Phage Therapy: Combating Evolution of Bacterial Resistance to Phages
Abstract
1. Introduction
- (i)
- Prior to resistance becoming problematic (lower frequency resistance);
- (ii)
- While resistance develops into a concern (higher frequency resistance);
- (iii)
- Only once resistance is present at a very high frequency.
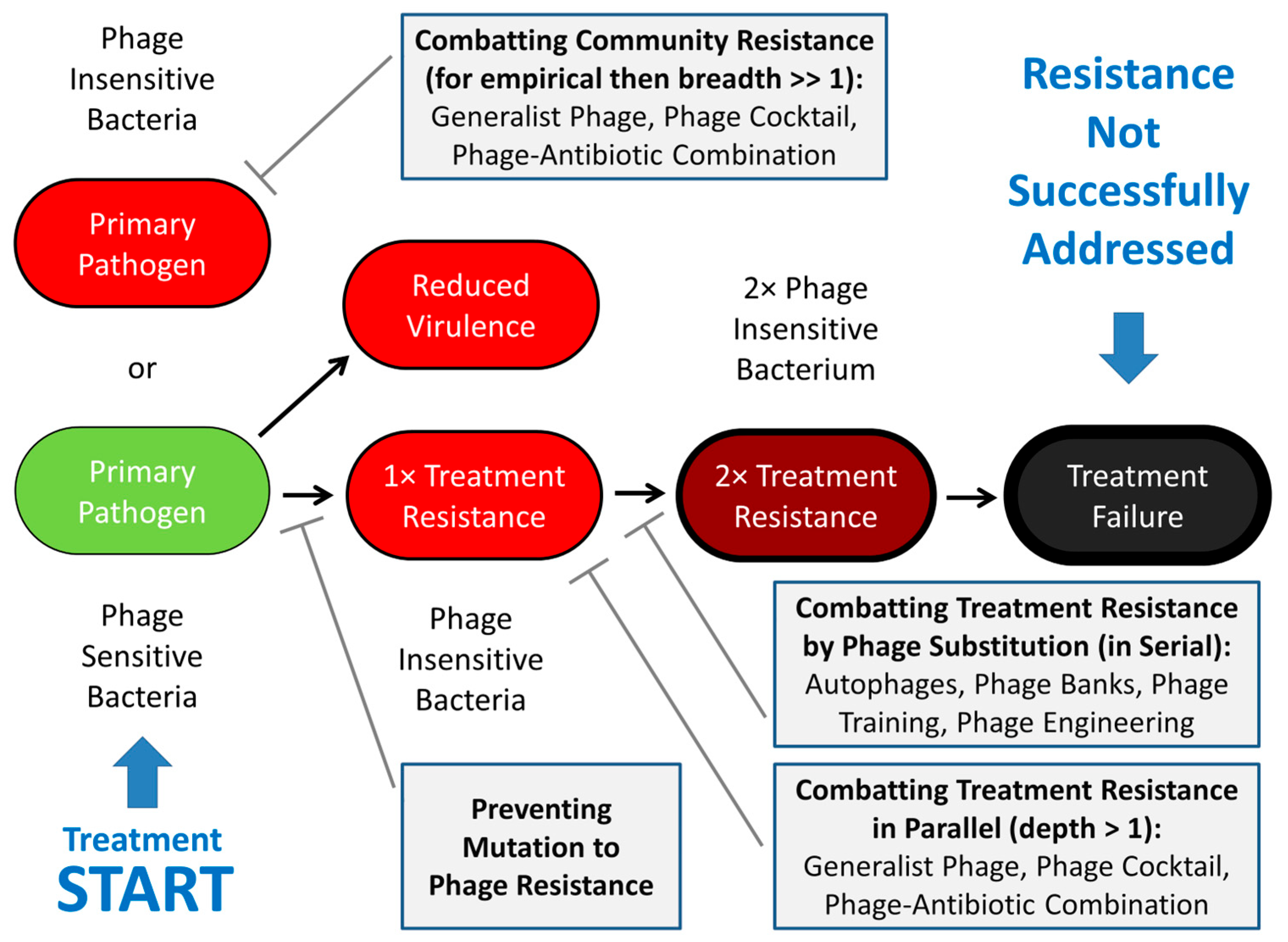
2. Treatment Resistance
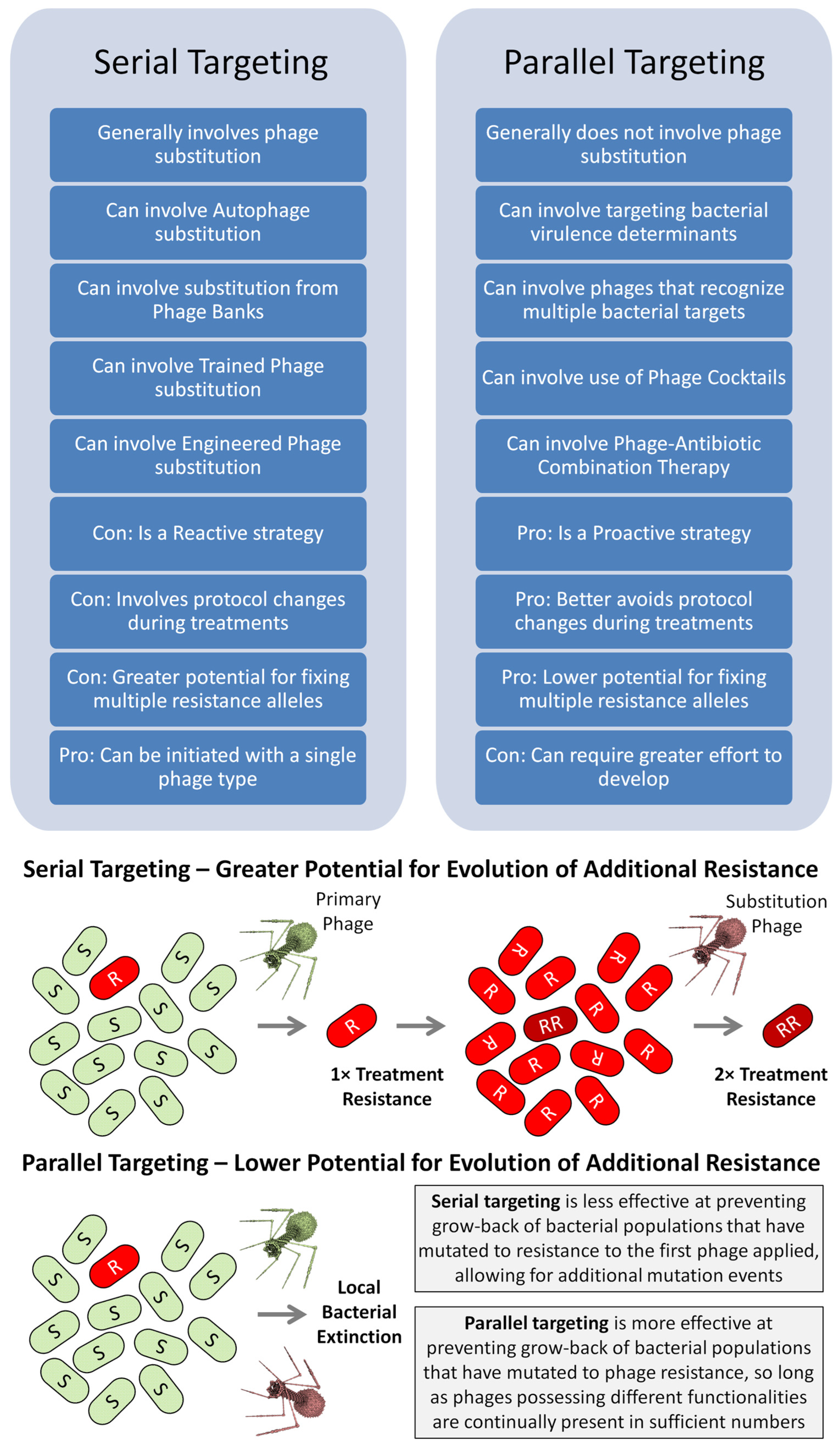
2.1. Serial vs. Parallel Anti-Resistance Strategies
2.2. Reactive vs. Proactive Anti-Resistance Strategies
2.3. Breadth vs. Depth of Activity
3. Reactively Addressing Treatment Resistance
3.1. Autophages
3.1.1. Explicit Avoidance of Cross-Resistance
3.1.2. Related Terms
3.2. Phage Banks
3.2.1. Maintaining an Optimal Phage Bank
3.2.2. Still Time Lags but Fewer Time Lags
3.3. Phage Training
3.3.1. Advantages of Training Phages
3.3.2. Disadvantages of Training Treatment Phages
3.3.3. More than Just Mutational Change
4. Proactively Addressing Treatment Resistance
4.1. Minimizing the Occurrence of Mutation to Resistance
4.1.1. An Advantage of Prophylactic Treatment
4.1.2. Less Applicable to Treatment of Established Bacterial Infections
4.1.3. Monitoring Treatment Resistance
The question then arises as to whether these variants were mere innocuous bystanders on the way of being eliminated by host defences, or whether they could still produce infection. In any case, the ideal experimental setting should be to apply the Koch postulate [sic] and inoculate the variants to the animals in order to re-evaluate their infectivity. Indeed, recovering phage-resistant variants from in vivo samples may not be automatically synonymous with therapeutic failure, a counter-intuitive concept that appears to apply to phage therapy.
4.2. Targeting Bacterial Fitness/Virulence Determinants
4.2.1. Collateral Sensitivity and Antagonistic Pleiotropy
4.2.2. Reciprocal Collateral Sensitivity
4.2.3. Phage Targeting of Bacterial Virulence Factors
4.2.4. Limitations to Collateral Sensitivity
4.3. Individual Phages Recognizing Multiple Receptors
4.3.1. Targeted Bacteria Must Display Both Receptors
4.3.2. Monophage Depth and Breadth
4.4. Phage Cocktails
4.4.1. Differentiating Breadth and Depth
4.4.2. Proactive Autophages
4.4.3. Breadth of Depth (Empirical Anti-Treatment-Resistance Phage Cocktails)
4.5. Phage–Antibiotic Combination Therapies
4.5.1. Combating Not Just Treatment Resistance
4.5.2. Mostly Avoids Cross-Resistance
4.5.3. Combating Also Antibiotic Resistance
4.5.4. Antibiotics as Back up Treatment
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Meanings of Autophage
…why try to prepare an auto-bacteriophage which will certainly be inferior, because the strain of bacteriophage will be not selected among hundreds of others, but simply found at random and therefore of variable virulence? And if, a contrario, the ‘coli- phage’ remains without activity against the bacterium infecting a given patient, barring a most remote chance of one in a thousand, it is pointless to try the preparation of an auto-bacteriophage because it would take months of assiduous research to discover a bacteriophage able to attack the resistant bacterial isolate.
| Year | Ref. | Quotation | Isolated * |
|---|---|---|---|
| 2023 | [249] | “bacteriophages isolated from the same environment as the pathogen” (p. 9 and citing [250]) | From |
| 2022 | [251] | “personalized phage (autophages) or standard formulation (fixed phage cocktails)” (p. 552) | Against? |
| 2022 | [252] | “phages can be obtained or isolated from the patient where the pathogenic agent is found, calling this virus autochthonous phage or autophage” (p. 105 and citing, in part, [250]) | From |
| 2021 | [253] | “A custom phage (autophage) was prepared… that was fully sensitive against the S. mitis isolated from the patient’s sample.” (p. 5) | Against |
| 2021 | [37] | “In cases when patients’ strains are not susceptible to the commercially available preparations, or if their infection is caused by an entirely different organism (on a species or genus level), an individualized phage preparation—custom phage †—is offered. Such tailored bacteriophages are targeted at specific strains that have been isolated and identified in patients’ biological samples.” (p. 3); “This is when she ordered her first autophage.” (p. 7) | Against? |
| 2020 | [84] | “the auto-phage preparation which is personalised for an individual patient” (p. 307) | Against |
| 2018 | [254] | “i.e., auto-phage specifically manufactured for the use of a particular patient” (p. 5) | Against |
| 2018 | [35] | “a tailored strategy by training phages on the patient’s strain as soon as it became available in a form of highly personalized medicine. Of note, this latter strategy is applied at the Eliava Institute in the process of development of so-called ‘autophages’.” (p. 7) | Against ‡ |
| 2018 | [250] | “autophage (bacteriophage isolated from the same environment where the pathogen is isolated)” (p. 4367 and citing [255] elsewhere which we can speculate is for the statement there, “When no active phage is present against a severe pathogen, the lytic phage may be found, isolated directly from environment”, though that reference [255] does not appear to actually use the term, “autophage”, nor “same” in front of “environment”) | From |
| 2011 | [256] | “we isolate specific ‘autophage’ against patient’s specific bacteria” (p. 646) | Against |
| 2011 | [63] | “Sometimes custom phage preparations are developed for a patient’s infection (autophage), a procedure that usually takes a few days to weeks.” (p. 936) | Against? |
| 2010 | [257] | “under extreme circumstances new ‘auto-phages’ may be isolated from environmental sources, using the patient’s own bacteria to select them” (p. 71 and see [69] as the citation) | Against |
| 2009 | [258] | “In problem cases, new phage specific to the patient’s bacteria are occasionally isolated from sewage, amplified and sent to the hospital; these are called ‘autophage’.” (p. 265) | Against |
References
- Acton, L.; Pye, H.V.; Thilliez, G.; Kolenda, R.; Matthews, M.; Turner, A.K.; Yasir, M.; Holden, E.; Al-Khanaq, H.; Webber, M.; et al. Collateral sensitivity increases the efficacy of a rationally designed bacteriophage combination to control Salmonella enterica. J. Virol. 2024, 98, e0147623. [Google Scholar] [CrossRef]
- Sanz-Gaitero, M.; Seoane-Blanco, M.; van Raaij, M.J. Structure and function of bacteriophages. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B., McConville, M., Eds.; Springer Nature Switzerland AG: New York, NY, USA, 2021; pp. 19–91. [Google Scholar]
- Lehman, S.M. Bacteriophage diversity. In Viruses of Microorganisms; Hyman, P., Abedon, S.T., Eds.; Caister Academic Press: Norwich, UK, 2018; pp. 145–165. [Google Scholar]
- Sulakvelidze, A.; Kutter, E. Bacteriophage therapy in humans. In Bacteriophages: Biology and Application; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 381–436. [Google Scholar]
- Knezevic, P.; Sabo, V.A. Combining bacteriophages with other antibacterial agents to combat bacteria. In Phage Therapy: A Practical Approach; Górski, A., Międzybrodzki, R., Borysowski, J., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 257–293. [Google Scholar]
- Bruynoghe, R.; Maisin, J. Essais de thérapeutique au moyen du bactériophage du Staphylocoque. Compt. Rend. Soc. Biol. 1921, 85, 1120–1121. [Google Scholar]
- Fleming, A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Brit. J. Exp. Path. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Brown, K. ‘That’s funny!’: The discovery and development of penicillin. Microbiol. Today 2009, 36, 12–15. [Google Scholar]
- Wainwright, M.; Swan, H.T.C.G. Paine and the earliest surviving clinical records of penicillin therapy. Med. Hist. 1986, 30, 42–56. [Google Scholar] [CrossRef]
- Innes, A.; Ellis, V.H. Battle casualties treated with penicllin. Lancet 1945, 245, 524–528. [Google Scholar] [CrossRef]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Quinn, R. Rethinking antibiotic research and development: World War II and the penicillin collaborative. Am. J. Public Health 2013, 103, 426–434. [Google Scholar] [CrossRef]
- Abedon, S.T. Kinetics of phage-mediated biocontrol of bacteria. Foodborne Pathog. Dis. 2009, 6, 807–815. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Borysowski, J. Phage Therapy: A Practical Approach; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Abedon, S.T.; Danis-Wlodarczyk, K.; Alves, D.R. Phage therapy in the 21st Century: Is there modern, clinical evidence of phage-mediated clinical efficacy? Pharmaceuticals 2021, 14, 1157. [Google Scholar] [CrossRef]
- Marongiu, L.; Burkard, M.; Lauer, U.M.; Hoelzle, L.E.; Venturelli, S. Reassessment of historical clinical trials supports the effectiveness of phage therapy. Clin. Microbiol Rev. 2022, 35, e0006222. [Google Scholar] [CrossRef] [PubMed]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.; et al. Considerations for the use of phage therapy in clinical practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef]
- Petrovic Fabijan, A.; Iredell, J.; Danis-Wlodarczyk, K.; Kebriaei, R.; Abedon, S.T. Translating phage therapy into the clinic: Recent accomplishments but continuing challenges. PLoS Biol. 2023, 21, e3002119. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Kim, M.K.; Suh, G.A.; Cullen, G.D.; Perez, R.S.; Dharmaraj, T.; Chang, T.H.W.; Li, Z.; Chen, Q.; Green, S.I.; Lavigne, R.; et al. Bacteriophage therapy for multidrug-resistant infections: Current technologies and therapeutic approaches. J. Clin. Investig. 2025, 135, e187996. [Google Scholar] [CrossRef]
- Bronfenbrenner, J.J.; Korb, C. On variants of B. pestis caviae resistant to lysis by bacteriophage. Proc. Soc. Exp. Biol. Med. 1925, 23, 3–5. [Google Scholar] [CrossRef]
- Harper, D.R. Biological control by microorganisms. In The Encyclopedia of Life Sciences; John Wiley & Sons: Chichester, West Sussex, UK, 2006; pp. 1–10. [Google Scholar]
- Harper, D.R. Biological control by microorganisms. In eLS; John Wiley & Sons: Chichester, West Sussex, UK, 2013. [Google Scholar] [CrossRef]
- Alves, D.R.; Clark, J.; Abedon, S.T. Viruses as biocontrol agents of microorganisms. In Viruses of Microorganisms; Hyman, P., Abedon, S.T., Eds.; Caister Academic Press: Norwich, UK, 2018; pp. 313–330. [Google Scholar]
- Gildea, L.; Ayariga, J.A.; Robertson, B.K. Bacteriophages as biocontrol agents in livestock food production. Microorganisms 2022, 10, 2126. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Rahim, K.; Paker, N.P. Exploring the historical roots, advantages and efficacy of phage therapy in plant diseases management. Plant Sci. 2024, 346, 112164. [Google Scholar] [CrossRef]
- Ul Haq, I.; Khan, M.; Khan, I. Phytopathological management through bacteriophages: Enhancing food security amidst climate change. J. Ind. Microbiol. Biotechnol. 2024, 51, kuae031. [Google Scholar] [CrossRef]
- Czajkowski, R.; Roca, A.; Matilla, M.A. Harnessing bacteriophages for sustainable crop protection in the face of climate change. Microb. Biotechnol. 2025, 18, e70108. [Google Scholar] [CrossRef] [PubMed]
- Teklemariam, A.D.; Al Hindi, R.; Qadri, I.; Alharbi, M.G.; Hashem, A.M.; Alrefaei, A.A.; Basamad, N.A.; Haque, S.; Alamri, T.; Harakeh, S. Phage cocktails—An emerging approach for the control of bacterial infection with major emphasis on foodborne pathogens. Biotechnol. Genet. Eng. Rev. 2023, 40, 36–64. [Google Scholar] [CrossRef]
- Akram, F.; Imtiaz, M.; Haq, I.U. Emergent crisis of antibiotic resistance: A silent pandemic threat to 21st century. Microb. Pathog. 2023, 174, 105923. [Google Scholar] [CrossRef] [PubMed]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Pawełczyk, Z.; Rogóz, P.; Kłak, M.; Wojtasik, E.; et al. Clinical aspects of phage therapy. Adv. Virus Res. 2012, 83, 73–121. [Google Scholar]
- Hong, Y.; Schmidt, K.; Marks, D.; Hatter, S.; Marshall, A.; Albino, L.; Ebner, P. Treatment of Salmonella-contaminated eggs and pork with a broad-spectrum, single bacteriophage: Assessment of efficacy and resistance development. Foodborne Pathog. Dis. 2016, 13, 679–688. [Google Scholar] [CrossRef]
- Zhvania, P.; Hoyle, N.S.; Nadareishvili, L.; Nizharadze, D.; Kutateladze, M. Phage therapy in a 16-year-old boy with Netherton syndrome. Front. Med. 2017, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Rohde, C.; Resch, G.; Pirnay, J.-P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert opinion on three phage therapy related topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 2018, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- McCallin, S.; Oechslin, F. Bacterial resistance to phage and its impact on clinical therapy. In Phage Therapy: A Practical Approach; Górski, A., Międzybrodzki, R., Borysowski, J., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 59–88. [Google Scholar]
- Zaldastanishvili, E.; Leshkasheli, L.; Dadiani, M.; Nadareishvili, L.; Askilashvili, L.; Kvatadze, N.; Goderdzishvili, M.; Kutateladze, M.; Balarjishvili, N. Phage therapy experience at the Eliava Phage Therapy Center: Three cases of bacterial persistence. Viruses 2021, 13, 1901. [Google Scholar] [CrossRef]
- Abedon, S.T. Ecology and evolutionary biology of hindering phage therapy: The phage tolerance vs. phage resistance of bacterial biofilms. Antibiotics 2023, 12, 245. [Google Scholar] [CrossRef]
- Oromi-Bosch, A.; Antani, J.D.; Turner, P.E. Developing phage therapy that overcomes the evolution of bacterial resistance. Annu. Rev. Virol. 2023, 10, 503–524. [Google Scholar] [CrossRef]
- Marchi, J.; Zborowsky, S.; Debarbieux, L.; Weitz, J.S. The dynamic interplay of bacteriophage, bacteria and the mammalian host during phage therapy. iScience 2023, 26, 106004. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Romalde, J.L.; Almeida, A. A game of resistance: War between bacteria and phages and how phage cocktails can be the solution. Virology 2024, 599, 110209. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; Djebara, S.; Steurs, G.; Griselain, J.; Cochez, C.; De, S.S.; Glonti, T.; Spiessens, A.; Vanden Berghe, E.; Green, S.; et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: A multicentre, multinational, retrospective observational study. Nat. Microbiol. 2024, 9, 1434–1453. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Abedon, S.T. Pharmacologically aware phage therapy: Pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol. Mol. Biol. Rev. 2019, 83, e00012-19. [Google Scholar] [CrossRef] [PubMed]
- Styles, K.M.; Brown, A.T.; Sagona, A.P. A review of using mathematical modeling to improve our understanding of bacteriophage, bacteria, and eukaryotic interactions. Front. Microbiol. 2021, 12, 724767. [Google Scholar] [CrossRef] [PubMed]
- Torres-Barceló, C.; Turner, P.E.; Buckling, A. Mitigation of evolved bacterial resistance to phage therapy. Curr. Opin. Virol. 2022, 53, 101201. [Google Scholar] [CrossRef]
- Williams, J.; Severin, J.; Temperton, B.; Mitchelmore, P.J. Phage therapy administration route, regimen, and need for supplementary antibiotics in patients with chronic suppurative lung disease. Phage 2023, 4, 4–10. [Google Scholar] [CrossRef]
- Bono, L.M.; Mao, S.; Done, R.E.; Okamoto, K.W.; Chan, B.K.; Turner, P.E. Advancing phage therapy through the lens of virus host-breadth and emergence potential. Adv. Virus Res. 2021, 111, 63–110. [Google Scholar]
- Laanto, E. Overcoming bacteriophage resistance in phage therapy. Methods Mol. Biol. 2024, 2738, 401–410. [Google Scholar]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage cocktail development for bacteriophage therapy: Toward improving spectrum of activity breadth and depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef]
- Baym, M.; Stone, L.K.; Kishony, R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 2016, 351, aad3292. [Google Scholar] [CrossRef]
- Azam, A.H.; Tanji, Y. Bacteriophage-host arm race: An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 2019, 103, 2121–2131. [Google Scholar] [CrossRef]
- Fineran, P.C. Resistance is not futile: Bacterial ‘innate’ and CRISPR-Cas ‘adaptive’ immune systems. Microbiology 2019, 165, 834–841. [Google Scholar] [CrossRef]
- Rostol, J.T.; Marraffini, L. (Ph)ighting phages: How bacteria resist their parasites. Cell Host. Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive infection: Bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Ambroa, A.; Blasco, L.; Lopez, M.; Pacios, O.; Bleriot, I.; Fernandez-Garcia, L.; de Aledo, M.G.; Ortiz-Cartagena, C.; Millard, A.; Tomas, M. Genomic analysis of molecular bacterial mechanisms of resistance to phage infection. Front. Microbiol. 2021, 12, 784949. [Google Scholar] [CrossRef]
- Egido, J.E.; Costa, A.R.; Aparicio-Maldonado, C.; Haas, P.J.; Brouns, S.J.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022, 46, fuab048. [Google Scholar] [CrossRef]
- Teklemariam, A.D.; Al-Hindi, R.R.; Qadri, I.; Alharbi, M.G.; Ramadan, W.S.; Ayubu, J.; Al-Hejin, A.M.; Hakim, R.F.; Hakim, F.F.; Hakim, R.F.; et al. The battle between bacteria and bacteriophages: A conundrum to their immune system. Antibiotics 2023, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Borin, J.M.; Lee, J.J.; Gerbino, K.R.; Meyer, J.R. Comparison of bacterial suppression by phage cocktails, dual-receptor generalists, and coevolutionarily trained phages. Evol. Appl. 2023, 16, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Bleriot, I.; Pacios, O.; Blasco, L.; Fernandez-Garcia, L.; Lopez, M.; Ortiz-Cartagena, C.; Barrio-Pujante, A.; Garcia-Contreras, R.; Pirnay, J.P.; Wood, T.K.; et al. Improving phage therapy by evasion of phage resistance mechanisms. JAC Antimicrob. Resist. 2024, 6, dlae017. [Google Scholar] [CrossRef]
- Ślopek, S.; Weber-Dąbrowska, B.; Dąbrowski, M.; Kucharewicz-Krukowska, A. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981–1986. Arch. Immunol. Ther. Exp. 1987, 35, 569–583. [Google Scholar]
- McCallin, S.; Brüssow, H. Clinical trials of bacteriophage therapeutics. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer Nature Switzerland AG: New York, NY, USA, 2021; pp. 1099–1129. [Google Scholar]
- Djebara, S.; Lavigne, R.; Pirnay, J.P. Implementation challenges of personalised phage therapy. Lancet 2025, 405, 1901–1903. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The phage therapy paradigm: Prêt-à-porter or sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Screening for lysogen activity in therapeutically relevant bacteriophages. Bio. Protoc. 2021, 11, e3997. [Google Scholar]
- Christie, G.E.; Allison, H.A.; Kuzio, J.; McShan, M.; Waldor, M.K.; Kropinski, A.M. Prophage-induced changes in cellular cytochemistry and virulence. In Bacteriophages in Health and Disease; Hyman, P., Abedon, S.T., Eds.; CABI Press: Wallingford, UK, 2012; pp. 33–60. [Google Scholar]
- McCallin, S.; Alam, S.S.; Barretto, C.; Sultana, S.; Berger, B.; Huq, S.; Krause, L.; Bibiloni, R.; Schmitt, B.; Reuteler, G.; et al. Safety analysis of a Russian phage cocktail: From metaGenomic analysis to oral application in healthy human subjects. Virology 2013, 443, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Cook, B.W.M.; Hynes, A.P. Re-evaluating what makes a phage unsuitable for therapy. NPJ Antimicrob. Resist. 2025, 3, 45. [Google Scholar] [CrossRef]
- Kuhl, S.; Mazure, H.F. d’Hérelle. Preparation of Therapeutic Bacteriophages, Appendix 1 from: Le Phénomène de la Guérison dans les maladies infectieuses. Bacteriophage 2011, 1, 55–65. [Google Scholar] [CrossRef]
- Wright, R.C.T.; Friman, V.P.; Smith, M.C.M.; Brockhurst, M.A. Cross-resistance is modular in bacteria-phage interactions. PLoS Biol. 2018, 16, e2006057. [Google Scholar] [CrossRef]
- Trudelle, D.M.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Cross-resistance to phage infection in Listeria monocytogenes serotype 1/2a mutants. Food Microbiol. 2019, 84, 103239. [Google Scholar] [CrossRef]
- Wright, R.C.T.; Friman, V.P.; Smith, M.C.M.; Brockhurst, M.A. Resistance evolution against phage combinations depends on the timing and order of exposure. MBio 2019, 10, e01652-19. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Bustamante, C.A.; Dedrick, R.M.; Garlena, R.A.; Russell, D.A.; Hatfull, G.F. Toward a phage cocktail for tuberculosis: Susceptibility and tuberculocidal action of mycobacteriophages against diverse Mycobacterium tuberculosis strains. MBio 2021, 12, e00973-21. [Google Scholar] [CrossRef] [PubMed]
- Adler, B.A.; Kazakov, A.E.; Zhong, C.; Liu, H.; Kutter, E.; Lui, L.M.; Nielsen, T.N.; Carion, H.; Deutschbauer, A.M.; Mutalik, V.K.; et al. The genetic basis of phage susceptibility, cross-resistance and host-range in Salmonella. Microbiology 2021, 167, 001126. [Google Scholar] [CrossRef]
- McGee, L.W.; Barhoush, Y.; Shima, R.; Hennessy, M. Phage-resistant mutations impact bacteria susceptibility to future phage infections and antibiotic response. Ecol. Evol. 2023, 13, e9712. [Google Scholar] [CrossRef]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE 2012, 7, e31698. [Google Scholar] [CrossRef]
- Yu, L.; Wang, S.; Guo, Z.; Liu, H.; Sun, D.; Yan, G.; Hu, D.; Du, C.; Feng, X.; Han, W.; et al. A guard-killer phage cocktail effectively lyses the host and inhibits the development of phage-resistant strains of Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 971–983. [Google Scholar] [CrossRef]
- Li, C.; Shi, T.; Sun, Y.; Zhang, Y. A novel method to create efficient phage cocktails via use of phage-resistant bacteria. Appl. Environ. Microbiol. 2022, 88, aem0232321. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, N.; Zhvaniya, P.; Balarjishvili, N.; Bolkvadze, D.; Nadareishvili, L.; Nizharadze, D.; Wittmann, J.; Rohde, C.; Kutateladze, M. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: A case report. Res. Microbiol. 2018, 169, 540–542. [Google Scholar] [CrossRef]
- Corbellino, M.; Kieffer, N.; Kutateladze, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Tsertsvadze, G.; Rimoldi, S.G.; Nizharadze, D.; Hoyle, N.; et al. Eradication of a multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae isolate following oral and intra-rectal therapy with a custom made, lytic bacteriophage preparation. Clin. Infect. Dis. 2020, 70, 1998–2001. [Google Scholar] [CrossRef]
- Balogh, B.; Jones, J.B.; Iriarte, F.B.; Momol, M.T. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 2010, 11, 48–57. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Sacher, J. Phage Banks Around the World. 2019. Available online: https://phage.directory/capsid/phage-banks (accessed on 11 February 2023).
- Anand, T.; Virmani, N.; Bera, B.C.; Vaid, R.K.; Kumar, A.; Tripathi, B.N. Applications of personalised phage therapy highlighting the importance of bacteriophage banks against emerging antimicrobial resistance. Def. Life Sci. J. 2020, 5, 305–314. [Google Scholar] [CrossRef]
- Yerushalmy, O.; Khalifa, L.; Gold, N.; Rakov, C.; Alkalay-Oren, S.; Adler, K.; Ben-Porat, S.; Kraitman, R.; Gronovich, N.; Shulamit, G.K.; et al. The Israeli phage bank (IPB). Antibiotics 2020, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Nagel, T.; Musila, L.; Muthoni, M.; Nikolich, M.; Nakavuma, J.L.; Clokie, M.R. Phage banks as potential tools to rapidly and cost-effectively manage antimicrobial resistance in the developing world. Curr. Opin. Virol. 2022, 53, 101208. [Google Scholar] [CrossRef]
- Rimon, A.; Gelman, D.; Yerushalmy, O.; Coppenhagen-Glazer, S.; Katvan, E.; Nir-Paz, R.; Hazan, R. Phage therapy in Israel, past, present, and future. Phage 2022, 3, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C. Phage therapy’s latest makeover. Nat. Biotech. 2019, 37, 581–586. [Google Scholar] [CrossRef]
- LeMieux, J. Phage therapy may combine efficacy and convenience: A father-son team rolls out PhageBank ATMs to help ensure that phage therapy gains currency. Genet. Eng. Biotechnol. News 2020, 40, 42–44. [Google Scholar] [CrossRef]
- Rotman, E.; McClure, S.; Glazier, J.; Fuerte-Stone, J.; Foldi, J.; Erani, A.; McGann, R.; Arnold, J.; Lin, H.; Valaitis, S.; et al. Rapid design of bacteriophage cocktails to suppress the burden and virulence of gut-resident carbapenem-resistant Klebsiella pneumoniae. Cell Host Microbe 2024, 32, 1988–2003. [Google Scholar] [CrossRef]
- Gibson, S.B.; Green, S.I.; Liu, C.G.; Salazar, K.C.; Clark, J.R.; Terwilliger, A.L.; Kaplan, H.B.; Maresso, A.W.; Trautner, B.W.; Ramig, R.F. Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front. Microbiol. 2019, 10, 2537. [Google Scholar] [CrossRef]
- Lin, R.C.; Sacher, J.C.; Ceyssens, P.J.; Zheng, J.; Khalid, A.; Iredell, J.R. Phage biobank: Present challenges and future perspectives. Curr. Opin. Biotechnol. 2021, 68, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Hejnowicz, M.S.; Gągała, U.; Weber-Dąbrowska, B.; Węgrzyn, G.; Dadlez, M. The first step to bacteriophage therapy: How to choose the correct phage. In Phage Therapy: Current Research and Applications; Borysowski, J., Międzybrodzki, R., Górski, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 23–67. [Google Scholar]
- Abdelsattar, A.; Dawoud, A.; Makky, S.; Nofal, R.; Aziz, R.; El-Shibiny, A. Bacteriophages: From isolation to application. Curr. Pharm. Biotechnol. 2022, 23, 337–360. [Google Scholar] [CrossRef]
- Sacher, J.C.; Zheng, J.; McCallin, S. Sourcing phages for compassionate use. Microbiol. Aust. 2019, 40, 24–27. [Google Scholar] [CrossRef]
- Sacher, J.C.; Zheng, J. Phage therapy collaboration and compassionate use. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M., Eds.; Springer Nature Switzerland AG: New York, NY, USA, 2021; pp. 1069–1098. [Google Scholar]
- Emslander, Q.; Vogele, K.; Braun, P.; Stender, J.; Willy, C.; Joppich, M.; Hammerl, J.A.; Abele, M.; Meng, C.; Pichlmair, A.; et al. Cell-free production of personalized therapeutic phages targeting multidrug-resistant bacteria. Cell Chem. Biol. 2022, 29, 1434–1445. [Google Scholar] [CrossRef]
- Levrier, A.; Bowden, S.; Nash, B.; Lindner, A.; Noireaux, V. Cell-free synthesis and quantitation of bacteriophages. Methods Mol. Biol. 2024, 2760, 447–461. [Google Scholar]
- Bretaudeau, L.; Tremblais, K.; Aubrit, F.; Meichenin, M.; Arnaud, I. Good manufacturing practice (GMP) compliance for phage therapy medicinal products. Front. Microbiol. 2020, 11, 1161. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.-P. Phage therapy in the year 2035. Front. Microbiol. 2020, 11, 1171. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, M.; Alkalay-Oren, S.; Boon, M.; Clokie, M.; Sicheritz-Pontén, T.; Dąbrowska, K.; Hatfull, G.F.; Hazan, R.; Jalasvuori, M.; Kiljunen, S. Phage therapy. Nature Rev. Methods Primers 2025, 5, 9. [Google Scholar] [CrossRef]
- Abdelsattar, A.S.; Dawooud, A.; Rezk, N.; Makky, S.; Safwat, A.; Richards, P.J.; El-Shibiny, A. How to train your phage: The recent efforts in phage training. Biologics 2021, 1, 70–88. [Google Scholar] [CrossRef]
- Borin, J.M.; Avrani, S.; Barrick, J.E.; Petrie, K.L.; Meyer, J.R. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2104592118. [Google Scholar] [CrossRef]
- Kunisch, F.; Campobasso, C.; Wagemans, J.; Yildirim, S.; Chan, B.K.; Schaudinn, C.; Lavigne, R.; Turner, P.E.; Raschke, M.J.; Trampuz, A.; et al. Targeting Pseudomonas aeruginosa biofilm with an evolutionary trained bacteriophage cocktail exploiting phage resistance trade-offs. Nat. Commun. 2024, 15, 8572. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; De Vos, D. Guidelines to compose an ideal bacteriophage cocktail. Meth. Mol. Biol. 2018, 1693, 99–110. [Google Scholar]
- Peters, T.L.; Song, Y.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Mutant and recombinant phages selected from in vitro coevolution conditions overcome phage-resistant Listeria monocytogenes. Appl. Environ. Microbiol. 2020, 86, e02138-20. [Google Scholar] [CrossRef]
- Loose, M.; Moreno, D.S.; Mutti, M.; Hitzenhammer, E.; Visram, Z.; Dippel, D.; Schertler, S.; Tisáková, L.P.; Wittmann, J.; Corsini, L.; et al. Natural bred ε2-phages have an improved host range and virulence against uropathogenic Escherichia coli over their ancestor phages. Antibiotics 2021, 10, 1337. [Google Scholar] [CrossRef]
- Zhang, Q.G.; Chu, X.L.; Buckling, A. Overcoming the growth-infectivity trade-off in a bacteriophage slows bacterial resistance evolution. Evol. Appl. 2021, 14, 2055–2063. [Google Scholar] [CrossRef]
- Sáez Moreno, D.; Visram, Z.; Mutti, M.; Restrepo-Córdoba, M.; Hartmann, S.; Kremers, A.I.; Tišáková, L.; Schertler, S.; Wittmann, J.; Kalali, B.; et al. ε2-Phages are naturally bred and have a vastly improved host range in Staphylococcus aureus over wild type phages. Pharmaceuticals 2021, 14, 325. [Google Scholar] [CrossRef]
- Warring, S.L.; Malone, L.M.; Jayaraman, J.; Easingwood, R.A.; Rigano, L.A.; Frampton, R.A.; Visnovsky, S.B.; Addison, S.M.; Hernandez, L.; Pitman, A.R. A lipopolysaccharide-dependent phage infects a pseudomonad phytopathogen and can evolve to evade phage resistance. Environ. Microbiol. 2022, 24, 4834–4852. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Wichman, H.A.; Krone, S.M. Modeling the directed evolution of broad host range phages. Antibiotics 2022, 11, 1709. [Google Scholar] [CrossRef] [PubMed]
- Friman, V.P.; Soanes-Brown, D.; Sierocinski, P.; Molin, S.; Johansen, H.K.; Merabishvili, M.; Pirnay, J.-P.; De Vos, D.; Buckling, A. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. J. Evol. Biol. 2016, 29, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Mapes, A.C.; Trautner, B.W.; Liao, K.S.; Ramig, R.F. Development of expanded host range phage active on biofilms of multi-drug resistant Pseudomonas aeruginosa. Bacteriophage 2016, 6, e1096995. [Google Scholar] [CrossRef]
- Korona, R.; Levin, B.R. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution 1993, 47, 556–575. [Google Scholar] [CrossRef]
- Kelly, D.; McAuliffe, O.; Ross, R.P.; O’Mahony, J.; Coffey, A. Development of a broad-host-range phage cocktail for biocontrol. Bioeng. Bugs. 2011, 2, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Kobayashi, I. Restriction-modification systems as mobile epigenetic elements. In Bacterial Integrative Mobile Genetic Elements; Landes Bioscience, 2011. Available online: https://www.ncbi.nlm.nih.gov/books/NBK63963/ (accessed on 9 November 2020).
- Knödlseder, N.; Nevot, G.; Fábrega, M.J.; Mir-Pedrol, J.; Sanvicente-García, M.; Campama-Sanz, N.; Paetzold, B.; Lood, R.; Güell, M. Engineering selectivity of Cutibacterium acnes phages by epigenetic imprinting. PLoS Pathog. 2022, 18, e1010420. [Google Scholar] [CrossRef] [PubMed]
- Hashemolhosseini, S.; Holmes, Z.; Mutschler, B.; Henning, U. Alterations of receptor specificities of coliphages of the T2 family. J. Mol. Biol. 1994, 240, 105–110. [Google Scholar] [CrossRef]
- Hyman, P. Phage receptor. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017; Available online: https://www.sciencedirect.com/science/article/abs/pii/B9780128096338068849 (accessed on 9 November 2020). [CrossRef]
- Żaczek, M.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Międzybrodzki, R.; Fortuna, W.; Rogóż, P.; Letkiewicz, S.; Górski, A. Humoral immune response to phage-based therapeutics. In Phage Therapy: A Practical Approach; Górski, A., Międzybrodzki, R., Borysowski, J., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 123–143. [Google Scholar]
- Dedrick, R.M.; Freeman, K.G.; Nguyen, J.A.; Bahadirli-Talbott, A.; Smith, B.E.; Wu, A.E.; Ong, A.S.; Lin, C.T.; Ruppel, L.C.; Parrish, N.M.; et al. Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat. Med. 2021, 27, 1357–1361. [Google Scholar] [CrossRef]
- Berkson, J.D.; Wate, C.E.; Allen, G.B.; Schubert, A.M.; Dunbar, K.E.; Coryell, M.P.; Sava, R.L.; Gao, Y.; Hastie, J.L.; Smith, E.M.; et al. Phage-specific immunity impairs efficacy of bacteriophage targeting Vancomycin Resistant Enterococcus in a murine model. Nat. Commun. 2024, 15, 2993. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Żaczek, M.; Weber-Dąbrowska, B.; Międzybrodzki, R.; Letkiewicz, S.; Fortuna, W.; Rogóż, P.; Szufnarowski, K.; Jończyk-Matysiak, E.; Olchawa, E.; et al. Antiphage activity of sera during phage therapy in relation to its outcome. Future Microbiol. 2017, 12, 109–117. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Fortuna, W.; Borysowski, J.; Górski, A. Anti-phage serum antibody responses and the outcome of phage therapy. Folia Microbiol. 2021, 66, 127–131. [Google Scholar] [CrossRef]
- Lehman, S.M.; Donlan, R.M. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model. Antimicrob. Agents Chemother. 2015, 59, 1127–1137. [Google Scholar] [CrossRef]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in vitro evolution of therapeutic bacteriophages: The Appelmans protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Blasco, L.; Bleriot, I.; González de Aledo, M.; Fernández-García, L.; Pacios, O.; Oliveira, H.; López, M.; Ortiz-Cartagena, C.; Fernández-Cuenca, F.; Pascual, Á.; et al. Development of an anti-Acinetobacter baumannii biofilm phage cocktail: Genomic adaptation to the host. Antimicrob. Agents Chemother. 2022, 66, e0192321. [Google Scholar] [CrossRef]
- Lossouarn, J.; Beurrier, E.; Bouteau, A.; Moncaut, E.; Sir, S.M.; Portalier, H.; Zouari, A.; Cattoir, V.; Serror, P.; Petit, M.-A. The virtue of training: Extending phage host spectra against vancomycin-resistant Enterococcus faecium strains using the Appelmans method. Antimicrob. Agents Chemother. 2024, 68, e0143923. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.N.; Clark, J.R.; Jang, E.; D’Souza, R.; Nguyen, L.P.; Pinto, N.A.; Yoo, S.; Abadie, R.; Maresso, A.W.; Yong, D. Appelmans protocol—A directed in vitro evolution enables induction and recombination of prophages with expanded host range. Virus Res. 2024, 339, 199272. [Google Scholar] [CrossRef]
- Jakob, N.; Hammerl, J.A.; Swierczewski, B.E.; Wurstle, S.; Bugert, J.J. Appelmans protocol for in vitro Klebsiella pneumoniae phage host range expansion leads to induction of the novel temperate linear plasmid prophage vB_KpnS-KpLi5. Virus Genes 2025, 61, 132–135. [Google Scholar] [CrossRef]
- Toure, I.M.; Stenz, E. Der Einfluß ausgewählter Herbicide auf Bakteriophagen und Escherichia coli [The action of selected herbicides on bacteriophages and Escherichia coli (author’s transl)]. Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene 1977, 132, 163–177. [Google Scholar]
- Appelmans, R.; Abedon, S.T. Le dosage du Bactériophage, Note de R. Appelmans, Présentée Par R. Bruynoghe, Réunion de la Société Belge de Biologie, 1921, pp. 1098–1099, a Google Translation. 2024. Available online: https://asmallerflea.org/2024/12/23/le-dosage-du-bacteriophage-note-de-r-appelmans-presentee-par-r-bruynoghe-reunion-de-la-societe-belge-de-biologie-1921-pp-1098-1099-a-google-translation/ (accessed on 29 June 2025).
- Lenneman, B.R.; Fernbach, J.; Loessner, M.J.; Lu, T.K.; Kilcher, S. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol. 2020, 68, 151–159. [Google Scholar] [CrossRef]
- Guo, D.; Chen, J.; Zhao, X.; Luo, Y.; Jin, M.; Fan, F.; Park, C.; Yang, X.; Sun, C.; Yan, J.; et al. Genetic and chemical engineering of phages for controlling multidrug-resistant bacteria. Antibiotics 2021, 10, 202. [Google Scholar] [CrossRef]
- Łobocka, M.; Dąbrowska, K.; Górski, A. Engineered bacteriophage therapeutics: Rationale, challenges and future. BioDrugs. 2021, 35, 255–280. [Google Scholar] [CrossRef]
- Payaslian, F.; Gradaschi, V.; Piuri, M. Genetic manipulation of phages for therapy using BRED. Curr. Opin. Biotechnol. 2021, 68, 8–14. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, H.; Tan, Y.L.; Zhao, H.; Ang, E.L. Directed evolution of replication-competent double-stranded DNA bacteriophage toward new host specificity. ACS Synth. Biol. 2022, 11, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Hussain, W.; Yang, X.; Ullah, M.; Wang, H.; Aziz, A.; Xu, F.; Asif, M.; Ullah, M.W.; Wang, S. Genetic engineering of bacteriophages: Key concepts, strategies, and applications. Biotechnol. Adv. 2023, 64, 108116. [Google Scholar] [CrossRef] [PubMed]
- Anastassopoulou, C.; Tsakri, D.; Panagiotopoulos, A.-P.; Saldari, C.; Sagona, A.P.; Tsakris, A. Armed phages: A new weapon in the battle against antimicrobial resistance. Viruses 2025, 17, 911. [Google Scholar] [CrossRef]
- d’Hérelle, F.; George, H.S. The Bacteriophage and its Clinical Application; Charles, C., Ed.; Thomas Publisher: Springfield, IL, USA, 1930. [Google Scholar]
- Danis-Wlodarczyk, K.; Dąbrowska, K.; Abedon, S.T. Phage therapy: The pharmacology of antibacterial viruses. In Exploitation of Bacteriophages for Biocontrol and Therapeutics; Coffey, A., Buttimer, C., Eds.; Caister Academic Press: Norwich, UK, 2020; pp. 49–132. [Google Scholar]
- Abedon, S.T. Use of phage therapy to treat long-standing, persistent, or chronic bacterial infections. Adv. Drug Deliv. Rev. 2019, 145, 18–39. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Turner, P.E. Introduction to phage evolutionary biology. In Bacteriophage Ecology; Abedon, S.T., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 147–176. [Google Scholar]
- Luria, S.E.; Delbrück, M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 1943, 28, 491–511. [Google Scholar] [CrossRef]
- Abedon, S.T. Monitoring the Ecology vs. Evolutionary Biology of Phage Resistance: A Tale of Two Precisions. 2022. Available online: https://asmallerflea.org/2022/07/21/monitoring-the-ecology-vs-evolutionary-biology-of-phage-resistance-a-tale-of-two-precisions/ (accessed on 29 June 2025).
- Cui, L.; Watanabe, S.; Miyanaga, K.; Kiga, K.; Sasahara, T.; Aiba, Y.; Tan, X.E.; Veeranarayanan, S.; Thitiananpakorn, K.; Nguyen, H.M.; et al. A comprehensive review on phage therapy and phage-based drug development. Antibiotics 2024, 13, 870. [Google Scholar] [CrossRef]
- Mayorga-Ramos, A.; Carrera-Pacheco, S.E.; Barba-Ostria, C.; Guaman, L.P. Bacteriophage-mediated approaches for biofilm control. Front. Cell Infect. Microbiol. 2024, 14, 1428637. [Google Scholar] [CrossRef] [PubMed]
- Vera-Mansilla, J.; Silva-Valenzuela, C.A.; Sanchez, P.; Molina-Quiroz, R.C. Bacteriophages potentiate the effect of antibiotics by eradication of persister cells and killing of biofilm-forming cells. Res. Microbiol. 2023, 174, 104083. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Sullivan, M.B. Improving phage-biofilm in vitro experimentation. Viruses 2021, 13, 1175. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating bacterial infections with bacteriophage-based enzybiotics: In vitro, in vivo and clinical application. Antibiotics 2021, 10, 1497. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Pub. Health 2018, 1, 60–66. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Chen, Q.; Echterhof, A.; Pennetzdorfer, N.; McBride, R.C.; Banaei, N.; Burgener, E.B.; Milla, C.E.; Bollyky, P.L. A blueprint for broadly effective bacteriophage-antibiotic cocktails against bacterial infections. Nat. Commun. 2024, 15, 9987. [Google Scholar] [CrossRef]
- North, O.I.; Brown, E.D. Phage-antibiotic combinations: A promising approach to constrain resistance evolution in bacteria. Ann. N. Y. Acad. Sci. 2021, 1496, 23–34. [Google Scholar] [CrossRef]
- Mu, Y.; Song, Y.; Tian, X.; Ding, Z.; Yao, S.; Li, Y.; Wang, C.; Wei, D.; Vollmer, W.; Zhang, G.; et al. Leveraging collateral sensitivity to counteract the evolution of bacteriophage resistance in bacteria. mLife 2025, 4, 143–154. [Google Scholar] [CrossRef]
- Abedon, S.T. Pleiotropic costs of phage resistance. In Bacteriophages as Drivers of Evolution: An Evolutionary Ecological Perspective; Springer: Cham, Switzerland, 2022; pp. 253–262. [Google Scholar]
- Bohannan, B.J.M.; Lenski, R.E. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecol. Lett. 2000, 3, 362–377. [Google Scholar] [CrossRef]
- Cairns, J.; Becks, L.; Jalasvuori, M.; Hiltunen, T. Sublethal streptomycin concentrations and lytic bacteriophage together promote resistance evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160040. [Google Scholar] [CrossRef]
- Mahmud, H.A.; Wakeman, C.A. Navigating collateral sensitivity: Insights into the mechanisms and applications of antibiotic resistance trade-offs. Front. Microbiol. 2024, 15, 1478789. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Markwitz, P.; Sosnowska, E.; Lood, C.; Lavigne, R.; Drulis-Kawa, Z. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 2021, 23, 7723–7740. [Google Scholar] [CrossRef]
- Smug, B.J.; Majkowska-Skrobek, G.; Drulis-Kawa, Z. PhREEPred: Phage resistance emergence prediction web tool to foresee encapsulated bacterial escape from phage cocktail treatment. J. Mol. Biol. 2022, 434, 167670. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.W.; Huggins, M.B. Successful treatment of experimental Escherichia coli infections in mice using phage: Its general superiority over antibiotics. J. Gen. Microbiol. 1982, 128, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Bull, J.J. Phage therapy revisited: The population biology of a bacterial infection and its treatment with bacteriophage and antibiotics. Am. Nat. 1996, 147, 881–898. [Google Scholar] [CrossRef]
- León, M.; Bastías, R. Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 2015, 6, 343. [Google Scholar] [CrossRef]
- Regeimbal, J.M.; Jacobs, A.C.; Corey, B.W.; Henry, M.S.; Thompson, M.G.; Pavlicek, R.L.; Quinones, J.; Hannah, R.M.; Ghebremedhin, M.; Crane, N.J.; et al. Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 2016, 60, 5806–5816. [Google Scholar] [CrossRef] [PubMed]
- Gaborieau, B.; Delattre, R.; Adiba, S.; Clermont, O.; Denamur, E.; Ricard, J.-D.; Debarbieux, L. Variable fitness effects of bacteriophage resistance mutations in Escherichia coli: Implications for phage therapy. J. Virol. 2024, 98, e0111324. [Google Scholar] [CrossRef]
- Tang, M.; Yao, Z.; Liu, Y.; Ma, Z.; Zhao, D.; Mao, Z.; Wang, Y.; Chen, L.; Zhou, T. Host immunity involvement in the outcome of phage therapy against hypervirulent Klebsiella pneumoniae infections. Antimicrob. Agents Chemother. 2024, 68, e0142923. [Google Scholar] [CrossRef]
- Fujiki, J.; Nakamura, T.; Kreimeyer, H.; Llorente, C.; Fouts, D.E.; Schnabl, B. Insertion sequence-mediated phage resistance contributes to attenuated colonization of cytolytic Enterococcus faecalis variants in the gut. Microbiol. Spectr. 2025, 13, e03303-24. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.; Huo, W.; Pas, S.; Dao, R.; Palmer, K.L. Loss-of-function mutations in epaR confer resistance to ϕNPV1 Infection in Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 2018, 62, e00758-18. [Google Scholar] [CrossRef] [PubMed]
- Gurney, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Public Health 2020, 2020, 148–157. [Google Scholar] [CrossRef]
- Canfield, G.S.; Chatterjee, A.; Espinosa, J.; Mangalea, M.R.; Sheriff, E.K.; Keidan, M.; McBride, S.W.; McCollister, B.D.; Hang, H.C.; Duerkop, B.A. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 2021, 65, e00143-21. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef]
- Wang, X.; Loh, B.; Gordillo Altamirano, F.; Yu, Y.; Hua, X.; Leptihn, S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes. Infect. 2021, 10, 2205–2219. [Google Scholar] [CrossRef]
- Coyne, A.J.K.; Stamper, K.; Kebriaei, R.; Holger, D.J.; El Ghali, A.; Morrisette, T.; Biswas, B.; Wilson, M.; Deschenes, M.V.; Canfield, G.S. Phage cocktails with daptomycin and ampicillin eradicates biofilm-embedded multidrug-resistant Enterococcus faecium with preserved phage susceptibility. Antibiotics 2022, 11, 1175. [Google Scholar]
- Diallo, K.; Dublanchet, A. Benefits of combined phage-antibiotic therapy for the control of antibiotic-resistant bacteria: A literature review. Antibiotics 2022, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Ji, H.; Wang, L.; Li, X.; Hu, D.; Zhao, J.; Wang, S.; Tao, P.; Li, X.; Qian, P. Fitness trade-offs in phage cocktail-resistant Salmonella enterica serovar Enteritidis results in increased antibiotic susceptibility and reduced virulence. Microbiol. Spectr. 2022, 10, e0291422. [Google Scholar] [CrossRef]
- Fujiki, J.; Nakamura, K.; Nakamura, T.; Iwano, H. Fitness trade-offs between phage and antibiotic sensitivity in phage-resistant variants: Molecular action and insights into clinical applications for phage therapy. Int. J. Mol. Sci. 2023, 24, 15628. [Google Scholar] [CrossRef]
- Chen, L.K.; Kuo, S.C.; Chang, K.C.; Cheng, C.C.; Yu, P.Y.; Chang, C.H.; Chen, T.Y.; Tseng, C.C. Clinical antibiotic-resistant Acinetobacter baumannii strains with higher susceptibility to environmental phages than antibiotic-sensitive strains. Sci. Rep. 2017, 7, 6319. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Chen, A.I.; Liu, M.; Zhou, H.; Egan, M.; Yang, X.; Kan, B.; Wang, H.; Goulian, M.; Zhu, J. Colistin-resistance-mediated bacterial surface modification sensitizes phage infection. Antimicrob. Agents Chemother. 2019, 63, e01609-19. [Google Scholar] [CrossRef]
- Demerec, M.; Fano, U. Bacteriophage-resistant mutants in Escherichia coli. Genetics 1945, 30, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.G.; Buckling, A. Phages limit the evolution of bacterial antibiotic resistance in experimental microcosms. Evol. Appl. 2012, 5, 575–582. [Google Scholar] [CrossRef]
- Zhang, Q.G. Exposure to phages has little impact on the evolution of bacterial antibiotic resistance on drug concentration gradients. Evol. Appl. 2014, 7, 394–402. [Google Scholar] [CrossRef]
- Wang, X.; Wei, Z.; Yang, K.; Wang, J.; Jousset, A.; Xu, Y.; Shen, Q.; Friman, V.P. Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotech. 2019, 37, 1513–1520. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Duerkop, B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 2020, 88, e00926-19. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E.; Levin, B.R. Constraints on the coevolution of bacteria and virulent phage: A model, some experiments, and predictions for natural communities. Am. Nat. 1985, 125, 585–602. [Google Scholar] [CrossRef]
- Gurney, J.; Brown, S.P.; Kaltz, O.; Hochberg, M.E. Steering phages to combat bacterial pathogens. Trends Microbiol. 2020, 28, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Czerwinski, S.; Gurney, J. Phage steering in the presence of a competing bacterial pathogen. Microbiol. Spectr. 2025, 13, e0288224. [Google Scholar] [CrossRef]
- Brockhurst, M.A.; Koskella, B.; Zhang, Q.G. Bacteria-phage antagonistic coevolution and the implications for phage therapy. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M., Eds.; Springer Nature Switzerland AG: New York, NY, USA, 2021; pp. 231–251. [Google Scholar]
- Fujiki, J.; Nakamura, K.; Ishiguro, Y.; Iwano, H. Using phage to drive selections toward restoring antibiotic sensitivity in Pseudomonas aeruginosa via chromosomal deletions. Front. Microbiol. 2024, 15, 1401234. [Google Scholar] [CrossRef]
- Olszak, T.; Augustyniak, D.; Garcia-Romero, I.; Markwitz, P.; Gula, G.; Molinaro, A.; Valvano, M.A.; Drulis-Kawa, Z. Phage treatment of Pseudomonas aeruginosa yields a phage-resistant population with different susceptibility to innate immune responses and mild effects on metabolic profiles. Microbiol. Res. 2024, 282, 127609. [Google Scholar] [CrossRef]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 2017, 22, 38–47. [Google Scholar] [CrossRef]
- Lenski, R.E. Two-step resistance by Escherichia coli B to bacteriophage T2. Genetics 1984, 107, 1–7. [Google Scholar] [CrossRef]
- Morona, R.; Henning, U. Host range mutants of bacteriophage Ox2 can use two different outer membrane proteins of Escherichia coli K-12 as receptors. J. Bacteriol. 1984, 159, 724–730. [Google Scholar] [CrossRef]
- Liu, M.; Deora, R.; Doulatov, S.R.; Gingery, M.; Eiserling, F.A.; Preston, A.; Maskell, D.J.; Simons, R.W.; Cotter, P.A.; Parkhill, J.; et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 2002, 295, 2091–2094. [Google Scholar] [CrossRef]
- Schwarzer, D.; Buettner, F.F.; Browning, C.; Nazarov, S.; Rabsch, W.; Bethe, A.; Oberbeck, A.; Bowman, V.D.; Stummeyer, K.; Muhlenhoff, M.; et al. A multivalent adsorption apparatus explains the broad host range of phage phi92: A comprehensive genomic and structural analysis. J. Virol. 2012, 86, 10384–10398. [Google Scholar] [CrossRef]
- de Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Yu, F.; Mizushima, S. Roles of lipopolysaccharide and outer membrane protein ompC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 1982, 151, 718–722. [Google Scholar] [CrossRef]
- Rojero, M.; Weaver-Rosen, M.; Serwer, P. Bypassing evolution of bacterial resistance to phages: The example of hyper-aggressive phage 0524phi7-1. Int. J. Mol. Sci. 2025, 26, 2914. [Google Scholar] [CrossRef]
- Meyer, J.R.; Dobias, D.T.; Weitz, J.S.; Barrick, J.E.; Quick, R.T.; Lenski, R.E. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 2012, 335, 428–432. [Google Scholar] [CrossRef]
- Harshey, R.M. Phage Mu. In The Bacteriophages. Volume 1, 1st ed.; Calendar, R., Ed.; Plenum Press: New York, NY, USA, 1988; pp. 193–234. [Google Scholar]
- Paolozzi, L.; Ghelardini, P. The bacteriophage Mu. In The Bacteriophages, 2nd ed.; Calendar, R., Abedon, S.T., Eds.; Oxford University Press: Oxford, UK, 2006; pp. 469–496. [Google Scholar]
- Abedon, S.T. Further considerations on how to improve phage therapy experimentation, practice, and reporting: Pharmacodynamics perspectives. Phage 2022, 3, 98–111. [Google Scholar] [CrossRef]
- Niu, Y.D.; Liu, H.; Du, H.; Meng, R.; Sayed, M.E.; Wang, G.; McAllister, T.A.; Stanford, K. Efficacy of individual bacteriophages does not predict efficacy of bacteriophage cocktails for control of Escherichia coli O157. Front. Microbiol. 2021, 12, 616712. [Google Scholar] [CrossRef] [PubMed]
- Tanji, Y.; Shimada, T.; Yoichi, M.; Miyanaga, K.; Hori, K.; Unno, H. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 2004, 64, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology: Phage cocktails. Adv. Appl. Microbiol. 2012, 78, 1–23. [Google Scholar] [PubMed]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Molina, F.; Simancas, A.; Ramirez, M.; Tabla, R.; Roa, I.; Rebollo, J.E. A new pipeline for designing phage cocktails based on phage-bacteria infection networks. Front. Microbiol. 2021, 12, 564532. [Google Scholar] [CrossRef]
- Lood, C.; Haas, P.J.; van Noort, V.; Lavigne, R. Shopping for phages? Unpacking design rules for therapeutic phage cocktails. Curr. Opin. Virol. 2022, 52, 236–243. [Google Scholar] [CrossRef]
- Mani, I. Phage and phage cocktails formulations. Prog. Mol. Biol. Transl. Sci. 2023, 200, 159–169. [Google Scholar]
- Hegarty, B. Making waves: Intelligent phage cocktail design, a pathway to precise microbial control in water systems. Water Res. 2025, 268, 122594. [Google Scholar] [CrossRef]
- Marchi, J.; Minh, C.N.N.; Debarbieux, L.; Weitz, J.S. Multi-strain phage induced clearance of bacterial infections. PLoS Comput. Biol. 2025, 21, e1012793. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Bull, J.J. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2004, 2, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Casey, E.; van Sinderen, D.; Mahony, J. In vitro characteristics of phages to guide ‘real life’ phage therapy suitability. Viruses 2018, 10, 163. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Cocktail Optimizer. 2020. Available online: http://www.phage-therapy.org/calculators/cocktail_optimizer.html (accessed on 29 June 2025).
- Díaz-Galián, M.V.; Vega-Rodríguez, M.A.; Molina, F. PhageCocktail: An R package to design phage cocktails from experimental phage-bacteria infection networks. Comput. Methods Programs Biomed. 2022, 221, 106865. [Google Scholar] [CrossRef] [PubMed]
- Menor-Flores, M.; Vega-Rodríguez, M.A.; Molina, F. Computational design of phage cocktails based on phage-bacteria infection networks. Comput. Biol. Med. 2022, 142, 105186. [Google Scholar] [CrossRef]
- Wright, R.C.T.; Friman, V.P.; Smith, M.C.M.; Brockhurst, M.A. Functional diversity increases the efficacy of phage combinations. Microbiology 2021, 167, 001110. [Google Scholar] [CrossRef]
- Chen, H.; Liu, H.; Gong, Y.; Dunstan, R.A.; Ma, Z.; Zhou, C.; Zhao, D.; Tang, M.; Lithgow, T.; Zhou, T. A Klebsiella-phage cocktail to broaden the host range and delay bacteriophage resistance both in vitro and in vivo. NPJ. Biofilms. Microbiomes 2024, 10, 127. [Google Scholar] [CrossRef]
- Abedon, S.T. Active bacteriophage biocontrol and therapy on sub-millimeter scales towards removal of unwanted bacteria from foods and microbiomes. AIMS Microbiol. 2017, 3, 649–688. [Google Scholar] [CrossRef]
- Li, G.; Leung, C.Y.; Wardi, Y.; Debarbieux, L.; Weitz, J.S. Optimizing the timing and composition of therapeutic phage cocktails: A control-theoretic approach. Bull. Math Biol. 2020, 82, 75. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; HE, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Dai, J.; Guo, M.; Li, J.; Zhou, X.; Li, F.; Gao, Y.; Qu, H.; Lu, H.; Jin, J.; et al. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microbes Infect. 2021, 10, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Phage Cross-Resistance Avoider. 2020. Available online: http://www.phage-therapy.org/calculators/xresistance_avoider.html (accessed on 29 June 2025).
- Bonhoeffer, S.; Lipsitch, M.; Levin, B.R. Evaluating treatment protocols to prevent resistance. Proc. Natl. Acad. Sci. USA 1997, 94, 12106–12111. [Google Scholar] [CrossRef]
- Fischbach, M.A. Combination therapies for combating antimicrobial resistance. Curr. Opin. Microbiol. 2011, 14, 519–523. [Google Scholar] [CrossRef]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef]
- REX Consortium. Heterogeneity of selection and the evolution of resistance. Trends Ecol. Evol. 2013, 28, 110–118. [Google Scholar] [CrossRef]
- Cadinanos, J.; Montejano, R.; de Miguel, B.R.; Marcelo, C.; Arribas, J.R. Risks and benefits of reducing the number of drugs to treat HIV-1 infection. Expert Opin. Drug Saf. 2021, 20, 397–409. [Google Scholar] [CrossRef]
- Torres-Barceló, C.; Hochberg, M.E. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016, 24, 249–256. [Google Scholar] [CrossRef]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.P. Fighting pathogenic bacteria on two fronts: Phages and antibiotics as combined strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef]
- Morrisette, T.; Kebriaei, R.; Lev, K.L.; Morales, S.; Rybak, M.J. Bacteriophage therapeutics: A primer for clinicians on phage-antibiotic combinations. Pharmacotherapy 2020, 40, 153–168. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, R.A.; Leung, C.Y.; Chan, B.K.; Turner, P.E.; Weitz, J.S. Quantitative models of phage-antibiotic combination therapy. mSystems 2020, 5, e00756-19. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Drulis-Kawa, Z.; Cater, K.; Knežević, P.; Winogradow, C.; Amaro, K.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Rękas, J.; et al. Bacteriophages and antibiotic interactions in clinical practice: What we have learned so far. J. Biomed. Sci. 2022, 29, 23. [Google Scholar] [CrossRef] [PubMed]
- Ferran, A.A.; Lacroix, M.Z.; Gourbeyre, O.; Huesca, A.; Gaborieau, B.; Debarbieux, L.; Bousquet-Melou, A. The selection of antibiotic- and bacteriophage-resistant Pseudomonas aeruginosa is prevented by their combination. Microbiol. Spectr. 2022, 10, e0287422. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Phage-antibiotic combination treatments: Antagonistic impacts of antibiotics on the pharmacodynamics of phage therapy? Antibiotics 2019, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Danis-Wlodarczyk, K.M.; Cai, A.; Chen, A.; Gittrich, M.R.; Sullivan, M.B.; Wozniak, D.J.; Abedon, S.T. Friends or foes? Rapid determination of dissimilar colistin and ciprofloxacin antagonism of Pseudomonas aeruginosa phages. Pharmaceuticals 2021, 14, 1162. [Google Scholar] [CrossRef]
- Allen, R.C.; Pfrunder-Cardozo, K.R.; Meinel, D.; Egli, A.; Hall, A.R. Associations among antibiotic and phage resistance phenotypes in natural and clinical Escherichia coli isolates. MBio 2017, 8, e01341-17. [Google Scholar] [CrossRef]
- Moulton-Brown, C.E.; Friman, V.P. Rapid evolution of generalized resistance mechanisms can constrain the efficacy of phage-antibiotic treatments. Evol. Appl. 2018, 11, 1630–1641. [Google Scholar] [CrossRef]
- Kortright, K.E.; Doss-Gollin, S.; Chan, B.K.; Turner, P.E. Evolution of bacterial cross-resistance to lytic phages and albicidin antibiotic. Front. Microbiol. 2021, 12, 658374. [Google Scholar] [CrossRef]
- McCallin, S.; Menzi, C.; Lassen, S.; Daraspe, J.; Oechslin, F.; Moreillon, P. Antibiotic exposure leads to reduced phage susceptibility in vancomycin intermediate Staphylococcus aureus (VISA). Antimicrob. Agents Chemother. 2022, 66, e0224721. [Google Scholar] [CrossRef]
- Rosas, N.C.; Lithgow, T. Targeting bacterial outer-membrane remodelling to impact antimicrobial drug resistance. Trends Microbiol 2022, 30, 544–552. [Google Scholar] [CrossRef]
- Pons, B.J.; Dimitriu, T.; Westra, E.R.; van Houte, S. Antibiotics that affect translation can antagonize phage infectivity by interfering with the deployment of counter-defenses. Proc. Natl. Acad. Sci. USA 2023, 120, e2216084120. [Google Scholar] [CrossRef] [PubMed]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.B.; Trampuz, A.; Di, L.M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef]
- Li, X.; He, Y.; Wang, Z.; Wei, J.; Hu, T.; Si, J.; Tao, G.; Zhang, L.; Xie, L.; Abdalla, A.E.; et al. A combination therapy of phages and antibiotics: Two is better than one. Int. J. Biol. Sci. 2021, 17, 3573–3582. [Google Scholar] [CrossRef]
- Liu, C.; Hong, Q.; Chang, R.Y.K.; Kwok, P.C.L.; Chan, H.K. Phage-antibiotic therapy as a promising strategy to combat multidrug-resistant infections and to enhance antimicrobial efficiency. Antibiotics 2022, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.-P.; Ferry, T.; Resch, G. Recent progress toward the implementation of phage therapy in Western medicine. FEMS Microbiol. Rev. 2022, 46, fuab040. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage adsorption: Likelihood of virion encounter with bacteria and other factors affecting rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef]
- Tang, Z.; Tang, N.; Wang, X.; Ren, H.; Zhang, C.; Zou, L.; Han, L.; Guo, L.; Liu, W. Characterization of a lytic Escherichia coli phage CE1 and its potential use in therapy against avian pathogenic Escherichia coli infections. Front. Microbiol. 2023, 14, 1091442. [Google Scholar] [CrossRef] [PubMed]
- Sevilla-Navarro, S.; Marin, C.; Cortes, V.; Garcia, C.; Vega, S.; Catala-Gregori, P. Autophage as a control measure for Salmonella in laying hens. Poult. Sci. 2018, 97, 4367–4373. [Google Scholar] [CrossRef]
- Tkhilaishvili, T.; Potapov, E.; Starck, C.; Mulzer, J.; Falk, V.; Trampuz, A.; Schoenrath, F. Bacteriophage therapy as a treatment option for complex cardiovascular implant infection: The German Heart Center Berlin experience. J. Heart Lung Transplant. 2022, 41, 551–555. [Google Scholar] [CrossRef]
- Vallenas-Sánchez, Y.P.A.; Bautista-Valles, M.F.; Llaque-Chávarri, F.; Mendoza-Coello, M.E. Bacteriophage cocktail as a substitute for antimicrobials in companion animal dermatology. J. Selva Andin. Anim. Sci. 2022, 9, 97–117. [Google Scholar] [CrossRef]
- Johri, A.V.; Johri, P.; Hoyle, N.; Pipia, L.; Nadareishvili, L.; Nizharadze, D. Case report: Chronic bacterial prostatitis treated with phage therapy after multiple failed antibiotic treatments. Front. Pharmacol. 2021, 12, 692614. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Hoyle, N.; Zhvaniya, F.; Lusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Lobocka, M.; Borysowski, J.; Alavidze, Z.; Górski, A.; Gogokhia, L. Current updates from the long-standing phage rsearch centers in Georgia, Poland, and Russia. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M., Eds.; Springer Nature Switzerland AG: New York, NY, USA, 2021; pp. 921–951. [Google Scholar]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B.; Delattre, A.S.; Lavigne, R. Learning from bacteriophages—Advantages and limitations of phage and phage-encoded protein applications. Curr. Protein Pept. Sci. 2012, 13, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Kvachadze, L.; Balarjishvili, N.; Meskhi, T.; Tevdoradze, E.; Skhirtladze, N.; Pataridze, T.; Adamia, R.; Topuria, T.; Kutter, E.; Rohde, C.; et al. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb. Biotechnol. 2011, 4, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E.; De Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S.T. Phage therapy in clinical practice: Treatment of human infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef]
- Kutter, E.M. Bacteriophage therapy: Past and present. In Encyclopedia of Microbiology; Schaecter, M., Ed.; Elsevier: Oxford, UK, 2009; pp. 258–266. [Google Scholar]


| Description | Pros | Cons |
|---|---|---|
| New-phage isolation using bacterial strains infecting a patient as isolation hosts | Phage host range specifically includes the targeted infection etiology | Requires time and expertise; In addressing treatment resistance, it is reactive rather than proactive; Can permit substantial replication of phage-resistant bacteria prior to phage substitution |
| Description | Pros | Cons |
|---|---|---|
| Previously isolated phage stocks are available for phage substitution | Phage characterization can take place ahead of time, allowing relatively rapid and safe phage substitution | Requires access to a phage bank, though phage crowdsourcing could serve as an alternative phage source; In addressing treatment resistance, it is reactive rather than proactive; Can permit substantial replication of phage-resistant bacteria prior to phage substitution |
| Description | Pros | Cons |
|---|---|---|
| In vitro evolution of a treatment phage especially toward new host range properties | Phages modified through mutation may require only minimal further characterization; Phages can also be modified in highly targeted manners using molecular techniques (phage engineering) | Requires time and expertise; In addressing treatment resistance, it is reactive rather than proactive; Can permit substantial replication of phage-resistant bacteria prior to phage substitution; Trained phages can possess similar or identical immunological characteristics to parental phages |
| Description | Pros | Cons |
|---|---|---|
| Rapidly bringing bacterial infections under control prior to their growing in cell number to a point where resistance mutations are present | Ideally, prevents mutations to resistance from occurring; Is by necessity proactive relative to the occurrence of resistance mutations | Unless treatments are prophylactic, or bacterial infections otherwise are caught very early, then this approach can be difficult or impossible to successfully implement |
| Description | Pros | Cons |
|---|---|---|
| Intentional selection by treatment phages for bacterial mutants that are unable to continue to support ongoing disease | Allows initiation of anti-treatment-resistance strategies with monophages; Combats bacterial evolution of phage resistance by harnessing natural selection; Can be proactive rather than reactive | Evidence is needed on a per-phage basis that reductions in bacterial fitness are seen across multiple potentially targeted bacterial strains; May not be as effective given bacterial infections of immunocompromised individuals |
| Description | Pros | Cons |
|---|---|---|
| Certain phages are able to adsorb using different receptor molecules displayed by the same bacterial strains | Allows initiation of anti-treatment-resistance strategies with monophages; Two independent mutations may be required of bacteria to achieve phage resistance rather than just one mutation; Can be proactive rather than reactive | It is uncertain how many phages possess this property; It is uncertain what fraction of bacterial hosts found within a phage’s host range will normally display both phage receptors |
| Description | Pros | Cons |
|---|---|---|
| Combination therapy involving only phages (in principle, though, phage cocktails can also be combined with non-phage antibacterial agents such as antibiotics) | Can prevent substantial growth of bacteria that have mutated to phage resistance; Can be proactive rather than reactive | Requires multiple phage types, each able to impact a targeted bacterium; Requires a low potential for bacteria to mutate to cross-resistance to those multiple phage types; Potential for phage antagonism; Greater cost and complexity |
| Description | Pros | Cons |
|---|---|---|
| Therapy involving phage combination especially with an antibiotic | Can prevent substantial growth of phage-resistant bacteria; Two independent mutations in most cases are required of bacteria to achieve co-resistance rather than just one mutation; Can be proactive rather than reactive | Antibiotics can be antagonistic to phage infection abilities; Antibiotics can possess side effects that would tend to be absent given treatments solely with phages; Any observed efficacy will be difficult to assign to phage action alone |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abedon, S.T. Phage Therapy: Combating Evolution of Bacterial Resistance to Phages. Viruses 2025, 17, 1094. https://doi.org/10.3390/v17081094
Abedon ST. Phage Therapy: Combating Evolution of Bacterial Resistance to Phages. Viruses. 2025; 17(8):1094. https://doi.org/10.3390/v17081094
Chicago/Turabian StyleAbedon, Stephen T. 2025. "Phage Therapy: Combating Evolution of Bacterial Resistance to Phages" Viruses 17, no. 8: 1094. https://doi.org/10.3390/v17081094
APA StyleAbedon, S. T. (2025). Phage Therapy: Combating Evolution of Bacterial Resistance to Phages. Viruses, 17(8), 1094. https://doi.org/10.3390/v17081094









