The Role of Chemical Modifications in the Genome of Negative-Sense RNA Viruses on the Innate Immune Response
Abstract
1. Introduction
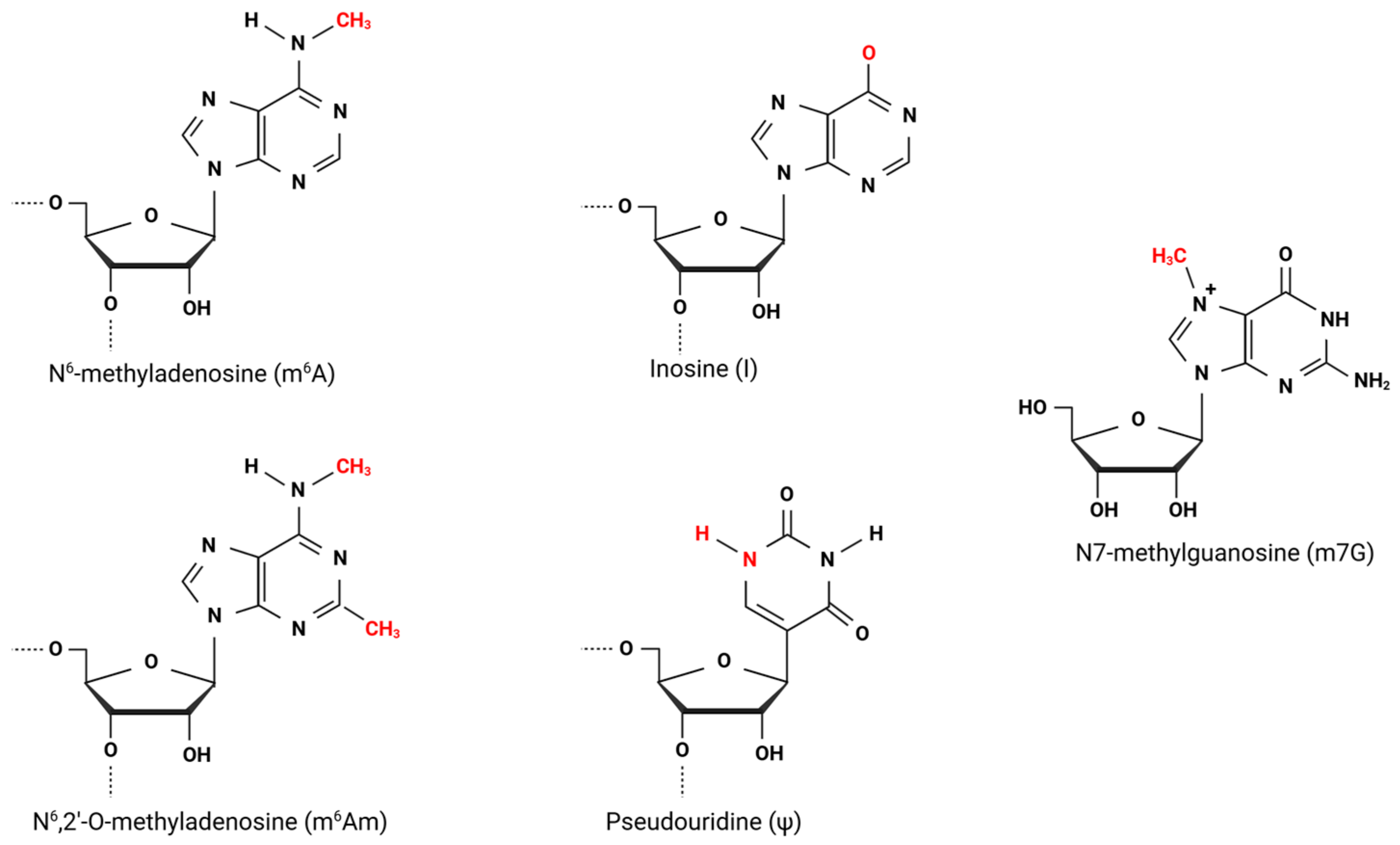
2. N-6 Methyladenosine
3. N6,2′-O-Dimethyladenosine
4. Inosine
5. Pseudouridine
6. Viral Methyltransferases, Direct Addition of N7-Methylguanosine (m7G), and 2′-O-Methylated Nucleotides
7. Other RNA Modifications
8. The Impact of Chemical Modifications on the Genome of Negative-Sense RNA Viruses Beyond the Immune Response
9. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kleiner, V.A.; Fearns, R. How Does the Polymerase of Non-Segmented Negative Strand RNA Viruses Commit to Transcription or Genome Replication? J. Virol. 2024, 98, e00332-24. [Google Scholar] [CrossRef]
- An, W.; Lakhina, S.; Leong, J.; Rawat, K.; Husain, M. Host Innate Antiviral Response to Influenza A Virus Infection: From Viral Sensing to Antagonism and Escape. Pathogens 2024, 13, 561. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Basler, C.F.; Amarasinghe, G.K.; Leung, D.W. Molecular Mechanisms of Innate Immune Inhibition by Non-Segmented Negative-Sense RNA Viruses. J. Mol. Biol. 2016, 428, 3467–3482. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen Recognition by the Innate Immune System. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Recognition of Viruses by Innate Immunity. Immunol. Rev. 2007, 220, 214–224. [Google Scholar] [CrossRef]
- Ma, D.Y.; Suthar, M.S. Mechanisms of Innate Immune Evasion in Re-Emerging RNA Viruses. Curr. Opin. Virol. 2015, 12, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Panne, D. The Enhanceosome. Curr. Opin. Struct. Biol. 2008, 18, 236–242. [Google Scholar] [CrossRef]
- Lazear, H.M.; Lancaster, A.; Wilkins, C.; Suthar, M.S.; Huang, A.; Vick, S.C.; Clepper, L.; Thackray, L.; Brassil, M.M.; Virgin, H.W.; et al. IRF-3, IRF-5, and IRF-7 Coordinately Regulate the Type I IFN Response in Myeloid Dendritic Cells Downstream of MAVS Signaling. PLoS Pathog. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Yoneyama, M.; Fujita, T. RIG-I Family RNA Helicases: Cytoplasmic Sensor for Antiviral Innate Immunity. Cytokine Growth Factor. Rev. 2007, 18, 545–551. [Google Scholar] [CrossRef]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral Innate Immunity Pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Pétrilli, V.; Dostert, C.; Muruve, D.A.; Tschopp, J. The Inflammasome: A Danger Sensing Complex Triggering Innate Immunity. Curr. Opin. Immunol. 2007, 19, 615–622. [Google Scholar] [CrossRef]
- Kanneganti, T.D.; Lamkanfi, M.; Núñez, G. Intracellular NOD-like Receptors in Host Defense and Disease. Immunity 2007, 27, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Randall, R.E.; Goodbourn, S. Interferons and Viruses: An Interplay between Induction, Signalling, Antiviral Responses and Virus Countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and CDNA Cloning of the AdoMet-Binding Subunit of the Human MRNA (N6-Adenosine)-Methyltransferase. RNA 1997, 3, 1233. [Google Scholar]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of M6A Writers Reveals Two Distinct Classes of MRNA Methylation at Internal and 5′ Sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 Is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The Dynamic Epitranscriptome: N6-Methyladenosine and Gene Expression Control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Brocard, M.; Ruggieri, A.; Locker, N. M6A RNA Methylation, a New Hallmark in Virus-Host Interactions. J. Gen. Virol. 2017, 98, 2207–2214. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Courtney, D.G.; Tsai, K.; Cullen, B.R. Viral Epitranscriptomics. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N6-Methyladenosine Modification Enables Viral RNA to Escape Recognition by RNA Sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Zhang, Y.; Wang, H.; Kairis, E.L.; Lu, M.; Ahmad, S.; Attia, Z.; Harder, O.; Zhang, Z.; Wei, J.; et al. Viral RNA N6-Methyladenosine Modification Modulates Both Innate and Adaptive Immune Responses of Human Respiratory Syncytial Virus. PLoS Pathog. 2021, 17, e1010142. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Marchena, L.; Martínez-Pérez, M.; Úbeda, J.R.; Pallas, V.; Aparicio, F. Impact of the Potential M6A Modification Sites at the 3′UTR of Alfalfa Mosaic Virus RNA3 in the Viral Infection. Viruses 2022, 14, 1718. [Google Scholar] [CrossRef]
- Lu, M.; Xue, M.; Wang, H.-T.; Kairis, E.L.; Ahmad, S.; Wei, J.; Zhang, Z.; Liu, Q.; Zhang, Y.; Gao, Y.; et al. Nonsegmented Negative-Sense RNA Viruses Utilize N6-Methyladenosine (M6A) as a Common Strategy To Evade Host Innate Immunity. J. Virol. 2021, 95, e01939-20. [Google Scholar] [CrossRef] [PubMed]
- Khan, O.; Tanuj, G.N.; Choravada, D.R.; Rajak, K.K.; Chandra Sekar, S.; Lingaraju, M.C.; Dhara, S.K.; Gupta, P.K.; Mishra, B.P.; Dutt, T.; et al. N6-Methyladenosine RNA Modification in Host Cells Regulates Peste Des Petits Ruminants Virus Replication. Microbiol. Spectr. 2023, 11, e02666-22. [Google Scholar] [CrossRef]
- Bayoumi, M.; Munir, M. Evolutionary Conservation of the DRACH Signatures of Potential N6-Methyladenosine (M6A) Sites among Influenza A Viruses. Sci. Rep. 2021, 11, 4548. [Google Scholar] [CrossRef]
- Hernández-Díaz, T.; Valiente-Echeverría, F.; Soto-Rifo, R. RNA Helicase DDX3: A Double-Edged Sword for Viral Replication and Immune Signaling. Microorganisms 2021, 9, 1206. [Google Scholar] [CrossRef]
- Courtney, D.G.; Kennedy, E.M.; Dumm, R.E.; Bogerd, H.P.; Tsai, K.; Heaton, N.S.; Cullen, B.R. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe 2017, 22, 377–386.e5. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Zhao, B.S.; Zhang, Z.; Lu, M.; Harder, O.; Chen, P.; Lu, Z.; Li, A.; Ma, Y.; Xu, Y.; et al. Viral N6-Methyladenosine Upregulates Replication and Pathogenesis of Human Respiratory Syncytial Virus. Nat. Commun. 2019, 10, 4595. [Google Scholar] [CrossRef]
- Durbin, A.F.; Wang, C.; Marcotrigiano, J.; Gehrke, L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7, e00833-16. [Google Scholar] [CrossRef]
- Dang, W.; Xie, Y.; Cao, P.; Xin, S.; Wang, J.; Li, S.; Li, Y.; Lu, J. N6-Methyladenosine and Viral Infection. Front. Microbiol. 2019, 10, 417. [Google Scholar] [CrossRef]
- Chen, J.; Wei, X.; Wang, X.; Liu, T.; Zhao, Y.; Chen, L.; Luo, Y.; Du, H.; Li, Y.; Liu, T.; et al. TBK1-METTL3 Axis Facilitates Antiviral Immunity. Cell Rep. 2022, 38, 110373. [Google Scholar] [CrossRef] [PubMed]
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza Viral MRNA Contains Internal N6-Methyladenosine and 5′-Terminal 7-Methylguanosine in Cap Structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, F.; Vega-Gibson, A.; Catrileo, J.; Gaete-Argel, A.; Riquelme-Barrios, S.; Alonso-Palomares, L.A.; Tapia, L.I.; Valiente-Echeverría, F.; Soto-Rifo, R.; Acevedo, M.L. N6-Methyladenosine Negatively Regulates Human Respiratory Syncytial Virus Replication. Front. Cell Dev. Biol. 2021, 9, 739445. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Zhang, L.; Tian, J.; Wang, H.; Ding, H.; Nie, J.; Pi, H.; Wang, B.; Liu, D. N 6-Methyladenosine Modification Contributes to Respiratory Syncytial Virus Infection. Heliyon 2023, 9, e15307. [Google Scholar] [CrossRef]
- Picavet, L.W.; van Vroonhoven, E.C.N.; Scholman, R.C.; Smits, Y.T.H.; Banerjee, R.; Besteman, S.B.; Viveen, M.C.; van der Vlist, M.M.; Tanenbaum, M.E.; Lebbink, R.J.; et al. M6A Reader YTHDC1 Impairs Respiratory Syncytial Virus Infection by Downregulating Membrane CX3CR1 Expression. Viruses 2024, 16, 778. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, M.; Li, K.; Bai, D.; Yi, C. Cap-Specific, Terminal N6-Methylation by a Mammalian M6Am Methyltransferase. Cell Res. 2018, 29, 80–82. [Google Scholar] [CrossRef]
- Wei, C.M.; Gershowitz, A.; Moss, B. N6, O2′-Dimethyladenosine a Novel Methylated Ribonucleoside next to the 5′ Terminal of Animal Cell and Virus MRNAs. Nature 1975, 257, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Boo, S.H.; Kim, Y.K. The Emerging Role of RNA Modifications in the Regulation of MRNA Stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-Specific Terminal N 6-Methylation of RNA by an RNA Polymerase II-Associated Methyltransferase. Science 2019, 363, eaav0080. [Google Scholar] [CrossRef]
- Sendinc, E.; Valle-Garcia, D.; Dhall, A.; Chen, H.; Henriques, T.; Navarrete-Perea, J.; Sheng, W.; Gygi, S.P.; Adelman, K.; Shi, Y. PCIF1 Catalyzes M6Am MRNA Methylation to Regulate Gene Expression. Mol. Cell 2019, 75, 620–630.e9. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible Methylation of M6Am in the 5′ Cap Controls MRNA Stability. Nature 2016, 541, 371–375. [Google Scholar] [CrossRef]
- Tartell, M.A.; Boulias, K.; Hoffmann, G.B.; Bloyet, L.M.; Greer, E.L.; Whelan, S.P.J. Methylation of Viral MRNA Cap Structures by PCIF1 Attenuates the Antiviral Activity of Interferon-β. Proc. Natl. Acad. Sci. USA 2021, 118, e2025769118. [Google Scholar] [CrossRef]
- Song, W.; Liu, L.; Liang, H.; Cheng, H.; He, W.; Yin, Q.; Zhang, Z.; Lin, W.; Li, H.; Li, Q.; et al. M6Am Methyltransferase PCIF1 Regulates Periodontal Inflammation. J. Dent. Res. 2024, 103, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- George, C.X.; John, L.; Samuel, C.E. An RNA Editor, Adenosine Deaminase Acting on Double-Stranded RNA (ADAR1). J. Interferon Cytokine Res. 2014, 34, 437–446. [Google Scholar] [CrossRef]
- Samuel, C.E. Adenosine Deaminases Acting on RNA (ADARs) Are Both Antiviral and Proviral. Virology 2011, 411, 180–193. [Google Scholar] [CrossRef]
- Slotkin, W.; Nishikura, K. Adenosine-to-Inosine RNA Editing and Human Disease. Genome Med. 2013, 5. [Google Scholar] [CrossRef]
- Zinshteyn, B.; Nishikura, K. Adenosine-to-Inosine RNA Editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ruan, G.-X.; Chen, W.; Huang, H.; Zhang, R.; Wang, J.; Li, Y.; Xu, S.; Ou, X. RNA-Editing Enzyme ADAR1 P150 Isoform Is Critical for Germinal Center B Cell Response. J. Immunol. 2022, 209, 1071–1082. [Google Scholar] [CrossRef]
- Ratcliff, J.; Simmonds, P. The Roles of Nucleic Acid Editing in Adaptation of Zoonotic Viruses to Humans. Curr. Opin. Virol. 2023, 60, 101326. [Google Scholar] [CrossRef] [PubMed]
- Netzband, R.; Pager, C.T. Epitranscriptomic Marks: Emerging Modulators of RNA Virus Gene Expression. Wiley Interdiscip. Rev. RNA 2020, 11, e1576. [Google Scholar] [CrossRef]
- Toth, A.M.; Zhang, P.; Das, S.; George, C.X.; Samuel, C.E. Interferon Action and the Double-Stranded RNA-Dependent Enzymes ADAR1 Adenosine Deaminase and PKR Protein Kinase. Prog. Nucleic Acid. Res. Mol. Biol. 2006, 81, 369–434. [Google Scholar] [CrossRef]
- O’Connell, M.A.; Mannion, N.M.; Keegan, L.P. The Epitranscriptome and Innate Immunity. PLoS Genet. 2015, 11, e1005687. [Google Scholar] [CrossRef]
- Zahn, R.C.; Schelp, I.; Utermöhlen, O.; von Laer, D. A-to-G Hypermutation in the Genome of Lymphocytic Choriomeningitis Virus. J. Virol. 2007, 81, 457–464. [Google Scholar] [CrossRef]
- Piontkivska, H.; Frederick, M.; Miyamoto, M.M.; Wayne, M.L. RNA Editing by the Host ADAR System Affects the Molecular Evolution of the Zika Virus. Ecol. Evol. 2017, 7, 4475–4485. [Google Scholar] [CrossRef]
- Ortín, J.; Martín-Benito, J. The RNA Synthesis Machinery of Negative-Stranded RNA Viruses. Virology 2015, 479, 532–544. [Google Scholar] [CrossRef]
- Gutsche, I.; Desfosses, A.; Effantin, G.; Ling, W.L.; Haupt, M.; Ruigrok, R.W.H.; Sachse, C.; Schoehn, G. Structural Virology. Near-Atomic Cryo-EM Structure of the Helical Measles Virus Nucleocapsid. Science 2015, 348, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Rima, B.K.; Gatherer, D.; Young, D.F.; Norsted, H.; Randall, R.E.; Davison, A.J. Stability of the Parainfluenza Virus 5 Genome Revealed by Deep Sequencing of Strains Isolated from Different Hosts and Following Passage in Cell Culture. J. Virol. 2014, 88, 3826–3836. [Google Scholar] [CrossRef]
- Bass, B.L.; Weintraub, H. An Unwinding Activity That Covalently Modifies Its Double-Stranded RNA Substrate. Cell 1988, 55, 1089–1098. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic Identification of Abundant A-to-I Editing Sites in the Human Transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.R.; Puig, M.; Darnell, M.E.R.; Mihalik, K.; Feinstone, S.M. New Antiviral Pathway That Mediates Hepatitis C Virus Replicon Interferon Sensitivity through ADAR1. J. Virol. 2005, 79, 6291–6298. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, X.; Qi, R.; Billiar, T. RNA Editing, ADAR1, and the Innate Immune Response. Genes 2017, 8, 41. [Google Scholar] [CrossRef]
- Liao, J.Y.; Thakur, S.A.; Zalinger, Z.B.; Gerrish, K.E.; Imani, F. Inosine-Containing RNA Is a Novel Innate Immune Recognition Element and Reduces RSV Infection. PLoS ONE 2011, 6, e26463. [Google Scholar] [CrossRef][Green Version]
- Sarvestani, S.T.; Tate, M.D.; Moffat, J.M.; Jacobi, A.M.; Behlke, M.A.; Miller, A.R.; Beckham, S.A.; McCoy, C.E.; Chen, W.; Mintern, J.D.; et al. Inosine-Mediated Modulation of RNA Sensing by Toll-Like Receptor 7 (TLR7) and TLR8. J. Virol. 2014, 88, 799–810. [Google Scholar] [CrossRef]
- Cattaneo, R.; Schmid, A.; Eschle, D.; Baczko, K.; ter Meulen, V.; Billeter, M.A. Biased Hypermutation and Other Genetic Changes in Defective Measles Viruses in Human Brain Infections. Cell 1988, 55, 255–265. [Google Scholar] [CrossRef]
- Kolakofsky, D. Isolation and Characterization of Sendai Virus DI-RNAs. Cell 1976, 8, 547–555. [Google Scholar] [CrossRef]
- Pfaller, C.K.; Mastorakos, G.M.; Matchett, W.E.; Ma, X.; Samuel, C.E.; Cattaneo, R. Measles Virus Defective Interfering RNAs Are Generated Frequently and Early in the Absence of C Protein and Can Be Destabilized by Adenosine Deaminase Acting on RNA-1-Like Hypermutations. J. Virol. 2015, 89, 7735–7747. [Google Scholar] [CrossRef]
- Furuse, Y. RNA Modifications in Genomic RNA of Influenza a Virus and the Relationship between RNA Modifications and Viral Infection. Int. J. Mol. Sci. 2021, 22, 9127. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Barberan-Soler, S.; Lowe, T.M. A Computational Screen for Mammalian Pseudouridylation Guide H/ACA RNAs. RNA 2006, 12, 15–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ge, J.; Yu, Y.T. RNA Pseudouridylation: New Insights into an Old Modification. Trends Biochem. Sci. 2013, 38, 210–218. [Google Scholar] [CrossRef]
- Karijolich, J.; Yu, Y.T. Converting Nonsense Codons into Sense Codons by Targeted Pseudouridylation. Nature 2011, 474, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.L.; Youssef, O.A.; Chastkofsky, M.I.R.; Dy, D.A.; Terns, R.M.; Terns, M.P. RNA-Guided RNA Modification: Functional Organization of the Archaeal H/ACA RNP. Genes. Dev. 2005, 19, 1238–1248. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, C.; Schattner, P.; Yu, Y.T. Functionality and Substrate Specificity of Human Box H/ACA Guide RNAs. RNA 2009, 15, 176–186. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-Wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of NcRNA and MRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef]
- Yu, Y.T.; Meier, U.T. RNA-Guided Isomerization of Uridine to Pseudouridine--Pseudouridylation. RNA Biol. 2014, 11, 1483–1494. [Google Scholar] [CrossRef]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine Profiling Reveals Regulated MRNA Pseudouridylation in Yeast and Human Cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef]
- Li, X.; Zhu, P.; Ma, S.; Song, J.; Bai, J.; Sun, F.; Yi, C. Chemical Pulldown Reveals Dynamic Pseudouridylation of the Mammalian Transcriptome. Nat. Chem. Biol. 2015, 11, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, Y.; Sekiya, T. Review Discovery of m 7 G-Cap in Eukaryotic MRNAs. Proc. Jpn. Acad. Ser. B 2015, 91, 394–409. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. MRNA Capping: Biological Functions and Applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Chiang, J.J.; Sparrer, K.M.J.; Van Gent, M.; Lässig, C.; Huang, T.; Osterrieder, N.; Hopfner, K.P.; Gack, M.U. Viral Unmasking of Cellular 5S RRNA Pseudogene Transcripts Induces RIG-I-Mediated Immunity Article. Nat. Immunol. 2018, 19, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.M.; Pichlmair, A.; Górna, M.W.; Superti-Furga, G.; Nagar, B. Structural Basis for Viral 5′-PPP-RNA Recognition by Human IFIT Proteins. Nature 2013, 494, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Avila-Bonilla, R.G.; Macias, S. The Molecular Language of RNA 5′ Ends: Guardians of RNA Identity and Immunity. RNA 2024, 30, 327. [Google Scholar] [CrossRef]
- Ferron, F.; Longhi, S.; Henrissat, B.; Canard, B. Viral RNA-Polymerases–a Predicted 2′-O-Ribose Methyltransferase Domain Shared by All Mononegavirales. Trends Biochem. Sci. 2002, 27, 222–224. [Google Scholar] [CrossRef]
- Sun, J.; Wei, Y.; Rauf, A.; Zhang, Y.; Ma, Y.; Zhang, X.; Shilo, K.; Yu, Q.; Saif, Y.M.; Lu, X.; et al. Methyltransferase-Defective Avian Metapneumovirus Vaccines Provide Complete Protection against Challenge with the Homologous Colorado Strain and the Heterologous Minnesota Strain. J. Virol. 2014, 88, 12348–12363. [Google Scholar] [CrossRef]
- Sutto-Ortiz, P.; Tcherniuk, S.; Ysebaert, N.; Abeywickrema, P.; Noël, M.; Decombe, A.; Debart, F.; Vasseur, J.J.; Canard, B.; Roymans, D.; et al. The Methyltransferase Domain of the Respiratory Syncytial Virus L Protein Catalyzes Cap N7 and 2′-O-Methylation. PLoS Pathog. 2021, 17, e1009562. [Google Scholar] [CrossRef]
- Valle, C.; Martin, B.; Debart, F.; Vasseur, J.-J.; Imbert, I.; Canard, B.; Coutard, B.; Decroly, E. The C-Terminal Domain of the Sudan Ebolavirus L Protein Is Essential for RNA Binding and Methylation. J. Virol. 2020, 94, e00833-16. [Google Scholar] [CrossRef]
- Ogino, T.; Banerjee, A.K. Unconventional Mechanism of MRNA Capping by the RNA-Dependent RNA Polymerase of Vesicular Stomatitis Virus. Mol. Cell 2007, 25, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Green, T.J. RNA Synthesis and Capping by Nonsegmented Negative Strand RNA Viral Polymerases: Lessons from a Prototypic Virus. Front. Microbiol. 2019, 10, 464386. [Google Scholar] [CrossRef]
- Ogino, T.; Banerjee, A.K. An Unconventional Pathway of MRNA Cap Formation by Vesiculoviruses. Virus Res. 2011, 162, 100. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; McElvain, L.E.; Whelan, S.P.J. Vesicular Stomatitis Virus MRNA Capping Machinery Requires Specific Cis-Acting Signals in the RNA. J. Virol. 2007, 81, 11499. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T. In Vitro Capping and Transcription of Rhabdoviruses. Methods 2012, 59, 188. [Google Scholar] [CrossRef]
- te Velthuis, A.J.W.; Grimes, J.M.; Fodor, E. Structural Insights into RNA Polymerases of Negative-Sense RNA Viruses. Nat. Rev. Microbiol. 2021, 19, 303–318. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Hiono, T.; Yamada, S.; Matsuno, K.; Faist, A.; Claff, T.; Hou, J.; Namasivayam, V.; vom Hemdt, A.; Sugimoto, S.; et al. Inhibition of Cellular RNA Methyltransferase Abrogates Influenza Virus Capping and Replication. Science 2023, 379, 586–591. [Google Scholar] [CrossRef]
- Jeeva, S.; Mir, S.; Velasquez, A.; Weathers, B.A.; Leka, A.; Wu, S.; Sevarany, A.T.; Mir, M. Hantavirus RdRp Requires a Host Cell Factor for Cap Snatching. J. Virol. 2019, 93, e02088-18. [Google Scholar] [CrossRef]
- Cheng, E.; Mir, M.A. Signatures of Host MRNA 5′ Terminus for Efficient Hantavirus Cap Snatching. J. Virol. 2012, 86, 10173–10185. [Google Scholar] [CrossRef]
- Olschewski, S.; Cusack, S.; Rosenthal, M. The Cap-Snatching Mechanism of Bunyaviruses. Trends Microbiol. 2020, 28, 293–303. [Google Scholar] [CrossRef]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O Methylation of the Viral MRNA Cap Evades Host Restriction by IFIT Family Members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-Methylation Provides a Molecular Signature for the Distinction of Self and Non-Self MRNA Dependent on the RNA Sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef]
- Abbas, Y.M.; Laudenbach, B.T.; Martínez-Montero, S.; Cencic, R.; Habjan, M.; Pichlmair, A.; Damha, M.J.; Pelletier, J.; Nagar, B. Structure of Human IFIT1 with Capped RNA Reveals Adaptable MRNA Binding and Mechanisms for Sensing N1 and N2 Ribose 2′-O Methylations. Proc. Natl. Acad. Sci. USA 2017, 114, E2106–E2115. [Google Scholar] [CrossRef]
- Fehrholz, M.; Kendl, S.; Prifert, C.; Weissbrich, B.; Lemon, K.; Rennick, L.; Duprex, P.W.; Rima, B.K.; Koning, F.A.; Holmes, R.K.; et al. The Innate Antiviral Factor APOBEC3G Targets Replication of Measles, Mumps and Respiratory Syncytial Viruses. J. Gen. Virol. 2012, 93, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Tiwarekar, V.; Wohlfahrt, J.; Fehrholz, M.; Scholz, C.-J.; Kneitz, S.; Schneider-Schaulies, J. APOBEC3G-Regulated Host Factors Interfere with Measles Virus Replication: Role of REDD1 and Mammalian TORC1 Inhibition. J. Virol. 2018, 92, e00835-18. [Google Scholar] [CrossRef] [PubMed]
- Huthoff, H.; Autore, F.; Gallois-Montbrun, S.; Fraternali, F.; Malim, M.H. RNA-Dependent Oligomerization of APOBEC3G Is Required for Restriction of HIV-1. PLoS Pathog. 2009, 5, e1000330. [Google Scholar] [CrossRef]
- Khan, M.A.; Kao, S.; Miyagi, E.; Takeuchi, H.; Goila-Gaur, R.; Opi, S.; Gipson, C.L.; Parslow, T.G.; Ly, H.; Strebel, K. Viral RNA Is Required for the Association of APOBEC3G with Human Immunodeficiency Virus Type 1 Nucleoprotein Complexes. J. Virol. 2005, 79, 5870–5874. [Google Scholar] [CrossRef]
- Wichroski, M.J.; Robb, G.B.; Rana, T.M. Human Retroviral Host Restriction Factors APOBEC3G and APOBEC3F Localize to MRNA Processing Bodies. PLoS Pathog. 2006, 2, 374–383. [Google Scholar] [CrossRef]
- Kozak, S.L.; Marin, M.; Rose, K.M.; Bystrom, C.; Kabat, D. The Anti-HIV-1 Editing Enzyme APOBEC3G Binds HIV-1 RNA and Messenger RNAs That Shuttle between Polysomes and Stress Granules. J. Biol. Chem. 2006, 281, 29105–29119. [Google Scholar] [CrossRef]
- Gallois-Montbrun, S.; Kramer, B.; Swanson, C.M.; Byers, H.; Lynham, S.; Ward, M.; Malim, M.H. Antiviral Protein APOBEC3G Localizes to Ribonucleoprotein Complexes Found in P Bodies and Stress Granules. J. Virol. 2007, 81, 2165–2178. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, Z.; Cai, J.; Li, H.; Qiu, M. N1-Methylpseudouridine Modification Level Correlates with Protein Expression, Immunogenicity, and Stability of MRNA. MedComm 2024, 5, e691. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.V.; Soto Acosta, R.; Pruitt, L.; Wasdin, P.T.; Kedarinath, K.; Hernandez, K.R.; Gonzales, K.A.; Hill, K.; Weidner, N.G.; Mire, C.; et al. Comparison of Uridine and N1-Methylpseudouridine MRNA Platforms in Development of an Andes Virus Vaccine. Nat. Commun. 2024, 15, 6421. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, R.; Zou, J.; Tian, S.; Yu, L.; Zhou, Y.; Ran, Y.; Jin, M.; Chen, H.; Zhou, H. N6-Methyladenosine Reader Protein YTHDC1 Regulates Influenza A Virus NS Segment Splicing and Replication. PLoS Pathog. 2023, 19, e1011305. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, S.; Shen, W.; Wei, Y.; Han, L.; Wang, Z.; Yu, Y.; Liu, M.; Liu, J.; Deng, G.; et al. N 6-Methyladnosine of VRNA Facilitates Influenza A Virus Replication by Promoting the Interaction of VRNA with Polymerase Proteins. Proc. Natl. Acad. Sci. USA 2025, 122, e2411554122. [Google Scholar] [CrossRef]
| Modifications | Enzymes | Virus Described | Role in Innate Immunity |
|---|---|---|---|
| m6A | METTL3/14* [15,16] FTO* [19] ALKBH5* [20] YTHDF1-3* [21] | Respiratory syncytial virus [32] Influenza virus [29,31] Sendai virus [27] Measles virus [27] Metapneumovirus [27] Vesicular stomatitis virus [27] | 🠯 RIG-I [33] 🠭 IL-6 [33] 🠭 TNFα [33] |
| m6Am | PCIF1* [44,46] FTO* [40] | Vesicular stomatitis virus [45] Rabies virus [46] Measles virus [46] | Pro-inflammatory [47] |
| Inosine | ADARs* [52] | Respiratory syncytial virus [60] Measles virus [59] Parainfluenza virus [60] Influenza virus [51] | 🠭 IL-6 [60] 🠭 IFN-β [60] 🠭 TNFα [60] 🠭 MAPK [60] |
| Pseudouridine | PUS* [76,77] | Influenza virus [71] | Unknown Evasion? [33] |
| m7G | L MTase domain** [85,86,87,88] L and PA** | All non-segmented Mononegavirus [89] Influenza virus [95] Hantavirus [96,97,98] | Evasion [81,98,100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceballos, M.-A.; Acevedo, M.L. The Role of Chemical Modifications in the Genome of Negative-Sense RNA Viruses on the Innate Immune Response. Viruses 2025, 17, 795. https://doi.org/10.3390/v17060795
Ceballos M-A, Acevedo ML. The Role of Chemical Modifications in the Genome of Negative-Sense RNA Viruses on the Innate Immune Response. Viruses. 2025; 17(6):795. https://doi.org/10.3390/v17060795
Chicago/Turabian StyleCeballos, María-Alejandra, and Mónica L. Acevedo. 2025. "The Role of Chemical Modifications in the Genome of Negative-Sense RNA Viruses on the Innate Immune Response" Viruses 17, no. 6: 795. https://doi.org/10.3390/v17060795
APA StyleCeballos, M.-A., & Acevedo, M. L. (2025). The Role of Chemical Modifications in the Genome of Negative-Sense RNA Viruses on the Innate Immune Response. Viruses, 17(6), 795. https://doi.org/10.3390/v17060795





