SARS-CoV-2 Nsp1 Is a Major Suppressor of HLA Class I and Class II Expression
Abstract
1. Introduction
2. Material and Methods
2.1. Molecular Cloning
2.2. Cell Culture and Protein Overexpression
2.3. SARS-CoV-2 Viruses and Infection
2.4. Antibodies
2.5. Flow Cytometry
2.6. Western Blotting
2.7. Analysis of Published RNA-Seq Datasets
3. Results
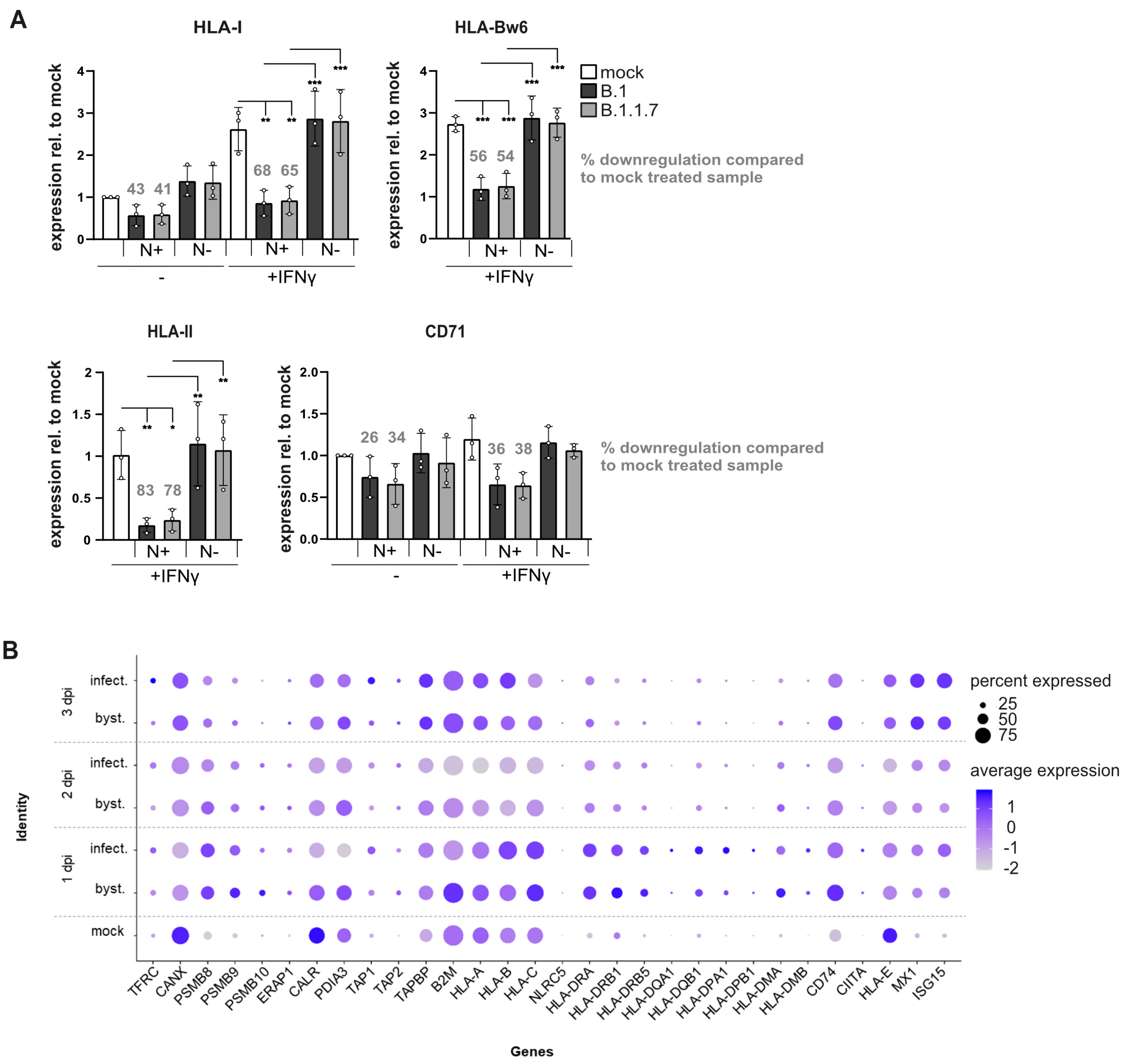
3.1. SARS-CoV-2 Efficiently Controls Surface HLA-I Levels on Infected Cells
3.2. SARS-CoV-2 Blocks IFNγ-Induced Surface Expression of HLA-I and HLA-II
3.3. Cells Stably Expressing ORF8 Do Not Regulate HLA-I
3.4. High-Level ORF8 Overexpression Leads to Downregulation of HLA-I
3.5. Nsp1 Effectively Inhibits Biosynthesis of HLA-I
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Peiris, J.S.; Lai, S.T.; Poon, L.L.; Guan, Y.; Yam, L.Y.; Lim, W.; Nicholls, J.; Yee, W.K.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Ibarrondo, F.J.; Fulcher, J.A.; Goodman-Meza, D.; Elliott, J.; Hofmann, C.; Hausner, M.A.; Ferbas, K.G.; Tobin, N.H.; Aldrovandi, G.M.; Yang, O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild COVID-19. N. Engl. J. Med. 2020, 383, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Geers, D.; Shamier, M.C.; Bogers, S.; den Hartog, G.; Gommers, L.; Nieuwkoop, N.N.; Schmitz, K.S.; Rijsbergen, L.C.; van Osch, J.A.T.; Dijkhuizen, E.; et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021, 6, eabj1750. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Rowntree, L.C.; Hensen, L.; Chua, B.Y.; van de Sandt, C.E.; Habel, J.R.; Zhang, W.; Jia, X.; Kedzierski, L.; Ashhurst, T.M.; et al. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. Cell Rep. Med. 2021, 2, 100208. [Google Scholar] [CrossRef]
- Bange, E.M.; Han, N.A.; Wileyto, P.; Kim, J.Y.; Gouma, S.; Robinson, J.; Greenplate, A.R.; Hwee, M.A.; Porterfield, F.; Owoyemi, O.; et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021, 27, 1280–1289. [Google Scholar] [CrossRef]
- Oberhardt, V.; Luxenburger, H.; Kemming, J.; Schulien, I.; Ciminski, K.; Giese, S.; Csernalabics, B.; Lang-Meli, J.; Janowska, I.; Staniek, J.; et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature 2021, 597, 268–273. [Google Scholar] [CrossRef]
- Yoo, J.S.; Sasaki, M.; Cho, S.X.; Kasuga, Y.; Zhu, B.; Ouda, R.; Orba, Y.; de Figueiredo, P.; Sawa, H.; Kobayashi, K.S. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021, 12, 6602. [Google Scholar] [CrossRef]
- Yuen, C.K.; Lam, J.Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.P.; Jin, D.Y.; To, K.K.; Chan, J.F.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Arshad, N.; Laurent-Rolle, M.; Ahmed, W.S.; Hsu, J.C.; Mitchell, S.M.; Pawlak, J.; Sengupta, D.; Biswas, K.H.; Cresswell, P. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to down-regulate MHC-I surface expression. Proc. Natl. Acad. Sci. USA 2023, 120, e2208525120. [Google Scholar] [CrossRef] [PubMed]
- Finkel, Y.; Gluck, A.; Nachshon, A.; Winkler, R.; Fisher, T.; Rozman, B.; Mizrahi, O.; Lubelsky, Y.; Zuckerman, B.; Slobodin, B.; et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 2021, 594, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.A.; Leibundgut, M.; Thiel, V.; Muhlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.; Gluck, A.; Narayanan, K.; Kuroda, M.; Nachshon, A.; Hsu, J.C.; Halfmann, P.J.; Yahalom-Ronen, Y.; Tamir, H.; Finkel, Y.; et al. Parsing the role of NSP1 in SARS-CoV-2 infection. Cell Rep. 2022, 39, 110954. [Google Scholar] [CrossRef]
- Banerjee, A.K.; Blanco, M.R.; Bruce, E.A.; Honson, D.D.; Chen, L.M.; Chow, A.; Bhat, P.; Ollikainen, N.; Quinodoz, S.A.; Loney, C.; et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020, 183, 1325–1339. [Google Scholar] [CrossRef]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef]
- Huang, C.; Lokugamage, K.G.; Rozovics, J.M.; Narayanan, K.; Semler, B.L.; Makino, S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011, 7, e1002433. [Google Scholar] [CrossRef]
- Kumar, A.; Ishida, R.; Strilets, T.; Cole, J.; Lopez-Orozco, J.; Fayad, N.; Felix-Lopez, A.; Elaish, M.; Evseev, D.; Magor, K.E.; et al. SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors. J. Virol. 2021, 95, e0026621. [Google Scholar] [CrossRef]
- Schubert, K.; Karousis, E.D.; Ban, I.; Lapointe, C.P.; Leibundgut, M.; Bäumlin, E.; Kummerant, E.; Scaiola, A.; Schönhut, T.; Ziegelmüller, J.; et al. Universal features of Nsp1-mediated translational shutdown by coronaviruses. Mol. Cell 2023, 83, 3546–3557. [Google Scholar] [CrossRef] [PubMed]
- Baumlin, E.; Andenmatten, D.; Luginbuhl, J.; Lalou, A.; Schwaller, N.; Karousis, E.D. The impact of Coronavirus Nsp1 on host mRNA degradation is independent of its role in translation inhibition. Cell Rep. 2025, 44, 115488. [Google Scholar] [CrossRef] [PubMed]
- Bujanic, L.; Shevchuk, O.; von Kügelgen, N.; Kalinina, A.; Ludwik, K.; Koppstein, D.; Zerna, N.; Sickmann, A.; Chekulaeva, M. The key features of SARS-CoV-2 leader and NSP1 required for viral escape of NSP1-mediated repression. Rna 2022, 28, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, C.; Kowalewski, D.; Bauersfeld, L.; Hildenbrand, A.; Gerke, C.; Schwarzmuller, M.; Le-Trilling, V.T.K.; Stevanovic, S.; Hengel, H.; Momburg, F.; et al. HLA-B locus products resist degradation by the human cytomegalovirus immunoevasin US11. PLoS Pathog. 2019, 15, e1008040. [Google Scholar] [CrossRef]
- Halenius, A.; Hauka, S.; Dölken, L.; Stindt, J.; Reinhard, H.; Wiek, C.; Hanenberg, H.; Koszinowski, U.H.; Momburg, F.; Hengel, H. Human Cytomegalovirus Disrupts the Major Histocompatibility Complex Class I Peptide-Loading Complex and Inhibits Tapasin Gene Transcription. J. Virol. 2011, 85, 3473–3485. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Kaleta, T.; Kern, L.; Hong, S.L.; Hölzer, M.; Kochs, G.; Beer, J.; Schnepf, D.; Schwemmle, M.; Bollen, N.; Kolb, P.; et al. Antibody escape and global spread of SARS-CoV-2 lineage A.27. Nat. Commun. 2022, 13, 1152. [Google Scholar] [CrossRef]
- Keskinen, P.; Ronni, T.; Matikainen, S.; Lehtonen, A.; Julkunen, I. Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology 1997, 91, 421–429. [Google Scholar] [CrossRef]
- Parham, P.; Barnstable, C.J.; Bodmer, W.F. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J. Immunol. 1979, 123, 342–349. [Google Scholar] [CrossRef]
- Stam, N.J.; Vroom, T.M.; Peters, P.J.; Pastoors, E.B.; Ploegh, H.L. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int. Immunol. 1990, 2, 113–125. [Google Scholar] [CrossRef]
- Ravindra, N.G.; Alfajaro, M.M.; Gasque, V.; Huston, N.C.; Wan, H.; Szigeti-Buck, K.; Yasumoto, Y.; Greaney, A.M.; Habet, V.; Chow, R.D.; et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021, 19, e3001143. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Wiertz, E.J.H.J.; Jones, T.R.; Sun, L.; Bogyo, M.; Geuze, H.J.; Ploegh, H.L. The Human Cytomegalovirus US11 Gene Product Dislocates MHC Class I Heavy Chains from the Endoplasmic Reticulum to the Cytosol. Cell 1996, 84, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, X.; Sun, Y.; Zhao, H.; Cheng, F.; Wang, J.; Yang, F.; Hu, J.; Zhang, H.; Wang, C.C.; et al. SARS-CoV-2 ORF8 reshapes the ER through forming mixed disulfides with ER oxidoreductases. Redox Biol. 2022, 54, 102388. [Google Scholar] [CrossRef]
- Rashid, F.; Dzakah, E.E.; Wang, H.; Tang, S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021, 296, 198350. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Wang, T.; Wang, J.; Jiang, Y.; Wang, X.; Qiu, Z.; Feng, N.; Sun, W.; Li, C.; et al. SARS-CoV-2 ORF8 Protein Induces Endoplasmic Reticulum Stress-like Responses and Facilitates Virus Replication by Triggering Calnexin: An Unbiased Study. J. Virol. 2023, 97, e0001123. [Google Scholar] [CrossRef]
- Tirosh, B.; Iwakoshi, N.N.; Lilley, B.N.; Lee, A.H.; Glimcher, L.H.; Ploegh, H.L. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 2005, 79, 2768–2779. [Google Scholar] [CrossRef][Green Version]
- Granados, D.P.; Tanguay, P.-L.; Hardy, M.-P.; Caron, É.; de Verteuil, D.; Meloche, S.; Perreault, C. ER stress affects processing of MHC class I-associated peptides. BMC Immunol. 2009, 10, 10. [Google Scholar] [CrossRef]
- Schuren, A.B.; Costa, A.I.; Wiertz, E.J. Recent advances in viral evasion of the MHC Class I processing pathway. Curr. Opin. Immunol. 2016, 40, 43–50. [Google Scholar] [CrossRef]
- Crozier, T.W.M.; Greenwood, E.J.D.; Williamson, J.C.; Guo, W.; Porter, L.M.; Gabaev, I.; Teixeira-Silva, A.; Grice, G.L.; Wickenhagen, A.; Stanton, R.J.; et al. Quantitative proteomic analysis of SARS-CoV-2 infection of primary human airway ciliated cells and lung epithelial cells demonstrates the effectiveness of SARS-CoV-2 innate immune evasion. Wellcome Open Res. 2022, 7, 224. [Google Scholar] [CrossRef]
- Banerjee, A.; El-Sayes, N.; Budylowski, P.; Jacob, R.A.; Richard, D.; Maan, H.; Aguiar, J.A.; Demian, W.L.; Baid, K.; D’Agostino, M.R.; et al. Experimental and natural evidence of SARS-CoV-2-infection-induced activation of type I interferon responses. iScience 2021, 24, 102477. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Kistler, K.E.; Perchetti, G.A.; Baker, N.; Frisbie, L.A.; Torres, L.M.; Aragona, F.; Yun, C.; Figgins, M.; Greninger, A.L.; et al. Positive selection underlies repeated knockout of ORF8 in SARS-CoV-2 evolution. Nat. Commun. 2024, 15, 3207. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Lucas, C.; Monteiro, V.; Iwasaki, A. Enhanced inhibition of MHC-I expression by SARS-CoV-2 Omicron subvariants. Proc. Natl. Acad. Sci. USA 2023, 18, e2221652120. [Google Scholar] [CrossRef] [PubMed]
- Junjappa, R.P.; Patil, P.; Bhattarai, K.R.; Kim, H.R.; Chae, H.J. IRE1α Implications in Endoplasmic Reticulum Stress-Mediated Development and Pathogenesis of Autoimmune Diseases. Front. Immunol. 2018, 9, 1289. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef]
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Rijsewijk, F.A.M.; Verschuren, S.B.E.; van Gaans-van den Brink, J.A.M.; Neisig, A.; Ressing, M.E.; Neefjes, J.; Wiertz, E. The UL41-encoded virion host shutoff (vhs) protein and vhs-independent mechanisms are responsible for down-regulation of MHC class I molecules by bovine herpesvirus 1. J. Gen. Virol. 2001, 82, 2071–2081. [Google Scholar] [CrossRef]
- Tigges, M.A.; Leng, S.; Johnson, D.C.; Burke, R.L. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 1996, 156, 3901–3910. [Google Scholar] [CrossRef]
- Trgovcich, J.; Johnson, D.; Roizman, B. Cell surface major histocompatibility complex class II proteins are regulated by the products of the gamma(1)34.5 and U(L)41 genes of herpes simplex virus 1. J. Virol. 2002, 76, 6974–6986. [Google Scholar] [CrossRef]




| Primer | Sequence |
|---|---|
| ORF8_for ORF8_rev | 5′-GCATGCTAGCATGAAATTTCTTGTTTTCTTAGG-3′ 5-GCATGGATCCTTAGATGAAATCTAAAACAACACG-3′ |
| ORF8-HA_for ORF8-HA_rev | 5′-GCATGCTAGCATGAAATTTCTTGTTTTCTTAGG-3′ 5′-GCATGGATCCTTATGCGTAATCTGGAACATCGTATGGGTAGATGAAATCTAAAACAACACG-3′ |
| ORF3a_for ORF3a_rev | 5′-GCATGCTAGC ATGGATTT GTTTATGAGA ATCTTCACAA-3′ 5′-GCATGGATCCTTACAAAGGCACGCTAGTAG-3′ |
| nsp1_for nsp1_rev | 5′-GCATGCTAGC ATGGAGAGCCTTGTCCCT-3′ 5′-GCATGGATCCTTACCCTCCGTTAAGCTCACG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schirmeister, I.; Eckert, N.; Weigang, S.; Fuchs, J.; Kern, L.; Kochs, G.; Halenius, A. SARS-CoV-2 Nsp1 Is a Major Suppressor of HLA Class I and Class II Expression. Viruses 2025, 17, 1083. https://doi.org/10.3390/v17081083
Schirmeister I, Eckert N, Weigang S, Fuchs J, Kern L, Kochs G, Halenius A. SARS-CoV-2 Nsp1 Is a Major Suppressor of HLA Class I and Class II Expression. Viruses. 2025; 17(8):1083. https://doi.org/10.3390/v17081083
Chicago/Turabian StyleSchirmeister, Ivo, Nicolas Eckert, Sebastian Weigang, Jonas Fuchs, Lisa Kern, Georg Kochs, and Anne Halenius. 2025. "SARS-CoV-2 Nsp1 Is a Major Suppressor of HLA Class I and Class II Expression" Viruses 17, no. 8: 1083. https://doi.org/10.3390/v17081083
APA StyleSchirmeister, I., Eckert, N., Weigang, S., Fuchs, J., Kern, L., Kochs, G., & Halenius, A. (2025). SARS-CoV-2 Nsp1 Is a Major Suppressor of HLA Class I and Class II Expression. Viruses, 17(8), 1083. https://doi.org/10.3390/v17081083





