Evolutionary Diversity of Bat Rabies Virus in São Paulo State, Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Antigenic Characterization
2.3. Sequencing and Genetic Characterization
2.4. Phylogenetic Analysis
2.5. Phylogenetic and Antigenic Site Amino Acid Visualization
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sequence ID | Group | Year | Host | Site Origin | Country | Genbank Access | Reference |
|---|---|---|---|---|---|---|---|
| AB201802_BR_AL4 | I | 2002 | Artibeus lituratus | Dracena, SP | Brazil | AB201802 | [24] |
| AB201803_BR_DR1 | I | 2000 | Desmodus rotundus | Lindóia, SP | Brazil | AB201803 | [24] |
| AB201805_BR_DR3 | I | 2001 | Desmodus rotundus | São José do Barreiro, SP | Brazil | AB201805 | [24] |
| AB201806_BR_NL1 | III | 1998 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | AB201806 | [24] |
| AB201807_BR_NL2 | II | 1999 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | AB201807 | [24] |
| AB201808_BR_NL3 | III | 2001 | Nyctinomops laticaudatus | Nova Granada, SP | Brazil | AB201808 | [24] |
| AB201812_BR_EF2 | II | 2001 | Eptesicus furinalis | Olimpia, SP | Brazil | AB201812 | [24] |
| AB201813_BR_EF3 | II | 2001 | Eptesicus furinalis | São José do Rio Preto, SP | Brazil | AB201813 | [24] |
| AB201814_BR_EF4 | II | 2002 | Eptesicus furinalis | Catanduva, SP | Brazil | AB201814 | [24] |
| AB201815_BR_MM1 | IV | 1999 | Molossus molossus | Jales, SP | Brazil | AB201815 | [24] |
| AB201816_BR_MM2 | IV | 2002 | Molossus molossus | Ilha Solteira, SP | Brazil | AB201816 | [24] |
| AB201817_BR_MR1 | I | 2002 | Molossus rufus | Presidente Venceslau, SP | Brazil | AB201817 | [24] |
| AB201818_BR_MA1 | IV | 2000 | Molossus abrasus | Itapira, SP | Brazil | AB201818 | [24] |
| AB297630_BR_AL6 | I | 2001 | Artibeus lituratus | Rio de Janeiro, RJ | Brazil | AB297630 | [48] |
| AB297631_BR_AL7 | I | 2004 | Artibeus lituratus | Vargem Grande Paulista, SP | Brazil | AB297631 | [48] |
| AB297647_BR_NL4 | III | 2004 | Nyctinomops laticaudatus | Rio de Janeiro, RJ | Brazil | AB297647 | [48] |
| AB618034_strain_MPVI | III | 2007 | Molossus molossus | Santo Antonio, Paraíba | Brazil | AB618034 | Unpublished |
| AF394886_2085 | V | 1986 | Lasiurus borealis | Walker County, Texas | USA | AF394886 | [49] |
| AF396064_cym3941_1995 | II | 1995 | Myotis chiloensis | - | Chile | AF396064 | [50] |
| AY233427_Batbbt123 | VII | 2001 | Tadarida brasiliensis | Buenos Aires | Argentine | AY233427 | [44] |
| AY233448_Stchmbt80 | III | 2000 | Histiotus montanus | Rio Turbio, Santa Cruz | Argentine | AY233448 | [44] |
| AY233451_Batbbt125 | V | 2001 | Tadarida brasiliensis | Buenos Aires | Argentine | AY233451 | [44] |
| AY654585_Brhm4097 | VI | 1998 | Human | Ceará | Brazil | AY654585 | [6] |
| AY654586_Brsg4108 | VI | 1998 | Callithrix jacchus jacchus | Ceará | Brazil | AY654586 | [6] |
| AY654587_Brhm4138 | VI | 1998 | Human | Ceará | Brazil | AY654587 | [6] |
| AY877435_V920 | I | 1993 | Bovine | Chiapas | Mexico | AY877435 | [51] |
| DQ631835_bref8150_05 | III | 2005 | Eptesicus furinalis | Jundiaí, SP | Brazil | DQ631835 | [41] |
| EF363743_IP306_Portel_PA_2004 | I | 2004 | Human | Portel, PA | Brazil | EF363743 | [52] |
| EF363751_IP5214Viseu_PA_2004 | I | 2004 | Human | Viseu, PA | Brazil | EF363751 | [52] |
| EF363757_IP7541Viseu_PA_2005 | I | 2005 | Human | Viseu, PA | Brazil | EF363757 | [52] |
| EU293113_9001FRA | I | 1990 | Fox | - | French Guyana | EU293113 | [53] |
| EU293116_9704ARG | VII | 1997 | bat | - | Argentine | EU293116 | [53] |
| EU873001_brctSP4551_96 | III | 1996 | cat | São Paulo State | Brazil | EU873001 | Unpublished, Favoretto |
| EU981922_IP6773U_2008 | II | 2008 | Myotis sp. | - | Uruguay | EU981922 | [43] |
| GU552788_IP2989_2007 | VII | 2007 | Nyctnomops laticaudatus | Joanópolis, SP | Brazil | GU552788 | [25] |
| GU552789_IP1779_2006 | IV | 2006 | Molossus rufus | Ribeirão Preto, SP | Brazil | GU552789 | [25] |
| GU552790_IP1992_2005 | III | 2005 | Histiotus velatus | Vargem Grande Paulista, SP | Brazil | GU552790 | [25] |
| GU552791_IP6883_2006 | III | 2006 | Histiotus velatus | Campo Limpo Paulista, SP | Brazil | GU552791 | [25] |
| GU552792_IP3321_2005 | III | 2005 | Histiotus sp. | Belo Horizonte, MG | Brazil | GU552792 | [25] |
| GU552795_IP10529_2005 | III | 2005 | Nyctnomops laticaudatus | Ribeirão Preto, SP | Brazil | GU552795 | [25] |
| GU552796_IP4359_2007 | III | 2007 | Molossus molossus | Campinas, SP | Brazil | GU552796 | [25] |
| GU552798_IP8089_2005 | III | 2005 | Nyctnomops laticaudatus | São Sebastião, SP | Brazil | GU552798 | [25] |
| GU552807_IP8061_2006 | II | 2006 | Eptesicus furinalis | Campinas, SP | Brazil | GU552807 | [25] |
| GU552810_IP3056_2007 | II | 2007 | Eptesicus furinalis | Barretos, SP | Brazil | GU552810 | [25] |
| GU552815_IP8665_2005 | II | 2005 | Myotis nigricans | Ribeirão Preto, SP | Brazil | GU552815 | [25] |
| GU552820_IP4157_2005 | II | 2005 | Myotis nigricans | Águas de Lindóia, SP | Brazil | GU552820 | [25] |
| GU552821_IP4896_2005 | II | 2005 | Myotis nigricans | Caçapava, SP | Brazil | GU552821 | [25] |
| GU552824_IP2654_2006 | V | 2006 | Lasiurus cinereus | Garça, SP | Brazil | GU552824 | [25] |
| GU592648_brdrusp100_07 | I | 2007 | Desmodus rotundus | São José do Barreiro, SP | Brazil | GU592648 | [33] |
| GU646775_brmn131_03 | II | 2003 | Myotis nigricans | Araçatuba, SP | Brazil | GU646775 | [13] |
| GU646776_brmn45_03 | II | 2003 | Myotis nigricans | Araçatuba, SP | Brazil | GU646776 | [13] |
| GU646777_brmm95_03 | V | 2003 | Molossus molossus | Nova Independência, SP | Brazil | GU646777 | [13] |
| GU646778_brmn38_03 | II | 2003 | Myotis nigricans | Penápolis, SP | Brazil | GU646778 | [13] |
| GU646779_bral268_98 | I | 1998 | Artibeus lituratus | Mirandópolis, SP | Brazil | GU646779 | [13] |
| GU646780_bral452_99 | I | 1999 | Artibeus lituratus | Ilha Solteira, SP | Brazil | GU646780 | [13] |
| GU646781_bref431_04 | II | 2004 | Eptesicus furinalis | Bilac, SP | Brazil | GU646781 | [13] |
| GU646782_brlb46_04 | II | 2004 | Lasiurus blossevillii | Valparaíso, SP | Brazil | GU646782 | [13] |
| GU646783_bral311_03 | I | 2003 | Artibeus lituratus | Penápolis, SP | Brazil | GU646783 | [13] |
| GU646784_bral304_03 | I | 2003 | Artibeus lituratus | Penápolis, SP | Brazil | GU646784 | [13] |
| GU646785_brmn150_03 | II | 2003 | Myotis nigricans | Araçatuba, SP | Brazil | GU646785 | [13] |
| GU646786_brmn234_02 | II | 2002 | Myotis nigricans | Bilac, SP | Brazil | GU646786 | [13] |
| GU646787_bral625_01 | I | 2001 | Artibeus lituratus | Birigui, SP | Brazil | GU646787 | [13] |
| GU646788_brmn610_01 | II | 2001 | Myotis nigricans | Sud Menucci, SP | Brazil | GU646788 | [13] |
| GU646789_bral499_01 | I | 2001 | Artibeus lituratus | Guararapes, SP | Brazil | GU646789 | [13] |
| GU646790_bref126_01 | II | 2001 | Eptesicus furinalis | Penápolis, SP | Brazil | GU646790 | [13] |
| GU646791_bral84_01 | I | 2001 | Artibeus lituratus | Guararapes, SP | Brazil | GU646791 | [13] |
| GU646792_bref40_01 | II | 2001 | Epitesicus furinalis | Bilac, SP | Brazil | GU646792 | [13] |
| GU646793_breg01_01 | II | 2001 | Eumops glaucinus | Bilac, SP | Brazil | GU646793 | [13] |
| GU646794_brmn839_00 | II | 2000 | Myotis nigricans | Bilac, SP | Brazil | GU646794 | [13] |
| GU646795_bral566_00 | I | 2000 | Artibeus lituratus | Penápolis, SP | Brazil | GU646795 | [13] |
| GU646796_bref213_00 | II | 2000 | Eptesicus furinalis | Araçatuba, SP | Brazil | GU646796 | [13] |
| GU646818_brbv119_03 | I | 2003 | Bovine/Cattle | José Bonifácio, SP | Brazil | GU646818 | [13] |
| GU646828_brbv356_97 | I | 1997 | Bovine/Cattle | Guararapes, SP | Brazil | GU646828 | [13] |
| GU646833_brbv32_94 | T | 1994 | Bovine/Cattle | Araçatuba, SP | Brazil | GU646833 | [13] |
| GU646835_brdg70_93 | T | 1993 | Dog | Araçatuba, SP | Brazil | GU646835 | [13] |
| GU646841_brhr308_00 | I | 2000 | Horse | Barbosa, SP | Brazil | GU646841 | [13] |
| GU646842_bral5341 | I | 1999 | Artibeus lituratus | Birigui, SP | Brazil | GU646842 | [13] |
| GU646843_brmm4105 | I | 1998 | Molossus molossus | Mirandópolis, SP | Brazil | GU646843 | [13] |
| GU646844_brle4132 | I | 1988 | Lasiurus ega | Glicério, SP | Brazil | GU646844 | [13] |
| GU646845_brmr4114 | I | 1998 | Molossus molossus | Penápolis, SP | Brazil | GU646845 | [13] |
| GU646846_brmr4095 | I | 1998 | Molossus rufus | Araçatuba, SP | Brazil | GU646846 | [13] |
| GU646849_brdg5356 | I | 2000 | Dog | Ilha Solteira, SP | Brazil | GU646849 | [13] |
| GU646855_brct60_92 | T | 1992 | Cat | Andradina, SP | Brazil | GU646855 | [13] |
| GU646856_brmr298_07 | I | 2007 | Molossus rufus | Ilha Solteira, SP | Brazil | GU646856 | [13] |
| GU646857_brmn182_07 | II | 2007 | Myotis nigricans | Bilac, SP | Brazil | GU646857 | [13] |
| GU646858_brmr350_06 | I | 2006 | Molossus rufus | Andradina, SP | Brazil | GU646858 | [13] |
| GU646859_bral309_06 | I | 2006 | Artibeus lituratus | Ilha Solteira, SP | Brazil | GU646859 | [13] |
| GU646860_bral195_06 | I | 2006 | Artibeus lituratus | Araçatuba, SP | Brazil | GU646860 | [13] |
| GU646861_bref341_02 | II | 2002 | Epitesicus furinalis | Birigui, SP | Brazil | GU646861 | [13] |
| HM173087_brmn100_05 | II | 2005 | Myotis nigricans | Guararapes, SP | Brazil | HM173087 | [13] |
| HM173088_bral239_05 | I | 2005 | Artibeus lituratus | Birigui, SP | Brazil | HM173088 | [13] |
| HM854029_brdg354_95 | I | 1995 | Dog | Birigui, SP | Brazil | HM854029 | [13] |
| HM854030_brct100_04 | I | 2004 | Cat | Guararapes, SP | Brazil | HM854030 | [13] |
| HM854031_brmr178_05 | IV | 2005 | Molossus rufus | Andradina, SP | Brazil | HM854031 | [13] |
| HM854032_brmn391_05 | II | 2005 | Myotis nigricans | Ilha Solteira, SP | Brazil | HM854032 | [13] |
| HM854033_bral540_05 | I | 2005 | Artibeus lituratus | Araçatuba, SP | Brazil | HM854033 | [13] |
| HM014315_brmm1994_08 | V | 2008 | Molossus molossus | São Paulo, SP | Brazil | HM014315 | [14] |
| HM014316_brmng2449_05 | III | 2005 | Molossops neglectus | São Paulo, SP | Brazil | HM014316 | [14] |
| HM014317_brmr6464_05 | II | 2005 | Myotis riparius | São Paulo, SP | Brazil | HM014317 | [14] |
| HQ666824_bral346_99 | I | 1999 | Artibeus lituratus | São José do Rio Preto, SP | Brazil | HQ666824 | This work |
| HQ666825_brma777_00 | IV | 2000 | Cynomops abrasus | Ipiguá, SP | Brazil | HQ666825 | This work |
| HQ666826_brnl249_01 | III | 2001 | Nyctinomops macrotis | São José do Rio Preto, SP | Brazil | HQ666826 | This work |
| HQ666827_brnl250_01 | III | 2001 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | HQ666827 | This work |
| HQ666828_bref636_01 | II | 2001 | Neoepitesicus furinalis | Olímpia, SP | Brazil | HQ666828 | This work |
| HQ666829_bral808_01 | I | 2001 | Artibeus lituratus | São José do Rio Preto, SP | Brazil | HQ666829 | This work |
| HQ666830_bresp1019_01 | II | 2001 | Neoepitesicus sp. | Cardoso, SP | Brazil | HQ666830 | This work |
| HQ666831_bref1070_01 | II | 2001 | Neoepitesicus furinalis | São José do Rio Preto, SP | Brazil | HQ666831 | This work |
| HQ666832_bref62_02 | II | 2002 | Neoepitesicus furinalis | Catanduva, SP | Brazil | HQ666832 | This work |
| HQ666833_brmm109_02 | IV | 2002 | Molossus molossus | Ilha Solteira, SP | Brazil | HQ666833 | This work |
| HQ666834_brmn835_02 | II | 2002 | Myotis nigricans | Cajobi, SP | Brazil | HQ666834 | This work |
| HQ666835_bral992_02 | I | 2002 | Artibeus lituratus | Dracena, SP | Brazil | HQ666835 | This work |
| HQ666836_brmr1021_02 | I | 2002 | Molossus fluminensis | Presidente Venceslau, SP | Brazil | HQ666836 | This work |
| HQ666837_bref1141_02 | II | 2002 | Neoepitesicus furinalis | Santo Anastácio, SP | Brazil | HQ666837 | This work |
| HQ666838_brbat1256_02 | I | 2002 | Non- hematophagous bat | Martinópolis, SP | Brazil | HQ666838 | This work |
| HQ666839_bral1371_02 | I | 2002 | Artibeus lituratus | Presidente Venceslau, SP | Brazil | HQ666839 | This work |
| HQ666840_bral1535_02 | I | 2002 | Artibeus lituratus | Taciba, SP | Brazil | HQ666840 | This work |
| HQ666841_brmm1539_02 | I | 2002 | Molossus molossus | Presidente Prudente, SP | Brazil | HQ666841 | This work |
| HQ666842_brle1782_02 | III | 2002 | Lasiurus ega | Presidente Prudente, SP | Brazil | HQ666842 | This work |
| HQ666843_braj349_03 | I | 2003 | Artibeus planirostris | Santa Fé do Sul, SP | Brazil | HQ666843 | This work |
| HQ666844_brbat350_03 | I | 2003 | Non-hematophagous bat | Catanduva, SP | Brazil | HQ666844 | This work |
| HQ666845_bral791_03 | I | 2003 | Artibeus lituratus | Presidente Prudente, SP | Brazil | HQ666845 | This work |
| HQ666846_brmn168_04 | II | 2004 | Myotis nigricans | Campinas, SP | Brazil | HQ666846 | This work |
| HQ666847_brnl184_04 | III | 2004 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | HQ666847 | This work |
| HQ666848_bral550_04 | I | 2004 | Artibeus lituratus | Caçapava, SP | Brazil | HQ666848 | This work |
| HQ666849_breg329_05 | I | 2005 | Eumops glaucinus | Araçatuba, SP | Brazil | HQ666849 | This work |
| HQ666850_bred43_09 | II | 2009 | Neoepitesicus diminutus | Pereira Barreto, SP | Brazil | HQ666850 | This work |
| HQ666851_bred84_09 | II | 2009 | Neoepitesicus diminutus | Araçatuba, SP | Brazil | HQ666851 | This work |
| HQ666852_brmn149_09 | II | 2009 | Myotis nigricans | Coroados, SP | Brazil | HQ666852 | This work |
| HQ666853_bral181_09 | I | 2009 | Artibeus lituratus | Penápolis, SP | Brazil | HQ666853 | This work |
| HQ666854_bref199_09 | II | 2009 | Neoeptesicus furinalis | Dracena, SP | Brazil | HQ666854 | This work |
| HQ666855_bref224_09 | I | 2009 | Neoeptesicus furinalis | Parapuã, SP | Brazil | HQ666855 | This work |
| HQ666856_bral325_09 | I | 2009 | Artibeus lituratus | Birigui, SP | Brazil | HQ666856 | This work |
| HQ666857_bral374_09 | I | 2009 | Artibeus lituratus | Guararapes, SP | Brazil | HQ666857 | This work |
| HQ666858_brmr389_09 | I | 2009 | Molossus fluminensis | Guararapes, SP | Brazil | HQ666858 | This work |
| HQ666859_brmn433_09 | II | 2009 | Myotis nigricans | Penápolis, SP | Brazil | HQ666859 | This work |
| HQ666860_bref589_09 | II | 2009 | Neoeptesicus furinalis | Penápolis, SP | Brazil | HQ666860 | This work |
| HQ666861_brbv672_09 | I | 2009 | Bovine | Narandiba, SP | Brazil | HQ666861 | This work |
| HQ666862_brmr17_10 | II | 2010 | Molossus fluminensis | Penápolis, SP | Brazil | HQ666862 | This work |
| HQ666863_breq28_10 | I | 2010 | Equine | Taciba, SP | Brazil | HQ666863 | This work |
| HQ666864_bred43_10 | I | 2010 | Neoeptesicus diminutus | Osvaldo Cruz, SP | Brazil | HQ666864 | This work |
| HQ666865_bref60_10 | II | 2010 | Neoeptesicus furinalis | Birigui, SP | Brazil | HQ666865 | This work |
| HQ666866_bref76_10 | II | 2010 | Neoeptesicus furinalis | Penápolis, SP | Brazil | HQ666866 | This work |
| HQ666867_brmm169_10 | V | 2010 | Molossus molossus | Araçatuba, SP | Brazil | HQ666867 | This work |
| HQ666868_brct171_10 | I | 2010 | Feline | Araçatuba, SP | Brazil | HQ666868 | This work |
| HQ666869_brmr177_10 | V | 2010 | Molossus fluminensis | Birigui, SP | Brazil | HQ666869 | This work |
| HQ666870_brlbl198_10 | V | 2010 | Lasiurus blossevillii | Teodoro Sampaio, SP | Brazil | HQ666870 | This work |
| HQ666871_bref299_10 | II | 2010 | Neoeptesicus furinalis | Penápolis, SP | Brazil | HQ666871 | This work |
| HQ666872_brmr300_10 | I | 2010 | Molossus fluminensis | Penápolis, SP | Brazil | HQ666872 | This work |
| JF916647_BRLB4096_1995 | V | 1995 | Lasiurus blossevillii | Unknown | Brazil | BRLB4096 | Unpublished, Favoretto |
| JF916650_brmyaSP4115_1998 | II | 1998 | Myotis albecens | Unknown | Brazil | JF916650 | Unpublished, Favoretto |
| JF916652_bralusp041_05 | I | 2005 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916652 | [26] |
| JF916655_brlbusp040_07 | V | 2007 | Lasiurus blossevillii | Presidente Prudente, SP | Brazil | JF916655 | [26] |
| JF916656_bralusp042_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916656 | [26] |
| JF916657_brmmusp043_07 | I | 2007 | Molossus molossus | Presidente Prudente, SP | Brazil | JF916657 | [26] |
| JF916659_bralusp047_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916659 | [26] |
| JF916661_bralusp049_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916661 | [26] |
| JF916662_bralusp050_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916662 | [26] |
| JF916663_bralusp052_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916663 | [26] |
| JF916664_bralusp054_07 | VII | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916664 | [26] |
| JF916667_brmnusp058_07 | II | 2007 | Myotis nigricans | Presidente Prudente, SP | Brazil | JF916667 | [26] |
| JF916668_brmnusp061_07 | II | 2007 | Myotis nigricans | Presidente Prudente, SP | Brazil | JF916668 | [26] |
| JF916669_bralusp062_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916669 | [26] |
| JF916671_brefusp064_07 | II | 2007 | Eptesicus furinalis | Presidente Prudente, SP | Brazil | JF916671 | [26] |
| JF916672_bralusp069_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916672 | [26] |
| JF916673_bralusp071_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916673 | [26] |
| JF916674_bralusp001_08 | I | 2008 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916674 | [26] |
| JF916678_brefusp008_09 | II | 2009 | Eptesicus furinalis | Presidente Prudente, SP | Brazil | JF916678 | [26] |
| KM594026_IP_512_09 | II | 2009 | Eptesicus furinalis | Ribeirão Preto, SP | Brazil | KM594026 | [42] |
| KM594027_IP_230_10 | II | 2010 | Eptesicus furinalis | Valinhos, SP | Brazil | KM594027 | [42] |
| KM594028_IP_346_10 | II | 2010 | Eptesicus furinalis | Tambaú, SP | Brazil | KM594028 | [42] |
| KM594029_IP_3208_06 | III | 2006 | Eptesicus furinalis | Vinhedo, SP | Brazil | KM594029 | [42] |
| KM594030_IP_1400_10 | II | 2010 | Myotis nigricans | Campinas, SP | Brazil | KM594030 | [42] |
| KM594031_IP_163_10 | II | 2010 | Myotis nigricans | Caieiras, SP | Brazil | KM594031 | [42] |
| KM594032_IP_497_10 | II | 2010 | Myotis nigricans | Campinas, SP | Brazil | KM594032 | [42] |
| KM594034_IP_350_10 | III | 2010 | Nyctinomops laticaudatus | Conchal, SP | Brazil | KM594034 | [42] |
| KM594035_IP_412_10 | III | 2010 | Nyctinomops laticaudatus | Barretos, SP | Brazil | KM594035 | [42] |
| KM594036_IP_542_10 | III | 2010 | Nyctinomops laticaudatus | Ribeirão Preto, SP | Brazil | KM594036 | [42] |
| KM594037_IP_3176_09 | VII | 2009 | Tadarida brasiliensis | Santo André, SP | Brazil | KM594037 | [42] |
| KM594038_IP_1586_10 | VII | 2010 | Tadarida brasiliensis | São Bernardo do Campo, SP | Brazil | KM594038 | [42] |
| MG458314_RV1789 | I | 1997 | Cow | Unknown | British Est Indies | MG458314 | [54] |
| PQ671596_RS61 | VII | 2022 | Tadarida brasiliensis | Rio Grande do Sul | Brazil | PQ671596 | unpublished |
| Gp_1 | Gp_3 | Gp_2 | Gp_4 | Gp_5 | Gp_7 | Gp_6 | |
|---|---|---|---|---|---|---|---|
| Gp_1 | |||||||
| Gp_3 | 0.140 | ||||||
| Gp_2 | 0.128 | 0.126 | |||||
| Gp_4 | 0.139 | 0.103 | 0.138 | ||||
| Gp_5 | 0.152 | 0.134 | 0.160 | 0.132 | |||
| Gp_7 | 0.087 | 0.117 | 0.118 | 0.111 | 0.136 | ||
| Gp_6 | 0.181 | 0.148 | 0.179 | 0.152 | 0.158 | 0.167 | |
| outgroup | 0.178 | 0.180 | 0.188 | 0.162 | 0.172 | 0.182 | 0.213 |
| Gp 1 | 0.0253 |
| Gp 3 | 0.0467 |
| Gp 2 | 0.0674 |
| Gp 4 | 0.0185 |
| Gp 5 | 0.0566 |
| Gp 7 | 0.0029 |
| Gp 6 | 0.0051 |
| outgroup | 0.0051 |
References
- de Almeida, M.F.; Queiroz, L.H. História Da Raiva No Brasil, 1st ed.; Editora UNESP Digital: São Paulo City, Brazil, 2023; ISBN 9786557144510. [Google Scholar]
- Favoretto, S.R.; Araújo, D.B.; Sacramento, D.V.; Martorelli, L.F.A. Raiva. In Microbiologia—Trabulsi-Alterthum; Trabulsi, A., Altherthum, F., Eds.; Atheneu: São Paulo City, Brazil, 2024; pp. 945–956. ISBN 9786555867985. [Google Scholar]
- Giménez, A.L.; Zabalza, M.J.; Novaro, L.P.; Centurion, G.A.; Barrios-Benito, M.Y.; Moncá, I.; Chaar Letourneau, F.; Casanovas, R.; Russo, S.E. Diversity of Rabies Virus Variants in Insectivorous Bats (Chiroptera: Vespertilionidae and Molossidae): An Epidemiological Study in Central Argentine Patagonia. Viruses 2025, 17, 788. [Google Scholar] [CrossRef]
- ICTV. ICTV-International Committee on Taxonomy of Viruses. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202401733&taxon_name=Lyssavirus rabies (accessed on 3 June 2025).
- Rupprecht, C.E.; Mshelbwala, P.P.; Reeves, R.G.; Kuzmin, I.V. Rabies in a Postpandemic World: Resilient Reservoirs, Redoubtable Riposte, Recurrent Roadblocks, and Resolute Recidivism. Anim. Dis. 2023, 3, 15. [Google Scholar] [CrossRef]
- Favoretto, S.R.; De Mattos, C.C.; Morais, N.B.; Araújo, F.A.A.; Mattos, C.C. De Rabies in Marmosets (Callithrix Jacchus) Ceará, Brazil. Emerg. Infect. Dis. 2001, 7, 1062–1065. [Google Scholar] [CrossRef]
- Favoretto, S.R.; De Mattos, C.C.; De Mattos, C.A.; Campos, A.C.A.; Sacramento, D.R.V.; Durigon, E.L. The Emergence of Wildlife Species as a Source of Human Rabies Infection in Brazil. Epidemiol. Infect. 2013, 141, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.-M.; Papo, S.; Rodriguez, A.; Smith, J.S. Antigenic Analysis of Rabies-virus Isolates from Latin America and the Caribbean. J. Vet. Med. Ser. B 1994, 41, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Villa, A.; Gómez-Sierra, M.; Hernández-Rodríguez, G.; Juárez-Islas, V.; Meléndez-Félix, A.; Vargas-Pino, F.; Velázquez-Monroy, O.; Flisser, A. Antigenic Diversity and Distribution of Rabies Virus in Mexico. J. Clin. Microbiol. 2002, 40, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Páez, A.; Nũñez, C.; García, C.; Bóshell, J. Molecular Epidemiology of Rabies Epizootics in Colombia: Evidence for Human and Dog Rabies Associated with Bats. J. Gen. Virol. 2003, 84, 795–802. [Google Scholar] [CrossRef]
- Favoretto, S.R.; Carrieri, M.L.; Cunha, E.M.S.; Aguiar, E.A.C.; Silva, L.H.Q.; Sodre, M.M.; Souza, M.C.A.M.; Kotait, I. Antigenic Typing of Brazilian Rabies Virus Samples Isolated from Animals and Humans, 1989–2000. Rev. Inst. Med. Trop. Sao Paulo 2002, 44, 91–95. [Google Scholar] [CrossRef]
- Castilho, J.G.; Canello, F.M.; Scheffer, K.C.; Achkar, S.M.; Carrieri, M.L.; Kotait, I. Antigenic and Genetic Characterization of the First Rabies Virus Isolated from the Bat Eumops Perotis in Brazil. Rev. Inst. Med. Trop. Sao Paulo 2008, 50, 95–99. [Google Scholar] [CrossRef]
- Queiroz, L.H.; Favoretto, S.R.; Cunha, E.M.S.; Campos, A.C.A.; Lopes, M.C.; de Carvalho, C.; Iamamoto, K.; Araújo, D.B.; Venditti, L.L.R.; Ribeiro, E.S.; et al. Rabies in Southeast Brazil: A Change in the Epidemiological Pattern. Arch. Virol. 2012, 157, 93–105. [Google Scholar] [CrossRef]
- da Rosa, A.R.; de Arruda Geraldes Kataoka, A.P.; Favoretto, S.R.; Sodré, M.M.; Netto, J.T.; de Almeida Campos, A.C.; Durigon, E.L.; Martorelli, L.F.A. First Report of Rabies Infection in Bats, Molossus Molossus, Molossops Neglectus and Myotis Riparius in the City of São Paulo, State of São Paulo, Southeastern Brazil. Rev. Soc. Bras. Med. Trop. 2011, 44, 146–149. [Google Scholar] [CrossRef]
- Menozzi, B.D.; de Novaes Oliveira, R.; Paiz, L.M.; Richini-Pereira, V.B.; Langoni, H. Antigenic and Genotypic Characterization of Rabies Virus Isolated from Bats (Mammalia: Chiroptera) from Municipalities in São Paulo State, Southeastern Brazil. Arch. Virol. 2017, 162, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Badrane, H.; Tordo, N. Host Switching in Lyssavirus History from the Chiroptera to the Carnivora Orders. J. Virol. 2001, 75, 8096–8104. [Google Scholar] [CrossRef] [PubMed]
- Kissi, B.; Tordo, N.; Bourhy, H. Genetic Polymorphism in the Rabies Virus Nucleoprotein Gene. Virology 1995, 209, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Nadin-Davis, S.A. Polymerase Chain Reaction Protocols for Rabies Virus Discrimination. J. Virol. Methods 1998, 75, 1–8. [Google Scholar] [CrossRef]
- Ito, M.; Arai, Y.T.; Itou, T.; Sakai, T.; Ito, F.H.; Takasaki, T.; Kurane, I. Genetic Characterization and Geographic Distribution of Rabies Virus Isolates in Brazil: Identification of Two Reservoirs, Dogs and Vampire Bats. Virology 2001, 284, 214–222. [Google Scholar] [CrossRef]
- Favoretto, S.R.; de Mattos, C.C.; De Morais, N.B.; Carrieri, M.L.; Rolim, B.N.; Silva, L.M.; Rupprecht, C.E.; Durigon, E.L.; de Mattos, C.A. Maintained by Dogs in Humans and Terrestrial Wildlife. Ceará State, Brazil. Emerg. Infect. Dis. 2006, 12, 1978–1981. [Google Scholar] [CrossRef]
- Carnieli, P.; Fahl, W.d.O.; Castilho, J.G.; Oliveira, R.d.N.; Macedo, C.I.; Durymanova, E.; Jorge, R.S.P.; Morato, R.G.; Spíndola, R.O.; Machado, L.M.; et al. Characterization of Rabies Virus Isolated from Canids and Identification of the Main Wild Canid Host in Northeastern Brazil. Virus Res. 2008, 131, 33–46. [Google Scholar] [CrossRef]
- Favoretto, S.R.; de Mattos, C.C.; Mattos, C.A.; Carrieri, M.L.; Durigon, E.L. Antigenic and Genetic Characterization of Rabies Virus Samples Isolated from Human in Brazil––Preliminary Results. In Proceedings of the XIII International Meeting on Research Advances and Rabies Control in the Américas–RITA, Oaxaca Municipality, Mexico, 3–8 November 2002; pp. 25–26. [Google Scholar]
- Martorelli, L.F.A. Diagnóstico Laboratorial e Diversidade Genética Do Vírus Rábico Isolado No Estado de São Paulo, 1989–2000; Universidade de São Paulo-USP: São Paulo, Brazil, 2004. [Google Scholar]
- Kobayashi, Y.; Sato, G.; Shoji, Y.; Sato, T.; Itou, T.; Cunha, E.M.S.; Samara, S.I.; Carvalho, A.A.B.; Nociti, D.P.; Ito, F.H.; et al. Molecular Epidemiological Analysis of Bat Rabies Viruses in Brazil. J. Vet. Med. Sci. 2005, 67, 647–652. [Google Scholar] [CrossRef]
- Oliveira, R.d.N.; de Souza, S.P.; Lobo, R.S.V.; Castilho, J.G.; Macedo, C.I.; Carnieli, P.; Fahl, W.O.; Achkar, S.M.; Scheffer, K.C.; Kotait, I.; et al. Rabies Virus in Insectivorous Bats: Implications of the Diversity of the Nucleoprotein and Glycoprotein Genes for Molecular Epidemiology. Virology 2010, 405, 352–360. [Google Scholar] [CrossRef]
- Albas, A.; Campos, A.C.D.A.; Araujo, D.B.; Rodrigues, C.S.; Sodré, M.M.; Durigon, E.L.; Favoretto, S.R. Molecular Characterization of Rabies Virus Isolated from Non-Haematophagous Bats in Brazil. Rev. Soc. Bras. Med. Trop. 2011, 44, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Dean, D.J.; Abelseth, M.K.; Atanasiu, P. The Fluorescent Antibody Test. In Laboratory Techniques in Rabies; Meslin, F., Kaplan, M.M., Koprowski, H., Eds.; CC BY-NC-SA 3.0 IGO; WHO—World Health Organization—Licence: Geneva, Switzerland, 1996; p. 47. ISBN 92 4 154479 1. [Google Scholar]
- Koprowski, H. The Mouse Inoculation Test. In Laboratory Techniques in Rabies; Meslin, F., Kaplan, M.M., Koprowski, H., Eds.; CC BY-NC-SA 3.0 IGO; WHO—World Health Organization—Licence: Geneva, Switzerland, 1996; p. 476. ISBN 92 4 154479 1. [Google Scholar]
- Cláudio, V.C.; Novaes, R.L.M.; Gardner, A.L.; Nogueira, M.R.; Wilson, D.E.; Maldonado, J.E.; Oliveira, J.A.; Moratelli, R. Taxonomic Re-Evaluation of New World Eptesicus and Histiotus (Chiroptera: Vespertilionidae), with the Description of a New Genus. Zoologia 2023, 40, e22029. [Google Scholar] [CrossRef]
- Yi, X.; Latch, E.K. Systematics of the New World Bats Eptesicus and Histiotus Suggest Trans-Marine Dispersal Followed by Neotropical Cryptic Diversification. Mol. Phylogenet. Evol. 2022, 175, 107582. [Google Scholar] [CrossRef]
- Chambi Velasquez, M.A.; Pavé, R.; Argoitia, M.A.; Schierloh, P.; Piccirilli, M.G.; Colombo, V.C.; Beltrán, F.J.; Cisterna, D.M.; Caraballo, D.A. Revisiting Molossus (Mammalia: Chiroptera: Molossidae) Diversity: Exploring Southern Limits and Revealing a Novel Species in Argentina. Vertebr. Zool. 2024, 74, 397–416. [Google Scholar] [CrossRef]
- Smith, J.S.; Orciari, L.A.; Yager, P. Molecular Epidemiology of Rabies in the US. Semin Virol. 1995, 6, 387–400. [Google Scholar] [CrossRef]
- Campos, A.C.d.A.; Melo, F.L.; Romano, C.M.; Araujo, D.B.; Cunha, E.M.S.; Sacramento, D.R.V.; de Andrade Zanotto, P.M.; Durigon, E.L.; Favoretto, S.R. One-Step Protocol for Amplification of near Full-Length CDNA of the Rabies Virus Genome. J. Virol. Methods 2011, 174, 1–6. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-Likelihood Phylodynamic Analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef] [PubMed]
- Albas, A.; de Souza, E.A.N.; Lourenço, R.A.; Favoretto, S.R.; Sodré, M.M. Antigen Profile of Rabies Virus Isolated from Different Species of Non-Hematophagous Bats in the Region of Presidente Prudente, State of São Paulo. Rev. Soc. Bras. Med. Trop. 2009, 42, 15–17. [Google Scholar] [CrossRef]
- Cunha, E.M.S.; da Silva, L.H.Q.; Lara, M.d.C.C.S.H.; Nassar, A.F.C.; Albas, A.; Sodré, M.M.; Pedro, W.A. Bat Rabies in the North-Northwestern Regions of the State of São Paulo, Brazil: 1997–2002. Rev. Saude Publica 2006, 40, 1082–1086. [Google Scholar] [CrossRef]
- Queiroz, L.H.; de Carvalho, C.; Buso, D.S.; Ferrari, C.I.d.L.; Pedro, W.A. Epidemiological Profile of Rabies in the Northwestern Region of São Paulo State, from 1993 to 2007. Rev. Soc. Bras. Med. Trop. 2009, 42, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.B.; de Carvalho, C.; Casagrande, D.; Picinato, M.A.d.C.; Pedro, W.A.; Marinho, M.; Queiroz, L.H. Rabies in Bats (Chiroptera, Mammalia) in Brazil: Prevalence and Potential Risk Factors Based on Twenty Years of Research in the Northwestern Region of São Paulo, Brazil. Vet. Sci. 2023, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- de Souza, D.N.; Oliveira, R.N.; Asprino, P.F.; Bettoni, F.; Macedo, C.I.; Achkar, S.M.; Fahl, W.O.; Brandão, P.E.; Castilho, J.G. Evolution and Divergence of the Genetic Lineage Desmodus rotundus/Artibeus lituratus of Rabies Virus in São Paulo State. Arch. Virol. 2023, 168, 266. [Google Scholar] [CrossRef]
- de Almeida, M.F.; Favoretto, S.R.; Martorelli, L.F.A.; Trezza-Netto, J.; Campos, A.C.d.A.; Ozahata, C.H.; Sodré, M.M.; Kataoka, A.P.A.G.; Veiga Sacramento, D.R.; Durigon, E.L. CHARACTERIZATION OF RABIES VIRUS ISOLATED FROM A COLONY OF Eptesicus Furinalis BATS IN BRAZIL. Rev. Inst. Med. Trop. Sao Paulo 2011, 53, 31–37. [Google Scholar] [CrossRef]
- Oliveira, R.N.; Freire, C.C.; Iamarino, A.; Zanotto, P.M.; Pessoa, R.; Sanabani, S.S.; de Souza, S.P.; Castilho, J.G.; Batista, H.B.C.R.; Carnieli, P.; et al. Rabies Virus Diversification in Aerial and Terrestrial Mammals. Genet. Mol. Biol. 2020, 43, e20190370. [Google Scholar] [CrossRef]
- Guarino, H.; Castilho, J.G.; Souto, J.; Oliveira, R.d.N.; Carrieri, M.L.; Kotait, I. Antigenic and Genetic Characterization of Rabies Virus Isolates from Uruguay. Virus Res. 2013, 173, 415–420. [Google Scholar] [CrossRef]
- Cisterna, D.; Bonaventura, R.; Caillou, S.; Pozo, O.; Andreau, M.L.; Dalla Fontana, L.; Echegoyen, C.; De Mattos, C.; De Mattos, C.; Russo, S.; et al. Antigenic and Molecular Characterization of Rabies Virus in Argentina. Virus Res. 2005, 109, 139–147. [Google Scholar] [CrossRef]
- Castilho, J.G.; Carnieli, P.; Oliveira, R.N.; Fahl, W.O.; Cavalcante, R.; Santana, A.A.; Rosa, W.L.G.A.; Carrieri, M.L.; Kotait, I. A Comparative Study of Rabies Virus Isolates from Hematophagous Bats in Brazil. J. Wildl. Dis. 2010, 46, 1335–1339. [Google Scholar] [CrossRef]
- Smith, J.S.; Orciari, L.A.; Yager, P.A.; Seidel, H.D.; Warner, C.K. Epidemiologic and Historical Relationships among 87 Rabies Virus Isolates as Determined by Limited Sequence Analysis. J. Infect. Dis. 1992, 166, 296–307. [Google Scholar] [CrossRef]
- de Sousa, L.L.F.; de Souza, T.L.; Tibo, L.H.S.; Moura, F.B.P.; Junior, F.A.S.; de Oliveira-Filho, E.F.; Ludwig-Begall, L.F.; Cabral-Miranda, G.; Andreata-Santos, R.; Janini, L.M.R.; et al. Rabies Virus Variants from Bats Closely Related to Variants Found in Marmosets (Callithrix Jacchus), a Neglected Source of Human Rabies Infection in Brazil. J. Med. Virol. 2023, 95, e29046. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Okuda, H.; Nakamura, K.; Sato, G.; Itou, T.; Carvalho, A.A.B.; Silva, M.V.; Mota, C.S.; Ito, F.H.; Sakai, T. Genetic Analysis of Phosphoprotein and Matrix Protein of Rabies Viruses Isolated in Brazil. J. Vet. Med. Sci. 2007, 69, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Rohde, R.E.; Mayes, B.C.; Smith, J.S.; Neill, S.U. Bat Rabies, Texas, 1996–2000. Emerg. Infect. Dis. 2004, 10, 948–952. [Google Scholar] [CrossRef]
- Favi, M.; de Mattos, C.A.; Yung, V.; Chala, E.; López, L.R.; de Mattos, C.C. First Case of Human Rabies in Chile Caused by an Insectivorous Bat Virus Variant. Emerg. Infect. Dis. 2002, 8, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Nadin-Davis, S.A.; Loza-Rubio, E. The Molecular Epidemiology of Rabies Associated with Chiropteran Hosts in Mexico. Virus Res. 2006, 117, 215–226. [Google Scholar] [CrossRef]
- Castilho, J.G.; Carnieli, P.; Durymanova, E.A.; Fahl, W.d.O.; Oliveira, R.d.N.; Macedo, C.I.; da Rosa, E.S.T.; Mantilla, A.; Carrieri, M.L.; Kotait, I. Human Rabies Transmitted by Vampire Bats: Antigenic and Genetic Characterization of Rabies Virus Isolates from the Amazon Region (Brazil and Ecuador). Virus Res. 2010, 153, 100–105. [Google Scholar] [CrossRef]
- Delmas, O.; Holmes, E.C.; Talbi, C.; Larrous, F.; Dacheux, L.; Bouchier, C.; Bourhy, H. Genomic Diversity and Evolution of the Lyssaviruses. PLoS ONE 2008, 3, e2057. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Freuling, C.M.; Müller, T.; Pfaff, F.; Bodenhofer, U.; Höper, D.; Fischer, M.; Marston, D.A.; Fooks, A.R.; Mettenleiter, T.C.; et al. Defining Objective Clusters for Rabies Virus Sequences Using Affinity Propagation Clustering. PLoS Negl. Trop. Dis. 2018, 12, e0006182. [Google Scholar] [CrossRef] [PubMed]
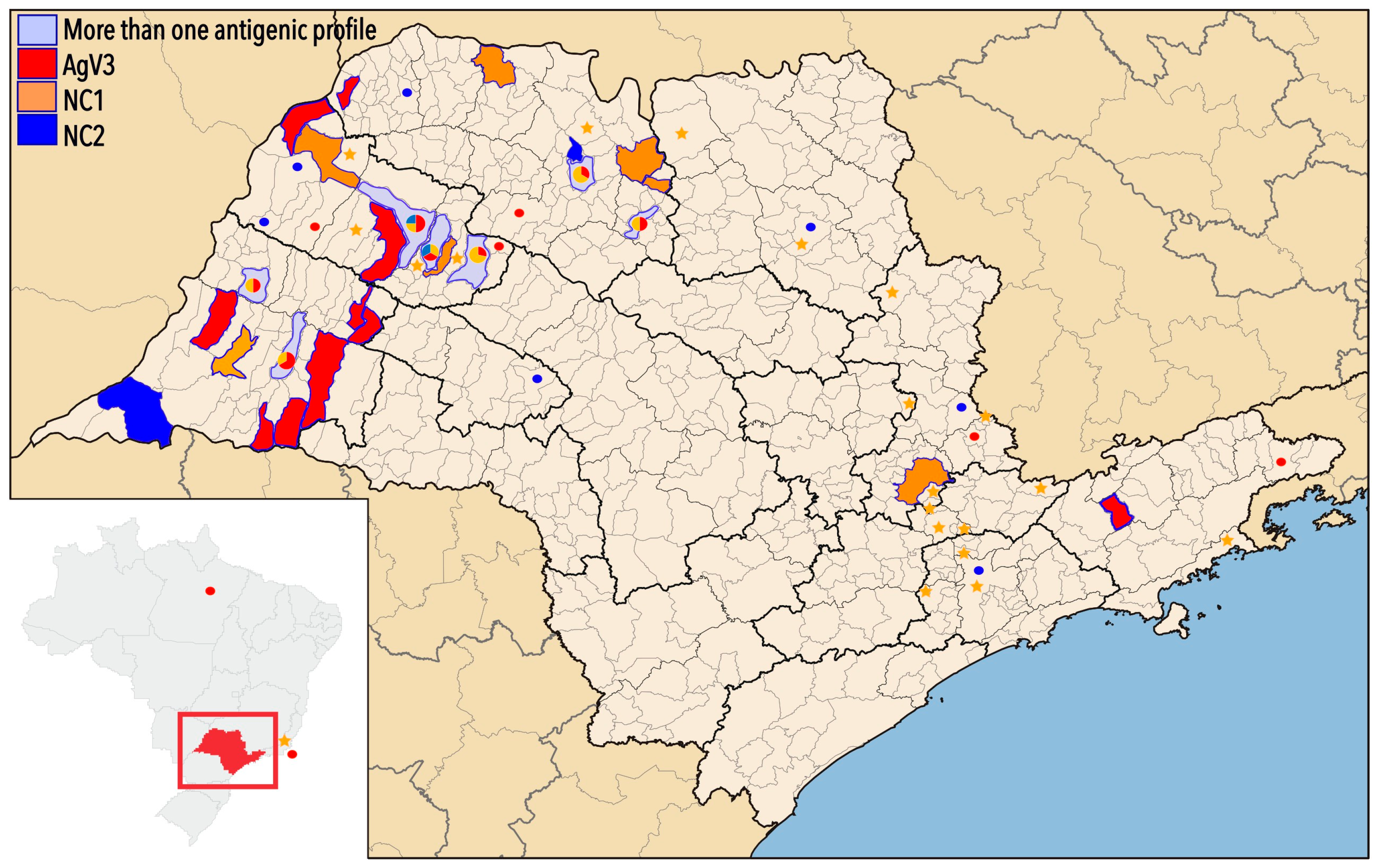
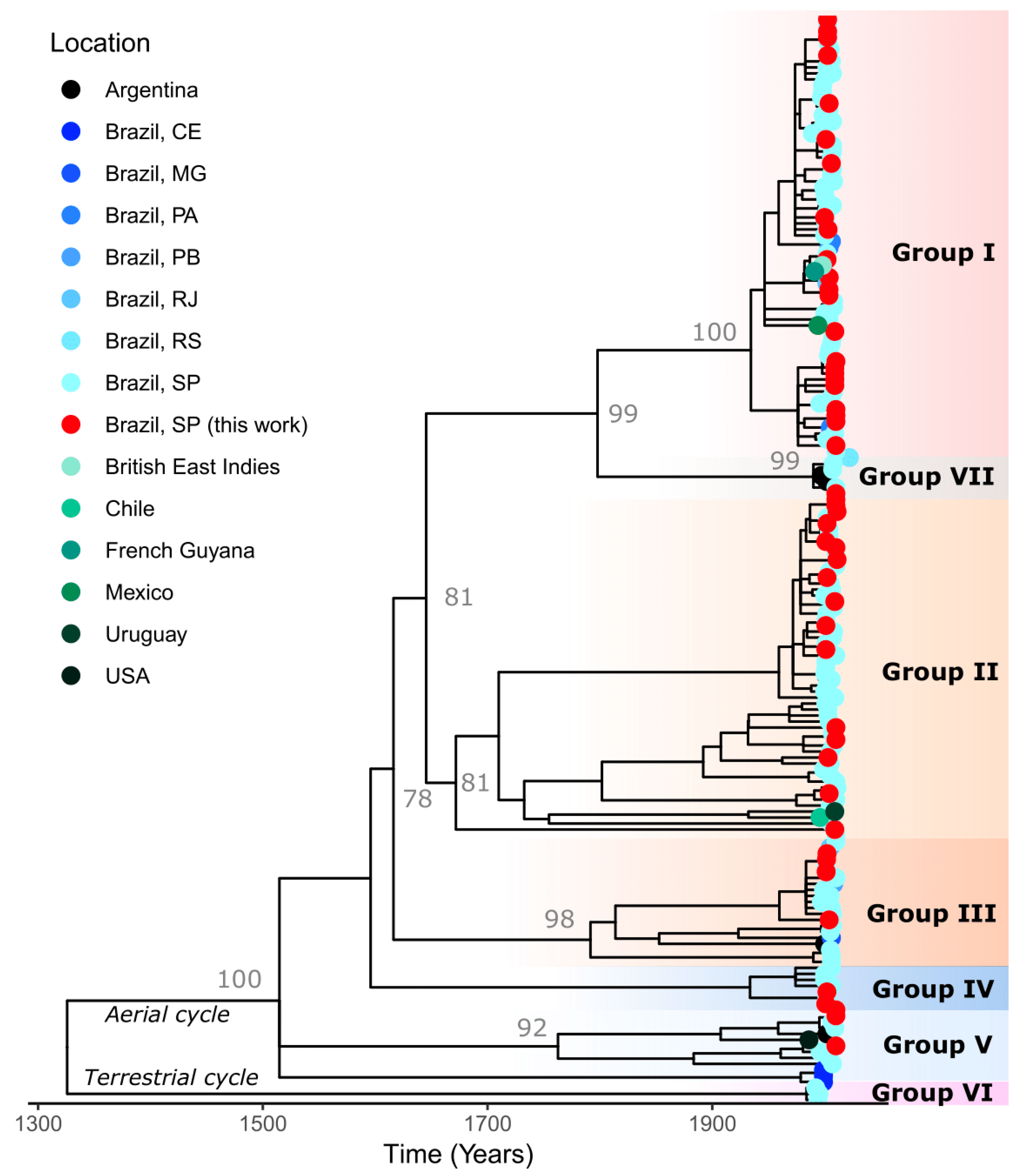

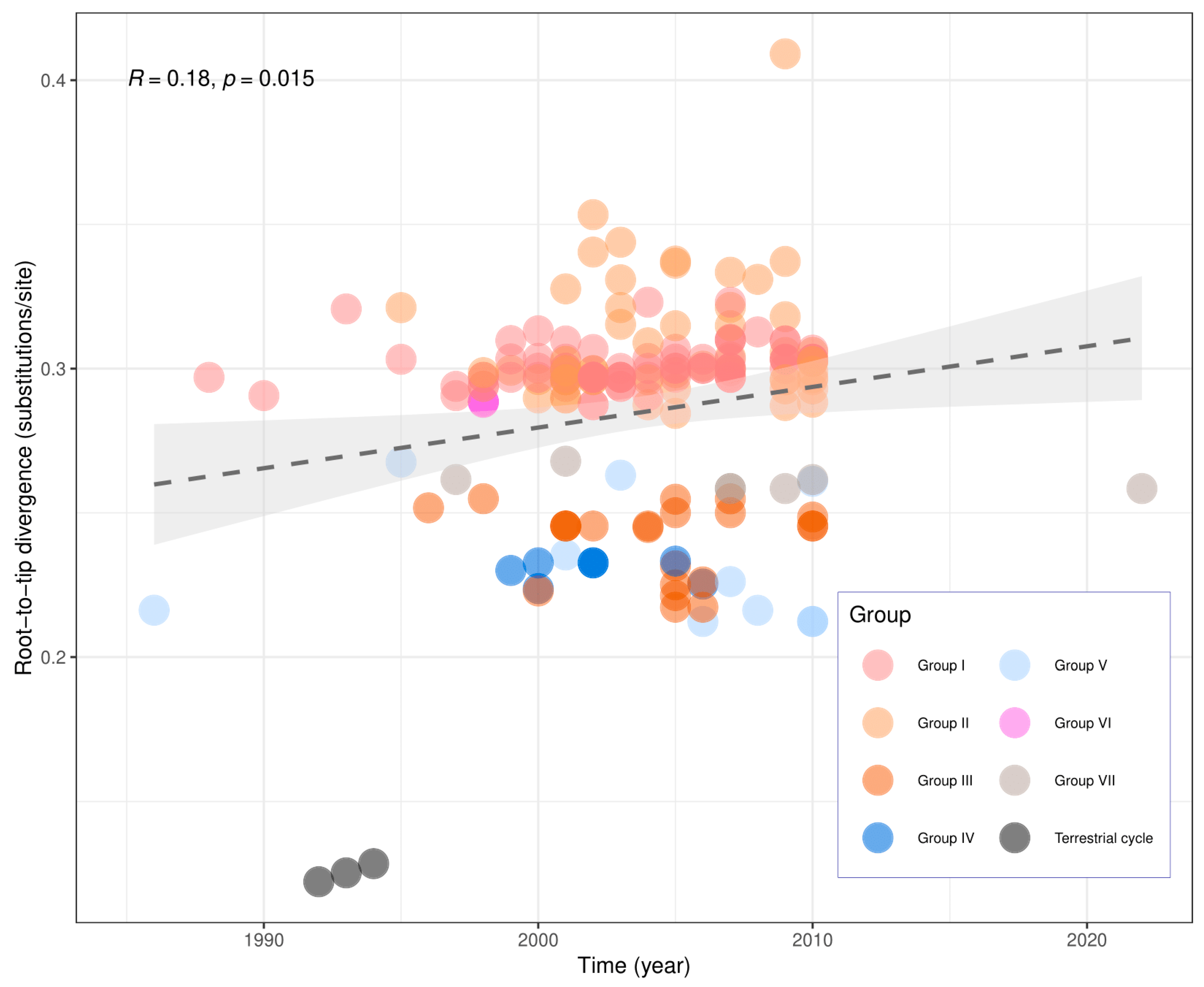
| GenBank Access | ID Sample/ Year | Species | Place of Origin | Antigenic Variant/Profile | Genetic Lineage |
|---|---|---|---|---|---|
| HQ666824 | IB 346/99 | Artibeus lituratus | São José do Rio Preto | V-3 | D. rotundus |
| HQ666825 | IB 777/00 | Cynomops abrasus | Ipiguá | NC2 | Insect. Bats |
| HQ666826 | IB 249/01 | Nyctinomops macrotis | São José do Rio Preto | NC1 | Insect. bats |
| HQ666827 | IB 250/01 | Nyctinomops laticaudatus | São José do Rio Preto | NC1 | Insect. Bats |
| HQ666828 | IB 636/01 | Neoeptesicus furinalis * | Olímpia | NC1 | Insect. Bats |
| HQ666829 | IB 808/01 | Artibeus lituratus | São José do Rio Preto | V-3 | D. rotundus |
| HQ666830 | IB 1019/01 | Neoeptesicus sp. * | Cardoso | NC1 | Insect. Bats |
| HQ666831 | IB 1070/01 | Neoeptesicus furinalis * | São José do Rio Preto | NC1 | Insect. Bats |
| HQ666832 | IB 62/02 | Neoeptesicus furinalis * | Catanduva | NC1 | Insect. Bats |
| HQ666833 | IB 109/02 | Molossus molossus | Ilha Solteira | NC2 | Insect. Bats |
| HQ666834 | IB 835/02 | Myotis nigricans | Cajobi | NC1 | Insect. Bats |
| HQ666835 | IB 992/02 | Artibeus lituratus | Dracena | V-3 | D. rotundus |
| HQ666836 | IB 1021/02 | Molossus fluminensis * | Presidente Venceslau | V-3 | D. rotundus |
| HQ666837 | IB 1141/02 | Neoeptesicus furinalis * | Santo Anastácio | NC1 | Insect. Bats |
| HQ666838 | IB 1256/02 | NH bat (not identified) | Martinópolis | V-3 | D. rotundus |
| HQ666839 | IB 1371B/02 | Artibeus lituratus | Presidente Venceslau | V-3 | D. rotundus |
| HQ666840 | IB 1535/02 | Artibeus lituratus | Taciba | V-3 | D. rotundus |
| HQ666841 | IB 1539/02 | Molossus molossus | Presidente Prudente | V-3 | D. rotundus |
| HQ666842 | IB 1782/02 | Lasiurus ega | Presidente Prudente | NC1 | D. rotundus |
| HQ666843 | IB 349/03 | Artibeus planirostris | Santa Fé do Sul | V-3 | D. rotundus |
| HQ666844 | IB 350/03 | NH bat (not identified) | Catanduva | V-3 | D. rotundus |
| HQ666845 | IB 791/03 | Artibeus lituratus | Presidente Prudente | V-3 | D. rotundus |
| - | IB 826/03 | Artibeus fimbriatus | São José do Rio Preto | V-3 | ND |
| HQ666846 | IB 168/04 | Myotis nigricans | Campinas | NC1 | Insect. Bats |
| HQ666847 | IB 184/04 | Nyctinomops laticaudatus | São José do Rio Preto | NC1 | Insect. Bats |
| HQ666848 | IB 550/04 | Artibeus lituratus | Caçapava | V-3 | D.rotundus |
| HQ666849 | LRU 329/05 | Eumops glaucinus | Araçatuba | V-3 | D.rotundus |
| - | LRU 397/05 | Artibeus lituratus | Araçatuba | V-3 | ND |
| HQ666850 | LRU 43/09 | Neoeptesicus diminutus * | Pereira Barreto | NC1 | Insect. Bats |
| HQ666851 | LRU 84/09 | Neoeptesicus diminutus * | Araçatuba | NC1 | Insect. Bats |
| HQ666852 | LRU 149/09 | Myotis nigricans | Coroados | NC1 | Insect. Bats |
| HQ666853 | LRU 181/09 | Artibeus lituratus | Penápolis | V-3 | D. rotundus |
| HQ666856 | LRU 325/09 | Artibeus lituratus | Birigui | V-3 | D. rotundus |
| HQ666857 | LRU 374/09 | Artibeus lituratus | Guararapes | V-3 | D. rotundus |
| HQ666858 | LRU 389/09 | Molossus fluminensis * | Guararapes | V-3 | D. rotundus |
| HQ666859 | LRU 433/09 | Myotis nigricans | Penápolis | NC1 | Insect. Bats |
| HQ666860 | LRU 589/09 | Neoeptesicus furinalis * | Penápolis | NC1 | Insect. Bats |
| HQ666854 | LRPP 199/09 | Neoeptesicus furinalis * | Dracena | NC1 | Insect. Bats |
| HQ666855 | LRPP 224/09 | Neoeptesicus furinalis * | Parapuã | V-3 | D. rotundus |
| HQ666861 | LRPP 672/09 | Bovine/Cattle | Narandiba | V-3 | D. rotundus |
| HQ666862 | LRU 17/10 | Molossus fluminensis * | Penápolis | NC1 | Insect. Bats |
| KU299782 | LRPP 28/10 | Horse | Taciba | V-3 | D. rotundus |
| HQ666864 | LRPP 43/10 | Neoeptesicus diminutus * | Osvaldo Cruz | V-3 | D. rotundus |
| HQ666865 | LRU 60/10 | Neoeptesicus furinalis * | Birigui | NC1 | Insect. Bats |
| HQ666866 | LRU 76/10 | Neoeptesicus furinalis * | Penápolis | NC1 | Insect. Bats |
| HQ666867 | LRU 169/10 | Molossus molossus | Araçatuba | NC2 | Insect. Bats |
| HQ666868 | LRU 171/10 | Cat | Araçatuba | V-3 | D. rotundus |
| HQ666869 | LRU 177/10 | Molossus fluminensis * | Birigui | NC2 | Insect. Bats |
| HQ666870 | LRPP 198/10 | Lasiurus blossevillii | Teodoro Sampaio | NC2 | Insect. Bats |
| HQ666871 | LRU 299/10 | Neoeptesicus furinalis * | Penápolis | NC1 | Insect. Bats |
| HQ666872 | LRU 300/10 | Molossus fluminensis * | Penápolis | V-3 | D. rotundus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Queiroz, L.H.; Campos, A.C.A.; Lopes, M.C.; Cunha, E.M.S.; Albas, A.; de Carvalho, C.; Pedro, W.A.; Silva, E.C.; Lot, M.S.; Inácio, S.V.; et al. Evolutionary Diversity of Bat Rabies Virus in São Paulo State, Brazil. Viruses 2025, 17, 1063. https://doi.org/10.3390/v17081063
Queiroz LH, Campos ACA, Lopes MC, Cunha EMS, Albas A, de Carvalho C, Pedro WA, Silva EC, Lot MS, Inácio SV, et al. Evolutionary Diversity of Bat Rabies Virus in São Paulo State, Brazil. Viruses. 2025; 17(8):1063. https://doi.org/10.3390/v17081063
Chicago/Turabian StyleQueiroz, Luzia H., Angélica C. A. Campos, Marissol C. Lopes, Elenice M. S. Cunha, Avelino Albas, Cristiano de Carvalho, Wagner A. Pedro, Eduardo C. Silva, Monique S. Lot, Sandra V. Inácio, and et al. 2025. "Evolutionary Diversity of Bat Rabies Virus in São Paulo State, Brazil" Viruses 17, no. 8: 1063. https://doi.org/10.3390/v17081063
APA StyleQueiroz, L. H., Campos, A. C. A., Lopes, M. C., Cunha, E. M. S., Albas, A., de Carvalho, C., Pedro, W. A., Silva, E. C., Lot, M. S., Inácio, S. V., Araújo, D. B., Cunha, M. P., Durigon, E. L., Góes, L. G. B., & Favoretto, S. R. (2025). Evolutionary Diversity of Bat Rabies Virus in São Paulo State, Brazil. Viruses, 17(8), 1063. https://doi.org/10.3390/v17081063






