrelated to described viral sequences. Virology 2021, 553, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Botella, L.; de la Peña, M.; Vainio, E.J.; Krupovic, M.; Lee, B.D.; Navarro, B.; Sabanadzovic, S.; Simmonds, P.; Turina, M. Ambiviricota, a novel ribovirian phylum for viruses with viroid-like properties. J. Virol. 2024, 98, e00831-24. [Google Scholar] [CrossRef] [PubMed]
- Sutela, S.; Forgia, M.; Vainio, E.J.; Chiapello, M.; Daghino, S.; Vallino, M.; Martino, E.; Girlanda, M.; Perotto, S.; Turina, M. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020, 6, veaa076. [Google Scholar] [CrossRef]
- Göker, M.; Scheuner, C.; Klenk, H.-P.; Stielow, J.B.; Menzel, W. Codivergence of mycoviruses with their hosts. PLoS ONE 2011, 6, e22252. [Google Scholar] [CrossRef]
- Koonin, E.V.; Lee, B.D. Diversity and evolution of viroids and viroid-like agents with circular RNA genomes revealed by metatranscriptome mining. Nucleic Acids Res. 2025, 53, 1278. [Google Scholar] [CrossRef]
- Sun, L.; Hadidi, A. Mycoviroids: Fungi as Hosts and Vectors of Viroids. Cells 2022, 11, 1335. [Google Scholar] [CrossRef]
- Lee, B.D.; Neri, U.; Roux, S.; Wolf, Y.I.; Camargo, A.P.; Krupovic, M.; Simmonds, P.; Kyrpides, N.; Gophna, U.; Dolja, V.V.; et al. Mining metatranscriptomes reveals a vast world of viroid-like circular RNAs. Cell 2023, 186, 646–661. [Google Scholar] [CrossRef]
- Friday, P.; Mukkara, R.A.; Owens, T.; Baumstark, M.F.; Bruist, M.F. Processing of potatospindle tuber viroid RNAs in yeast, a nonconventional host. J. Virol. 2017, 91, e01078-17. [Google Scholar] [CrossRef]
- Latifi, L.; Bernard, C.S. Replication of avocado sunblotch viroid in the cyanobacterium Nostocsp. PCC 7120. J. Plant Pathol. Microbiol. 2016, 7, 341. [Google Scholar] [CrossRef]
- Tian, M.; Wei, S.; Bian, R.; Luo, J.; Khan, H.A.; Tai, H.; Kondo, H.; Hadidi, A.; Andika, I.B.; Sun, L. Natural cross-kingdom spread of apple Scar Skin viroid from apple trees to fungi. Cells 2022, 11, 3686. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Bian, R.; Andika, I.B.; Niu, E.; Liu, Q.; Kondo, H.; Yang, L.; Zhou, H.; Pang, T.; Lian, Z.; et al. Symptomatic plant viroid infections in phytopathogenic fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 13042–13050. [Google Scholar] [CrossRef] [PubMed]
- Afanasenko, O.S.; Khiutti, A.V.; Mironenko, N.V.; Lashina, N.M. Transmission of potato spindle tuber viroid between Phytophthora infestans and host plants. Vavilovskii Zhurnal Genet. Sel. 2022, 26, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Xu, C.; Kotta-Loizou, I.; Jiang, J.; Lv, R.; Kong, L.; Li, S.; Hong, N.; Wang, G.; Coutts, R.H.A.; et al. Novel viroid-like RNAs naturally infect a filamentous fungus. Adv. Sci. 2023, 10, e2204308. [Google Scholar] [CrossRef]
- Navarro, B.; Turina, M. Viroid and viroid-like elements in plants and plant-associated microbiota: A new layer of biodiversity for plant holobionts. New Phytol. 2024, 244, 1216–1222. [Google Scholar] [CrossRef]
- Hermanns, K.; Marklewitz, M.; Zirkel, F.; Kopp, A.; Kramer-Schadt, S.; Junglen, S. Mosquito community composition shapes virus prevalence patterns along anthropogenic disturbance gradients. eLife 2023, 12, e66550. [Google Scholar] [CrossRef]
- Tirera, S.; de Thoisy, B.; Donato, D.; Bouchier, C.; Lacoste, V.; Franc, A.; Lavergne, A. The influence of habitat on viral diversity in neotropical rodent hosts. Viruses 2021, 13, 1690. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dálya, L.B.; Hejna, O.; de la Peña, M.; Stanivuković, Z.; Kudláček, T.; Botella, L. Diversity of RNA Viruses and Circular Viroid-like Elements in Heterobasidion spp. in Near-Natural Forests of Bosnia and Herzegovina. Viruses 2025, 17, 1144. https://doi.org/10.3390/v17081144
Dálya LB, Hejna O, de la Peña M, Stanivuković Z, Kudláček T, Botella L. Diversity of RNA Viruses and Circular Viroid-like Elements in Heterobasidion spp. in Near-Natural Forests of Bosnia and Herzegovina. Viruses. 2025; 17(8):1144. https://doi.org/10.3390/v17081144
Chicago/Turabian StyleDálya, László Benedek, Ondřej Hejna, Marcos de la Peña, Zoran Stanivuković, Tomáš Kudláček, and Leticia Botella. 2025. "Diversity of RNA Viruses and Circular Viroid-like Elements in Heterobasidion spp. in Near-Natural Forests of Bosnia and Herzegovina" Viruses 17, no. 8: 1144. https://doi.org/10.3390/v17081144
APA StyleDálya, L. B., Hejna, O., de la Peña, M., Stanivuković, Z., Kudláček, T., & Botella, L. (2025). Diversity of RNA Viruses and Circular Viroid-like Elements in Heterobasidion spp. in Near-Natural Forests of Bosnia and Herzegovina. Viruses, 17(8), 1144. https://doi.org/10.3390/v17081144








