Determining the Importance of Carbohydrate-Based Structures in Murine Norovirus Binding to Commensal Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Growth and Virus Production
2.2. Glycan Microarray
2.3. Competitive Inhibition Assays
2.4. Murine Norovirus–Bacterial Attachment Assay
2.5. Flow Cytometry Quantification of HuNoV VLPs
2.6. RT-qPCR Quantification of MNV
3. Results
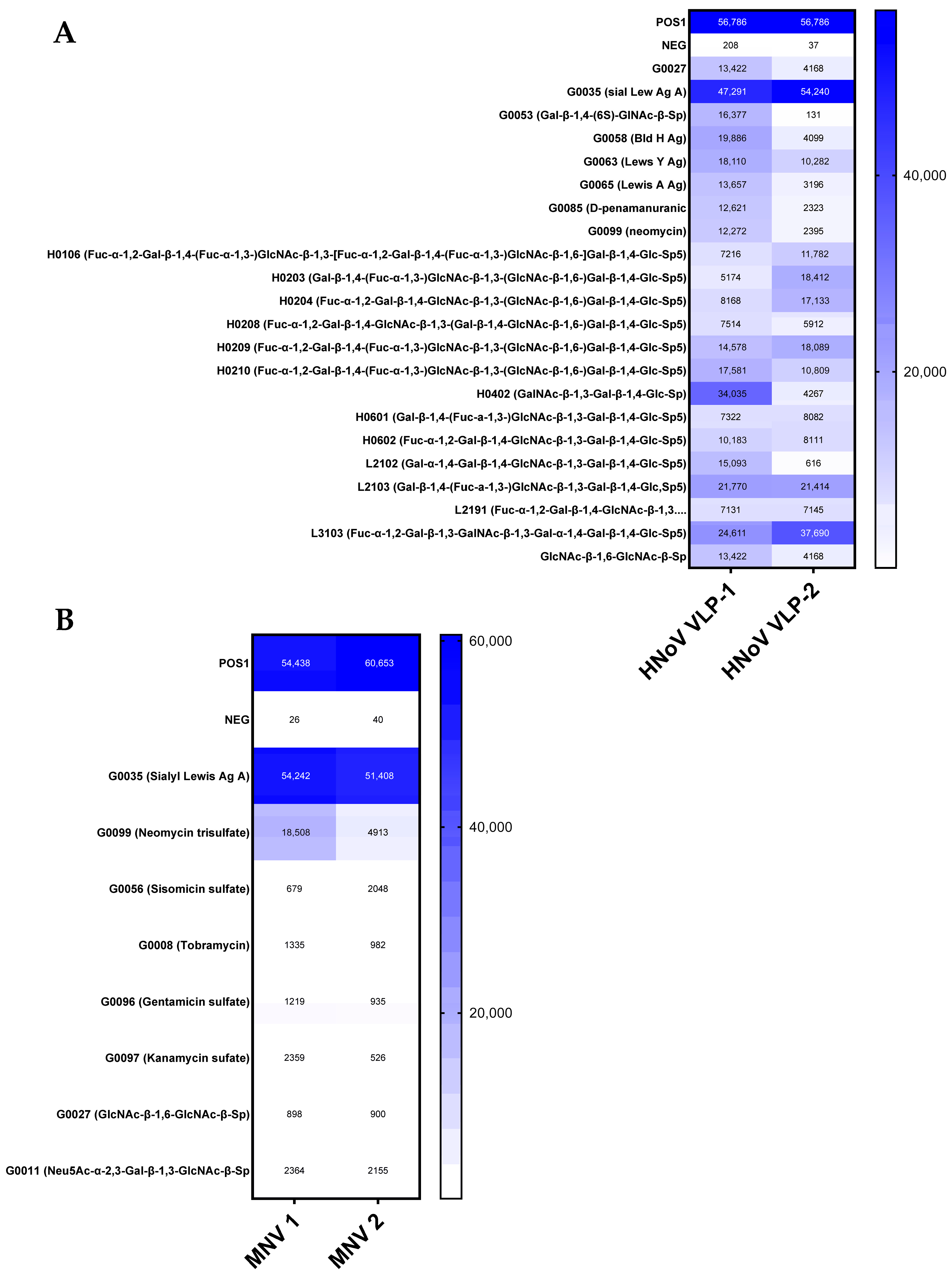
3.1. Glycan Microarray
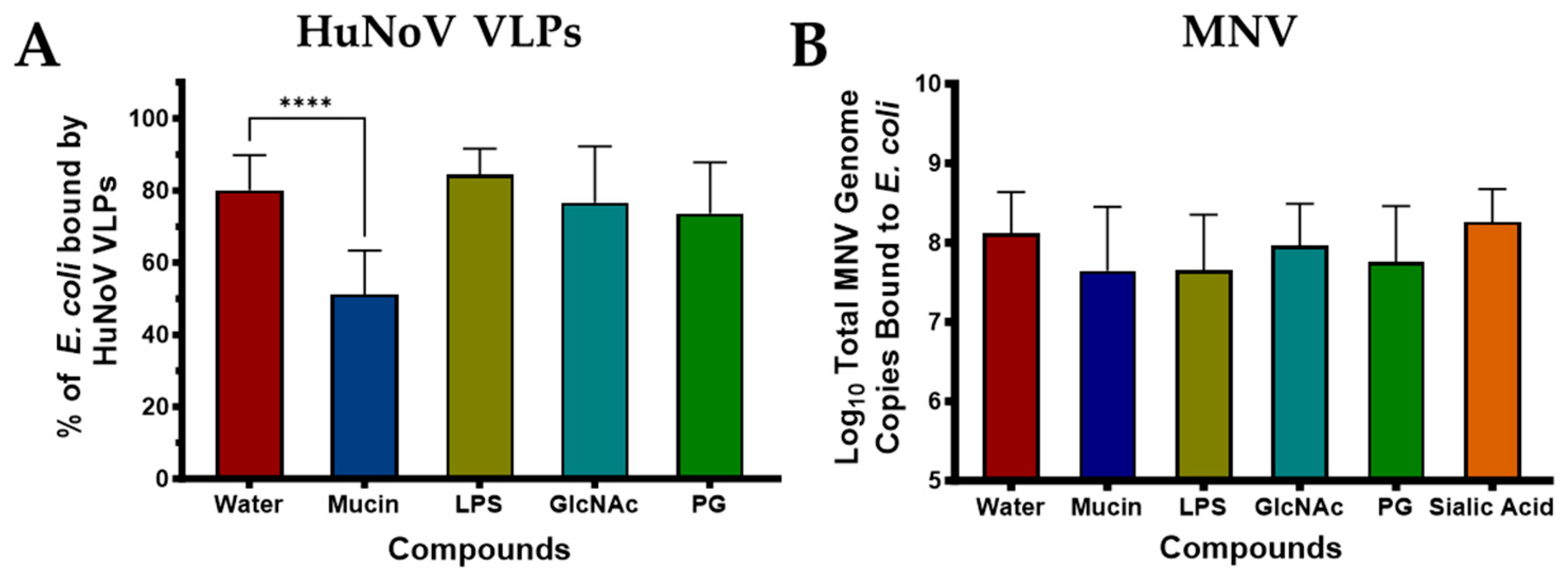
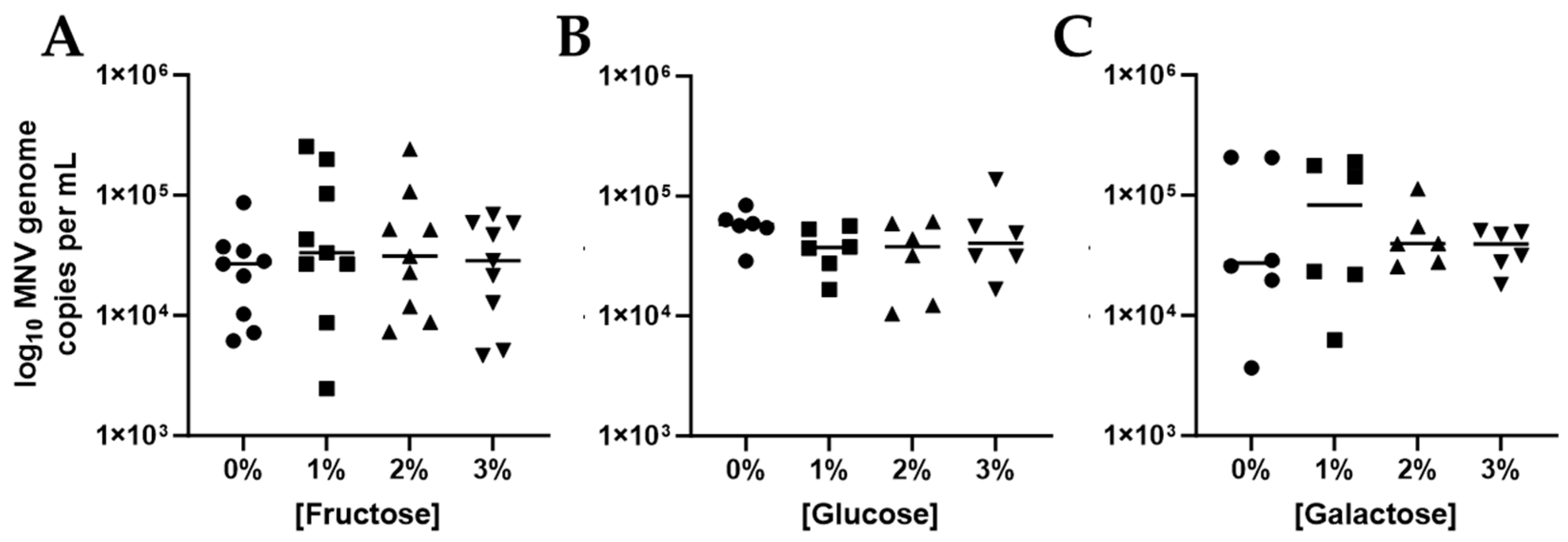
3.2. Competitive Inhibition Assays
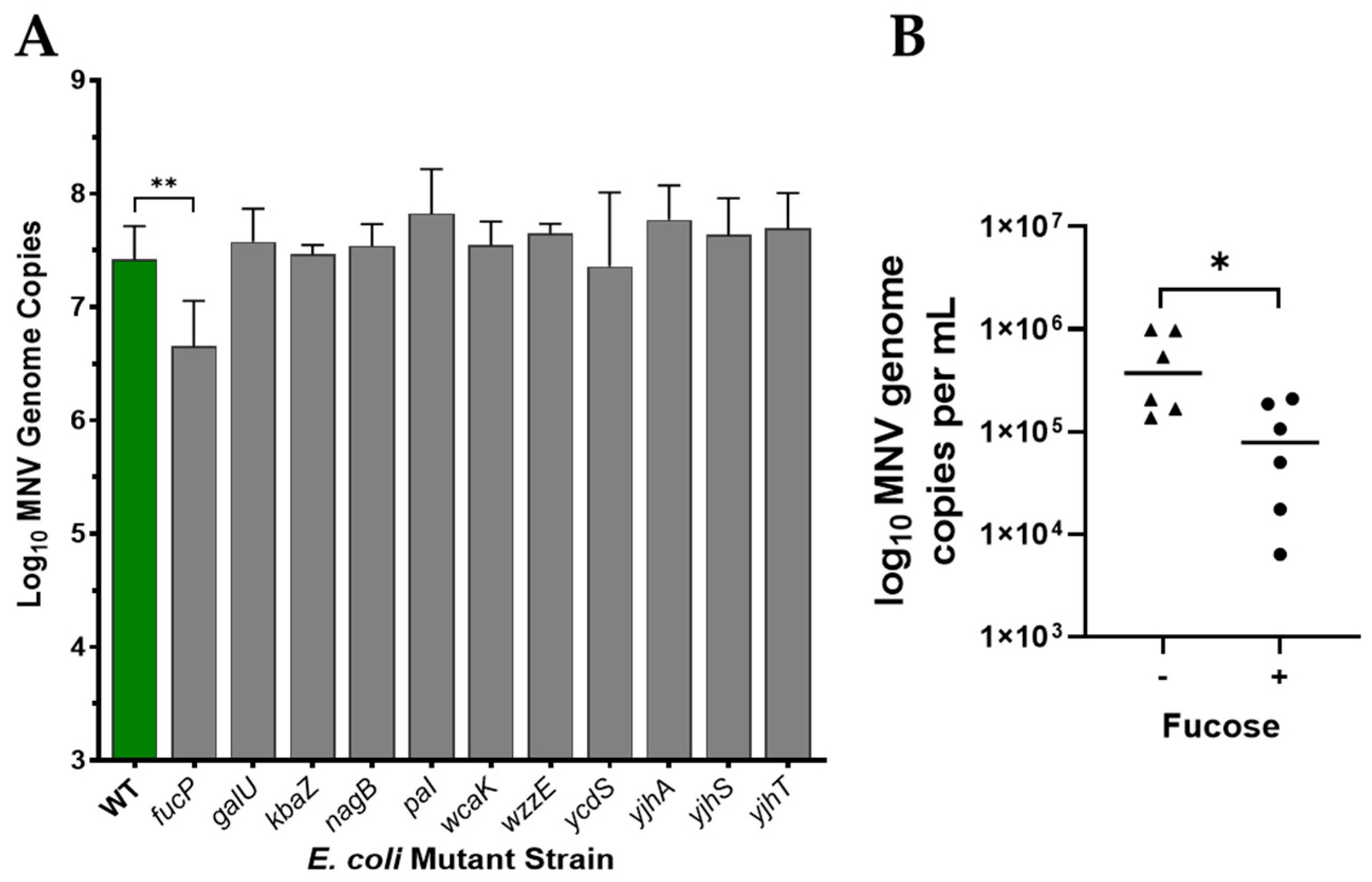
3.3. Role of Fucose in MNV Binding to E. coli
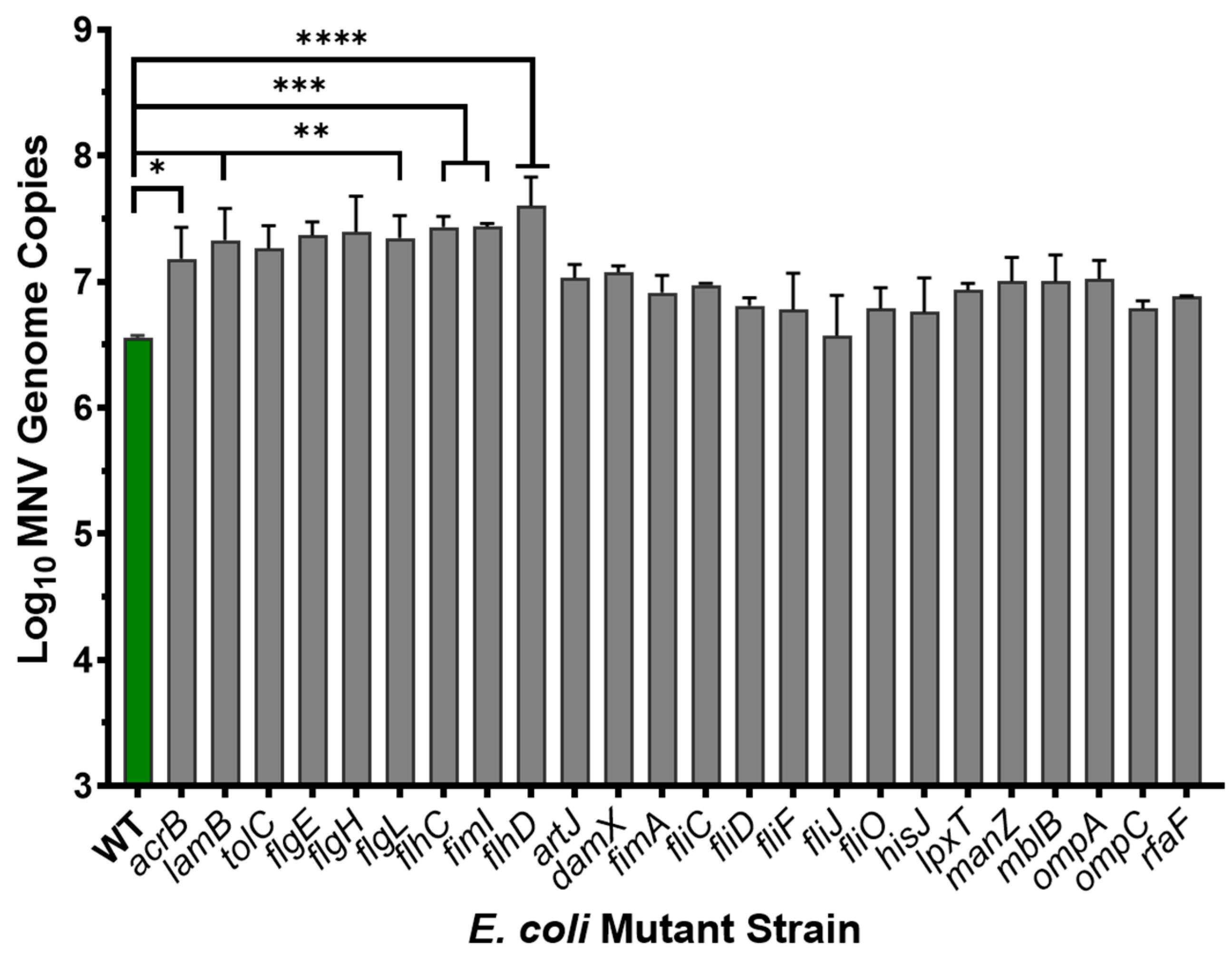
3.4. Role of Proteinaceous Structures in MNV Binding to E. coli
3.5. Binding of MNV Strains to E. coli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartsch, S.M.; Lopman, B.A.; Ozawa, S.; Hall, A.J.; Lee, B.Y. Global Economic Burden of Norovirus Gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar] [CrossRef]
- Pires, S.M.; Fischer-Walker, C.L.; Lanata, C.F.; Devleesschauwer, B.; Hall, A.J.; Kirk, M.D.; Duarte, A.S.; Black, R.E.; Angulo, F.J. Aetiology-Specific Estimates of the Global and Regional Incidence and Mortality of Diarrhoeal Diseases Commonly Transmitted through Food. PLoS ONE 2015, 10, e0142927. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Bhar, S.; Jones, M.K. In Vitro Replication of Human Norovirus. Viruses 2019, 11, 547. [Google Scholar] [CrossRef]
- Todd, K.V.; Tripp, R.A. Vero Cells as a Mammalian Cell Substrate for Human Norovirus. Viruses 2020, 12, 439. [Google Scholar] [CrossRef]
- Kuss, S.K.; Best, G.T.; Etheredge, C.A.; Pruijssers, A.J.; Frierson, J.M.; Hooper, L.V.; Dermody, T.S.; Pfeiffer, J.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 2011, 334, 249–252. [Google Scholar] [CrossRef]
- Robinson, C.M.; Woods Acevedo, M.A.; McCune, B.T.; Pfeiffer, J.K. Related enteric viruses have different requirements for host microbiota in mice. J. Virol. 2019, 93, e01339-19. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, R.; Chassaing, B.; Zhang, B.; Gewirtz, A.T. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 2014, 210, 171–182. [Google Scholar] [CrossRef]
- Budicini, M.R.; Pfeiffer, J.K. Stabilization of Murine Norovirus by Bacteria. mSphere 2022, 7, e00046-22. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.K.; Jesudhasan, P.R.; Mayer, M.J.; Narbad, A.; Winter, S.E.; Pfeiffer, J.K. Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe 2018, 23, 77–88.e5. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.M.; Jesudhasan, P.R.; Pfeiffer, J.K. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 2014, 15, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Grau, K.R.; Zhu, S.; Peterson, S.T.; Helm, E.W.; Philip, D.; Phillips, M.; Hernandez, A.; Turula, H.; Frasse, P.; Graziano, V.R.; et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol. 2020, 5, 84–92. [Google Scholar] [CrossRef]
- Miura, T.; Sano, D.; Suenaga, A.; Yoshimura, T.; Fuzawa, M.; Nakagomi, T.; Nakagomi, O.; Okabe, S. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J. Virol. 2013, 87, 9441–9451. [Google Scholar] [CrossRef]
- Madrigal, J.L.; Bhar, S.; Hackett, S.; Engelken, H.; Joseph, R.; Keyhani, N.O.; Jones, M.K. Attach Me If You Can: Murine Norovirus Binds to Commensal Bacteria and Fungi. Viruses 2020, 12, 759. [Google Scholar] [CrossRef]
- Almand, E.A.; Moore, M.D.; Jaykus, L.A. Norovirus Binding to Ligands Beyond Histo-Blood Group Antigens. Front. Microbiol. 2017, 8, 2549. [Google Scholar] [CrossRef]
- Almand, E.A.; Moore, M.D.; Jaykus, L.-A. Characterization of human norovirus binding to gut-associated bacterial ligands. BMC Res. Notes 2019, 12, 607. [Google Scholar] [CrossRef]
- Mosby, C.A.; Bhar, S.; Phillips, M.B.; Edelmann, M.J.; Jones, M.K. Interaction with mammalian enteric viruses alters outer membrane vesicle production and content by commensal bacteria. J. Extracell. Vesicles 2022, 11, e12172. [Google Scholar] [CrossRef]
- Bhar, S.; Zhao, G.; Bartel, J.D.; Sterchele, H.; Del Mazo, A.; Emerson, L.E.; Edelmann, M.J.; Jones, M.K. Bacterial extracellular vesicles control murine norovirus infection through modulation of antiviral immune responses. Front. Immunol. 2022, 13, 909949. [Google Scholar] [CrossRef]
- Almand, E.A.; Moore, M.D.; Outlaw, J.; Jaykus, L.A. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS ONE 2017, 12, e0173124. [Google Scholar] [CrossRef]
- Hutson, A.M.; Atmar, R.L.; Marcus, D.M.; Estes, M.K. Norwalk virus-like particle hemagglutination by binding to h histo-blood group antigens. J. Virol. 2003, 77, 405–415. [Google Scholar] [CrossRef]
- Shanker, S.; Choi, J.M.; Sankaran, B.; Atmar, R.L.; Estes, M.K.; Prasad, B.V. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: Implications for epochal evolution. J. Virol. 2011, 85, 8635–8645. [Google Scholar] [CrossRef]
- Tarris, G.; de Rougemont, A.; Estienney, M.; Charkaoui, M.; Mouillot, T.; Bonnotte, B.; Michiels, C.; Martin, L.; Belliot, G. Specific Norovirus Interaction with Lewis x and Lewis a on Human Intestinal Inflammatory Mucosa during Refractory Inflammatory Bowel Disease. mSphere 2021, 6, e01185-20. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. Histo-blood group antigens: A common niche for norovirus and rotavirus. Expert Rev. Mol. Med. 2014, 16, e5. [Google Scholar] [CrossRef]
- Subramanian, S.; Geng, H.; Wu, L.; Du, C.; Peiper, A.M.; Bu, H.F.; Chou, P.M.; Wang, X.; Tan, S.C.; Iyer, N.R.; et al. Microbiota regulates neonatal disease tolerance to virus-evoked necrotizing enterocolitis by shaping the STAT1-NLRC5 axis in the intestinal epithelium. Cell Host Microbe 2024, 32, 1805–1821.e10. [Google Scholar] [CrossRef]
- Helm, E.W.; Peiper, A.M.; Phillips, M.; Williams, C.G.; Sherman, M.B.; Kelley, T.; Smith, H.Q.; Jacobs, S.O.; Shah, D.; Tatum, S.M.; et al. Environmentally-triggered contraction of the norovirus virion determines diarrheagenic potential. Front. Immunol. 2022, 13, 1043746. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Watanabe, M.; Kirkpatrick, E.; Murray Akilah, B.; Sok, R.; Karst Stephanie, M. Regulation of Norovirus Virulence by the VP1 Protruding Domain Correlates with B Cell Infection Efficiency. J. Virol. 2016, 90, 2858–2867. [Google Scholar] [CrossRef] [PubMed]
- Madrigal, J.L.; Jones, M.K. Quantifying human norovirus VLP binding to commensal bacteria using flow cytometry. J. Vis. Exp. 2020, 158, e61048. [Google Scholar]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Taube, S.; Perry, J.W.; Yetming, K.; Patel, S.P.; Auble, H.; Shu, L.; Nawar, H.F.; Lee, C.H.; Connell, T.D.; Shayman, J.A.; et al. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 2009, 83, 4092–4101. [Google Scholar] [CrossRef]
- Wang, J.; Ma, W.; Wang, X. Insights into the structure of Escherichia coli outer membrane as the target for engineering microbial cell factories. Microb. Cell Factories 2021, 20, 73. [Google Scholar] [CrossRef]
- Mosby, C.A.; Edelmann, M.J.; Jones, M.K. Murine Norovirus Interaction with Enterobacter cloacae Leads to Changes in Membrane Stability and Packaging of Lipid and Metabolite Vesicle Content. Microbiol. Spectr. 2023, 11, e0469122. [Google Scholar] [CrossRef] [PubMed]
- Renkonen, O. Enzymatic in vitro synthesis of I-branches of mammalian polylactosamines: Generation of scaffolds for multiple selectin-binding saccharide determinants. Cell Mol. Life Sci. 2000, 57, 1423–1439. [Google Scholar] [CrossRef]
- Itoh, Y.; Wang, X.; Hinnebusch, B.J.; Preston, J.F., 3rd; Romeo, T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 2005, 187, 382–387. [Google Scholar] [CrossRef]
- Kotra, L.P.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256. [Google Scholar] [CrossRef]
- Yang, J.; Xie, D.; Ma, X. Recent Advances in Chemical Synthesis of Amino Sugars. Molecules 2023, 28, 4724. [Google Scholar] [CrossRef]
- Magnet, S.; Blanchard, J.S. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 2005, 105, 477–498. [Google Scholar] [CrossRef] [PubMed]
- Zaczek-Moczydlowska, M.A.; Beizaei, A.; Dillon, M.; Campbell, K. Current state-of-the-art diagnostics for Norovirus detection: Model approaches for point-of-care analysis. Trends Food Sci. Technol. 2021, 114, 684–695. [Google Scholar] [CrossRef]
- Parra, G.I. Emergence of norovirus strains: A tale of two genes. Virus Evol. 2019, 5, vez048. [Google Scholar] [CrossRef]

| Reference Name | Gene Deletion | Encoded Product |
|---|---|---|
| BW25113 | Wild type | N/A |
| Carbohydrate Mutant Strains | ||
| JW2773 | fucI | Converts aldose L-fucose to ketose L-fuculose |
| JW2772 | fucP | Mediates cellular uptake of L-fucose |
| JW1224 | galU | Galactose catabolic process |
| JW3101 | kbaZ | Carbohydrate metabolism |
| JW0664 | nagB | N-acetylglucosamine degradation |
| JW0731 | pal | Peptidoglycan-associated lipoprotein |
| JW2030 | wcaK | Biosynthesis of colonic acid |
| JW5601 | wzzE | Enterobacterial common antigen assembly |
| JW1010 | ycdS | PGA export across the outer membrane |
| JW5778 | yjhA | Outer membrane protein for Neu5Ac entry |
| JW4272 | yjhS | Catalyzes hydrolysis of alternative sialic acid |
| JW5777 | yjhT | Neu5Ac α- and β-anomers conversion |
| Protein Mutant Strains | ||
| JW0451 | acrB | Efflux pump membrane transporter |
| JW0844 | artJ | Arginine-binding periplasmic protein 2 |
| JW3351 | damX | Cellular division protein |
| JW4277 | fimA | Pilus/cell adhesion |
| JW4278 | fimI | Pilus/cell adhesion |
| JW0940 | flgE | Flagella hook protein |
| JW1066 | flgH | Cytoplasmic flagellar protein |
| JW1067 | flgI | Flagellar assembly outer membrane protein |
| JW1880 | flhC | Master regulator of flagella assembly |
| JW1881 | flhD | Master regulator of flagella assembly |
| JW1908 | fliC | Extracellular flagellar component |
| JW1909 | fliD | Regulator and cap of flagella |
| JW1922 | fliF | Membrane flagellar component |
| JW1926 | fliJ | Membrane flagellar component |
| JW1931 | fliO | Membrane flagellar component |
| JW2306 | hisJ | Outer membrane protein |
| JW3996 | lamB | Ion transport outer membrane protein |
| JW2162 | lpxT | Involved in modification of Lipid A of LPS |
| JW1808 | manZ | Intracellular protein |
| JW2137 | mglB | Pilus/cell adhesion protein |
| JW2203 | ompC | Ion transport outer membrane protein |
| JW3595 | rfaF | Involved in lipopolysaccharide formation |
| JW3003 | tolC | Outer membrane TolC type 1 secretion protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madrigal, J.L.; Sullivan, J.P.; Mathew, F.; Bland, M.; Jones, M.K. Determining the Importance of Carbohydrate-Based Structures in Murine Norovirus Binding to Commensal Bacteria. Viruses 2025, 17, 1142. https://doi.org/10.3390/v17081142
Madrigal JL, Sullivan JP, Mathew F, Bland M, Jones MK. Determining the Importance of Carbohydrate-Based Structures in Murine Norovirus Binding to Commensal Bacteria. Viruses. 2025; 17(8):1142. https://doi.org/10.3390/v17081142
Chicago/Turabian StyleMadrigal, Jasmine L., Joseph P. Sullivan, Feba Mathew, Melanie Bland, and Melissa K. Jones. 2025. "Determining the Importance of Carbohydrate-Based Structures in Murine Norovirus Binding to Commensal Bacteria" Viruses 17, no. 8: 1142. https://doi.org/10.3390/v17081142
APA StyleMadrigal, J. L., Sullivan, J. P., Mathew, F., Bland, M., & Jones, M. K. (2025). Determining the Importance of Carbohydrate-Based Structures in Murine Norovirus Binding to Commensal Bacteria. Viruses, 17(8), 1142. https://doi.org/10.3390/v17081142






