Complete Sequence Analysis of Grapevine Leafroll-Associated Virus 4 and Interactions Between the Encoded Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA-Seq
2.3. Amplification of GLRaV-4 and Sequencing Analysis
2.4. Construction of Vectors Containing Candidate Genes
2.5. Y2H Assay and BIFC Assay
3. Results
3.1. HTS Data Analysis and Validation of High-Throughput Sequencing Results
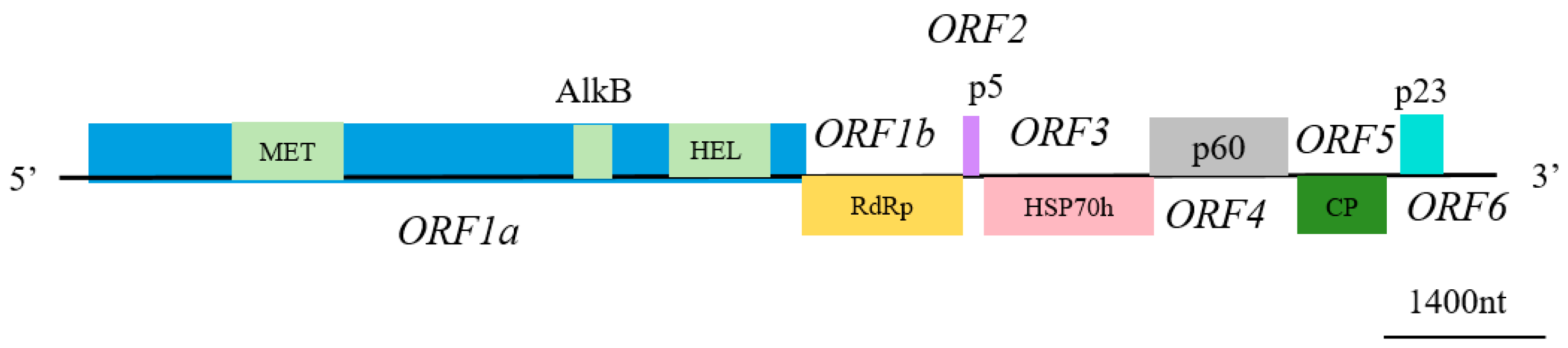
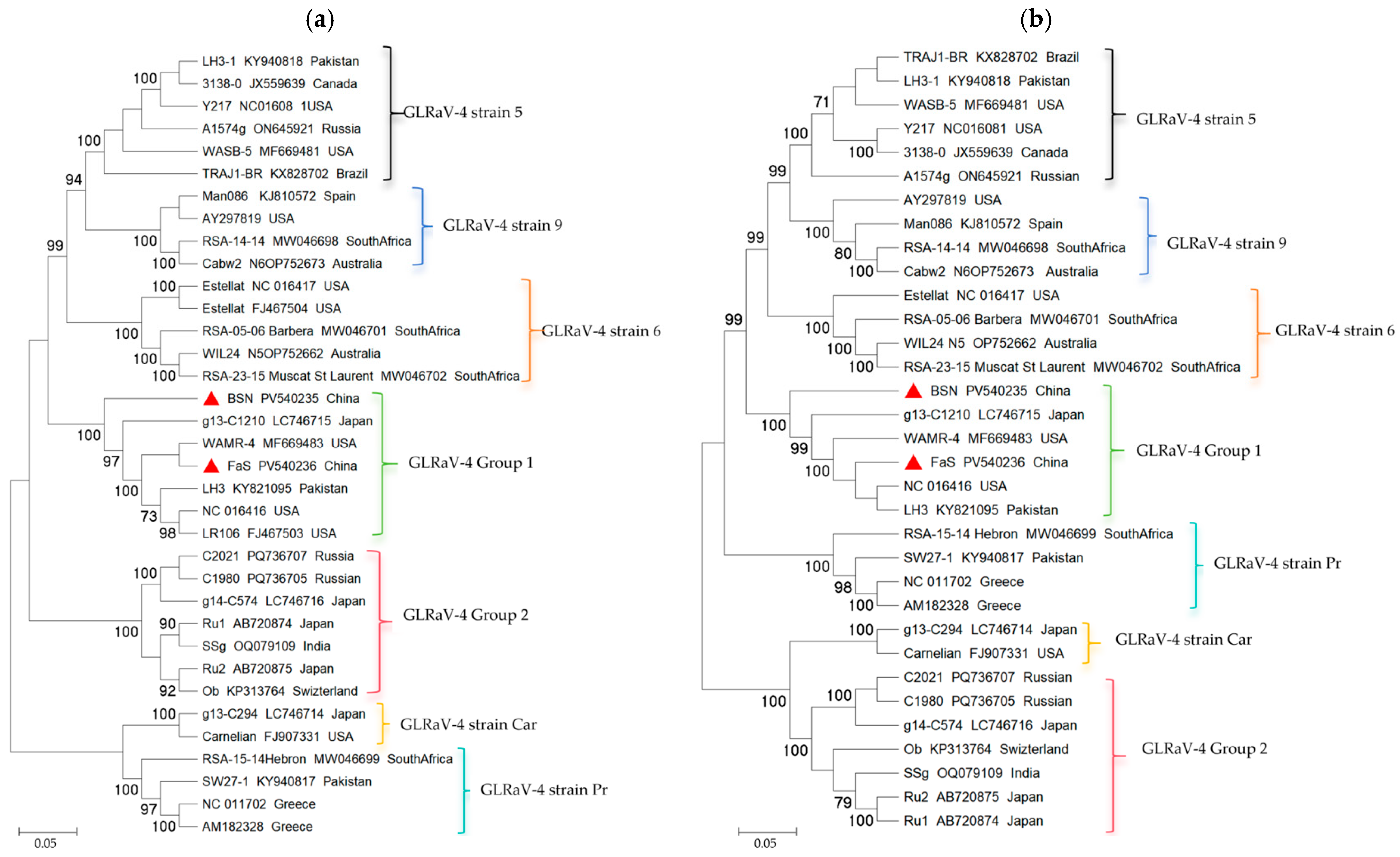
3.2. Genome Sequence and Phylogenetic Analysis
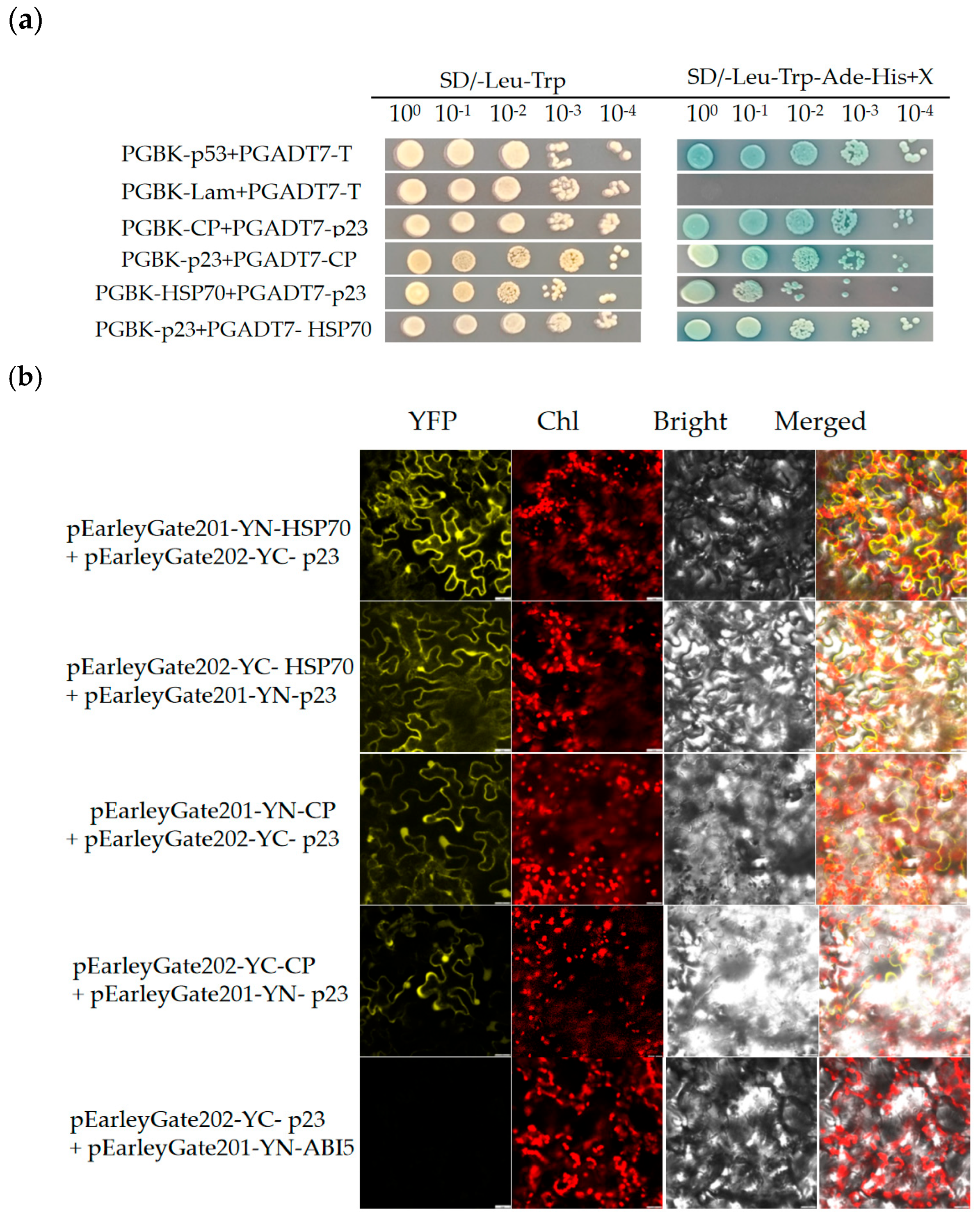
3.3. Interactions Between Proteins Encoded by GLRaV-4
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fajardo, T.V.M.; Silva, F.N.; Eiras, M.; Nickel, O. High-Throughput Sequencing Applied for the Identification of Viruses Infecting Grapevines in Brazil and Genetic Variability Analysis. Trop. Plant Pathol. 2017, 42, 250–260. [Google Scholar] [CrossRef]
- Glasa, M.; Predajňa, L.; Sihelská, N.; Šoltys, K.; Ruiz-García, A.-B. Analysis of Virome by High-Throughput Sequencing Revealed Multiple Infection and Intra-Virus Diversity in a Single Grapevine Plant. Acta Hortic. Regiotect. 2020, 23, 35–39. [Google Scholar] [CrossRef]
- Reynolds, A.G. The Grapevine, Viticulture, and Winemaking: A Brief Introduction. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–29. [Google Scholar]
- Fuchs, M.; Rwahnih, M.A.; Blouin, A.G.; Burger, J.; Chooi, K.M.; Constable, F.; Ertunc, F.; Fiore, N.; Habili, N.; Hily, J.-M.; et al. A List of Eclectic Viruses, Virus-like Diseases and Viroids of Grapevines That Should Not Be Considered for Regulatory Oversight: A Global Plea from Virologists. J. Plant Pathol. 2025, 107, 847–858. [Google Scholar] [CrossRef]
- Beuve, M.; Hily, J.-M.; Alliaume, A.; Reinbold, C.; Le Maguet, J.; Candresse, T.; Herrbach, E.; Lemaire, O. A Complex Virome Unveiled by Deep Sequencing Analysis of RNAs from a French Pinot Noir Grapevine Exhibiting Strong Leafroll Symptoms. Arch. Virol. 2018, 163, 2937–2946. [Google Scholar] [CrossRef]
- Jarugula, S.; Alabi, O.J.; Martin, R.R.; Naidu, R.A. Genetic Variability of Natural Populations of Grapevine Leafroll-Associated Virus 2 in Pacific Northwest Vineyards. Phytopathology 2010, 100, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Martinson, T.E.; Loeb, G.M.; Hoch, H.C. Survey for the Three Major Leafroll Disease-Associated Viruses in Finger Lakes Vineyards in New York. Plant Dis. 2009, 93, 395–401. [Google Scholar] [CrossRef]
- Martin, R.R.; Eastwell, K.C.; Wagner, A.; Lamprecht, S.; Tzanetakis, I.E. Survey for Viruses of Grapevine in Oregon and Washington. Plant Dis. 2005, 89, 763–766. [Google Scholar] [CrossRef]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine Leafroll-Associated Virus 3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef]
- Wolpert, J.A.; Vilas, E.P. Effect of Mild Leafroll Disease on Growth, Yield, and Fruit Maturity Indices of Riesling and Zinfandel. Am. J. Enol. Vitic. 1992, 43, 367–369. [Google Scholar] [CrossRef]
- Cabaleiro, C.; Segura, A.; García-Berrios, J.J. Effects of Grapevine Leafroll-Associated Virus 3 on the Physiology and Must of Vitis Vinifera, L. Cv. Albariño Following Contamination in the Field. Am. J. Enol. Vitic. 1999, 50, 40–44. [Google Scholar] [CrossRef]
- Kovacs, L.G.; Hanami, H.; Fortenberry, M.; Kaps, M.L. Latent Infection by Leafroll Agent GLRaV-3 Is Linked to Lower Fruit Quality in French-American Hybrid Grapevines Vidal Blanc and St. Vincent. Am. J. Enol. Vitic. 2001, 52, 254–259. [Google Scholar] [CrossRef]
- Rasool, S.; Naz, S. Detection and Elimination of Grapevine Leafroll-Associated Viruses (GLRaVs) from Vitis Vinifera Cultivars of Pakistan by Meristem Tip Culture. Eur. J. Plant Pathol. 2024, 168, 697–707. [Google Scholar] [CrossRef]
- Fan, X.; Dong, Y.; Zhang, Z.P.; Ren, F.; Hu, G.; Zhou, J. Detection and Sequence Analysis of Grapevine Leafroll-Associated Virus 2 Isolates from China. J. Phytopathol. 2015, 163, 978–986. [Google Scholar] [CrossRef]
- Martelli, G.P.; Aboughanem, N.; Agranovsky, A.; Al Rwahnih, M.; Dolja, V.V.; Dovas, C.; Fuchs, M.; Gugerli, P.; Hu, J.S.; Jelkmann, W.; et al. Taxonomic Revision of the Family Closteroviridae with Special Reference to the Grapevine Leafroll-Associated Members of the Genus Ampleovirus and the Putative Species Unassigned to the Family. J. Plant Pathol. 2012, 94, 7–19. [Google Scholar]
- Maliogka, V.I.; Dovas, C.I.; Katis, N.I. Evolutionary Relationships of Virus Species Belonging to a Distinct Lineage within the Ampelovirus Genus. Virus Res. 2008, 135, 125–135. [Google Scholar] [CrossRef]
- Maliogka, V.I.; Dovas, C.I.; Lotos, L.; Efthimiou, K.; Katis, N.I. Complete Genome Analysis and Immunodetection of a Member of a Novel Virus Species Belonging to the Genus Ampelovirus. Arch Virol. 2009, 154, 209–218. [Google Scholar] [CrossRef]
- Adiputra, J.; Jarugula, S.; Naidu, R.A. Intra-Species Recombination among Strains of the Ampelovirus Grapevine Leafroll-Associated Virus 4. Virol. J. 2019, 16, 13. [Google Scholar] [CrossRef]
- Aboughanem-Sabanadzovic, N.; Maliogka, V.; Sabanadzovic, S. Grapevineleafroll-Associated Virus 4. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 197–220. [Google Scholar]
- Gomez Talquenca, S.; Lanza Volpe, M.; Setien, N.; Gracia, O.; Grau, O. Las Relaciones Serológicas y La Identidad Molecular de Variantes de Grapevine Leafroll-Associated Virus 4 Reflejan El Comportamiento Evolutivo Del Gen de Su Proteína de Cubierta. Rev. Fac. Cienc. Agrar. Univ. Nac. Cuyo 2023, 55, 104–114. [Google Scholar]
- Abou Ghanem-Sabanadzovic, N.; Sabanadzovic, S.; Uyemoto, J.K.; Golino, D.; Rowhani, A. A Putative New Ampelovirus Associated with Grapevine Leafroll Disease. Arch. Virol. 2010, 155, 1871–1876. [Google Scholar] [CrossRef]
- Reynard, J.-S.; Schneeberger, P.H.H.; Frey, J.E.; Schaerer, S. Biological, Serological, and Molecular Characterization of a Highly Divergent Strain of Grapevine Leafroll-Associated Virus 4 Causing Grapevine Leafroll Disease. Phytopathology 2015, 105, 1262–1269. [Google Scholar] [CrossRef]
- Alkowni, R.; Rowhani, A.; Daubert, S.; Golino, D. Partial Characterization of a New Ampelovirus Associated with Grapevine Leafroll Disease. J. Plant Pathol. 2004, 86, 123–133. [Google Scholar]
- Yuan, Q.; Zhang, Y.; Ren, F.; Hu, G.; Fan, X.; Dong, Y. Complete Genome Sequencing and Infectious cDNA Clone Construction of a Chinese Isolate of Grapevine Pinot Gris Virus (GPGV). J. Plant Pathol. 2024, 107, 327–334. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Z.; Li, C.; Ren, F.; Hu, G.; Zhang, B.; Dong, Y. High-Throughput Sequencing Indicates a Novel Marafivirus in Grapevine Showing Vein-Clearing Symptoms. Plants 2021, 10, 1487. [Google Scholar] [CrossRef]
- Zheng, Y.; Gao, S.; Padmanabhan, C.; Li, R.; Galvez, M.; Gutierrez, D.; Fuentes, S.; Ling, K.-S.; Kreuze, J.; Fei, Z. VirusDetect: An Automated Pipeline for Efficient Virus Discovery Using Deep Sequencing of Small RNAs. Virology 2017, 500, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fan, X.; Dong, Y.; Zhang, Z.P.; Ren, F.; Hu, G. Detection and Genetic Variation Analysis of Grapevine Fanleaf Virus (GFLV) Isolates in China. Arch. Virol. 2015, 160, 2661–2667. [Google Scholar] [CrossRef]
- Holland, L.A.; Kaelin, E.A.; Maqsood, R.; Estifanos, B.; Wu, L.I.; Varsani, A.; Halden, R.U.; Hogue, B.G.; Scotch, M.; Lim, E.S. An 81-Nucleotide Deletion in SARS-CoV-2 ORF7a Identified from Sentinel Surveillance in Arizona (January to March 2020). J. Virol. 2020, 94, e00711-20. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Delport, W.; Poon, A.F.Y.; Frost, S.D.W.; Kosakovsky Pond, S.L. Datamonkey 2010: A Suite of Phylogenetic Analysis Tools for Evolutionary Biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef]
- Brückner, A.; Polge, C.; Lentze, N.; Auerbach, D.; Schlattner, U. Yeast Two-Hybrid, a Powerful Tool for Systems Biology. Int. J. Mol. Sci. 2009, 10, 2763–2788. [Google Scholar] [CrossRef]
- Fields, S.; Song, O. A Novel Genetic System to Detect Protein–Protein Interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Kerppola, T.K. Bimolecular Fluorescence Complementation (BiFC) Analysis as a Probe of Protein Interactions in Living Cells. Annu. Rev. Biophys. 2008, 37, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Rowhani, A. Deep Sequencing Analysis of RNAs from a Grapevine Showing Syrah Decline Symptoms Reveals a Multiple Virus Infection That Includes a Novel Virus. Virology 2009, 387, 395–401. [Google Scholar] [CrossRef]
- Donda, B.P.; Jarugula, S.; Naidu, R.A. An Analysis of the Complete Genome Sequence and Subgenomic RNAs Reveals Unique Features of the Ampelovirus, Grapevine Leafroll-Associated Virus 1. Phytopathology 2017, 107, 1069–1079. [Google Scholar] [CrossRef]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and Functional Genomics of Closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Suppressors of RNA Silencing Encoded by Plant Viruses and Their Role in Viral Infections. FEBS Lett. 2005, 579, 5958–5964. [Google Scholar] [CrossRef]
- Mostert, I.; Bester, R.; Burger, J.T.; Maree, H.J. Identification of Interactions between Proteins Encoded by Grapevine Leafroll-Associated Virus 3. Viruses 2023, 15, 208. [Google Scholar] [CrossRef]
- Mayer, M.P. Recruitment of Hsp70 Chaperones: A Crucial Part of Viral Survival Strategies. Rev. Physiol. Biochem. Pharmacol. 2005, 153, 1–46. [Google Scholar] [PubMed]
- Prokhnevsky, A.I.; Peremyslov, V.V.; Napuli, A.J.; Dolja, V.V. Interaction between Long-Distance Transport Factor and Hsp70-Related Movement Protein of Beet Yellows Virus. J. Virol. 2002, 76, 11003–11011. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C.; Kasschau, K.D.; Prokhnevsky, A.I.; Gopinath, K.; Pogue, G.P.; Carrington, J.C.; Dolja, V.V. Suppressor of RNA Silencing Encoded by Beet Yellows Virus. Virology 2003, 306, 203–209. [Google Scholar] [CrossRef]
- Tian, T.; Rubio, L.; Yeh, H.-H.; Crawford, B.; Falk, B.W. Lettuce Infectious Yellows Virus: In Vitro Acquisition Analysis Using Partially Purified Virions and the Whitefly Bemisia Tabaci. J. Gen. Virol. 1999, 80, 1111–1117. [Google Scholar] [CrossRef]
- Agranovsky, A.A.; Lesemann, D.E.; Maiss, E.; Hull, R.; Atabekov, J.G. “Rattlesnake” Structure of a Filamentous Plant RNA Virus Built of Two Capsid Proteins. Proc. Natl. Acad. Sci. USA 1995, 92, 2470–2473. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.-X.; Falk, B.W.; Dawson, W.O.; Ding, S.-W. Three Distinct Suppressors of RNA Silencing Encoded by a 20-Kb Viral RNA Genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-W.; Liu, Q.; Zeng, Q.; Huang, W.-T.; Wang, Q.; Cheng, Y.-Q. p24G1 Encoded by Grapevine Leafroll-Associated Virus 1 Suppresses RNA Silencing and Elicits Hypersensitive Response-Like Necrosis in Nicotiana Species. Viruses 2020, 12, 1111. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, J.; Feng, M.; Wang, X.; Luo, C.; Wang, Q.; Cheng, Y. Characterization of Silencing Suppressor P24 of Grapevine Leafroll-Associated Virus 2. Mol. Plant Pathol. 2018, 19, 355–368. [Google Scholar] [CrossRef]
| Phylogroup | Between Groups | Within Groups | |||||
|---|---|---|---|---|---|---|---|
| Group 1 | 0.048 ± 0.004 | ||||||
| Group 2 | 0.282 ± 0.011 | 0.115 ± 0.007 | |||||
| Strain 5 | 0.262 ± 0.010 | 0.294 ± 0.014 | 0.044 ± 0.004 | ||||
| Strain 6 | 0.270 ± 0.010 | 0.289 ± 0.014 | 0.223 ± 0.013 | 0.078 ± 0.007 | |||
| Strain 9 | 0.271 ± 0.010 | 0.304 ± 0.015 | 0.184 ± 0.013 | 0.216 ± 0.013 | 0.027 ± 0.004 | ||
| Strain Car | 0.291 ± 0.015 | 0.310 ± 0.014 | 0.286 ± 0.014 | 0.284 ± 0.014 | 0.295 ± 0.014 | 0.224 ± 0.015 | |
| Strain Pr | 0.284 ± 0.015 | 0.306 ± 0.015 | 0.289 ± 0.015 | 0.291 ± 0.015 | 0.300 ± 0.016 | 0.301 ± 0.015 | 0.063 ± 0.006 |
| Phylogroup | Between Groups | Within Groups | |||||
|---|---|---|---|---|---|---|---|
| Group 1 | 0.062 ± 0.003 | ||||||
| Group 2 | 0.357 ± 0.014 | 0.118 ± 0.005 | |||||
| Strain 5 | 0.265 ± 0.015 | 0.358 ± 0.011 | 0.056 ± 0.003 | ||||
| Strain 6 | 0.258 ± 0.014 | 0.364 ± 0.010 | 0.240 ± 0.010 | 0.093 ± 0.005 | |||
| Strain 9 | 0.259 ± 0.015 | 0.356 ± 0.011 | 0.209 ± 0.010 | 0.240 ± 0.010 | 0.039 ± 0.003 | ||
| Strain Car | 0.383 ± 0.010 | 0.364 ± 0.010 | 0.378 ± 0.011 | 0.372 ± 0.010 | 0.378 ± 0.010 | 0.260 ± 0.011 | |
| Strain Pr | 0.311 ± 0.011 | 0.371 ± 0.011 | 0.311 ± 0.011 | 0.310 ± 0.010 | 0.308 ± 0.011 | 0.384 ± 0.010 | 0.079 ± 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, T.; Hao, Y.; Gao, J.; Qiao, S.; Hu, G.; Ren, F.; Fan, X.; Dong, Y. Complete Sequence Analysis of Grapevine Leafroll-Associated Virus 4 and Interactions Between the Encoded Proteins. Viruses 2025, 17, 952. https://doi.org/10.3390/v17070952
Du T, Hao Y, Gao J, Qiao S, Hu G, Ren F, Fan X, Dong Y. Complete Sequence Analysis of Grapevine Leafroll-Associated Virus 4 and Interactions Between the Encoded Proteins. Viruses. 2025; 17(7):952. https://doi.org/10.3390/v17070952
Chicago/Turabian StyleDu, Tingting, Yuxin Hao, Jie Gao, Shane Qiao, Guojun Hu, Fang Ren, Xudong Fan, and Yafeng Dong. 2025. "Complete Sequence Analysis of Grapevine Leafroll-Associated Virus 4 and Interactions Between the Encoded Proteins" Viruses 17, no. 7: 952. https://doi.org/10.3390/v17070952
APA StyleDu, T., Hao, Y., Gao, J., Qiao, S., Hu, G., Ren, F., Fan, X., & Dong, Y. (2025). Complete Sequence Analysis of Grapevine Leafroll-Associated Virus 4 and Interactions Between the Encoded Proteins. Viruses, 17(7), 952. https://doi.org/10.3390/v17070952







