Evaluation of RT-LAMP for SARS-CoV-2 Detection in Animal Feces
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral Isolates
2.2. Fecal Suspension Preparation and Nucleic Acid Extraction
2.3. RT-LAMP Testing
2.4. rRT-PCR Testing
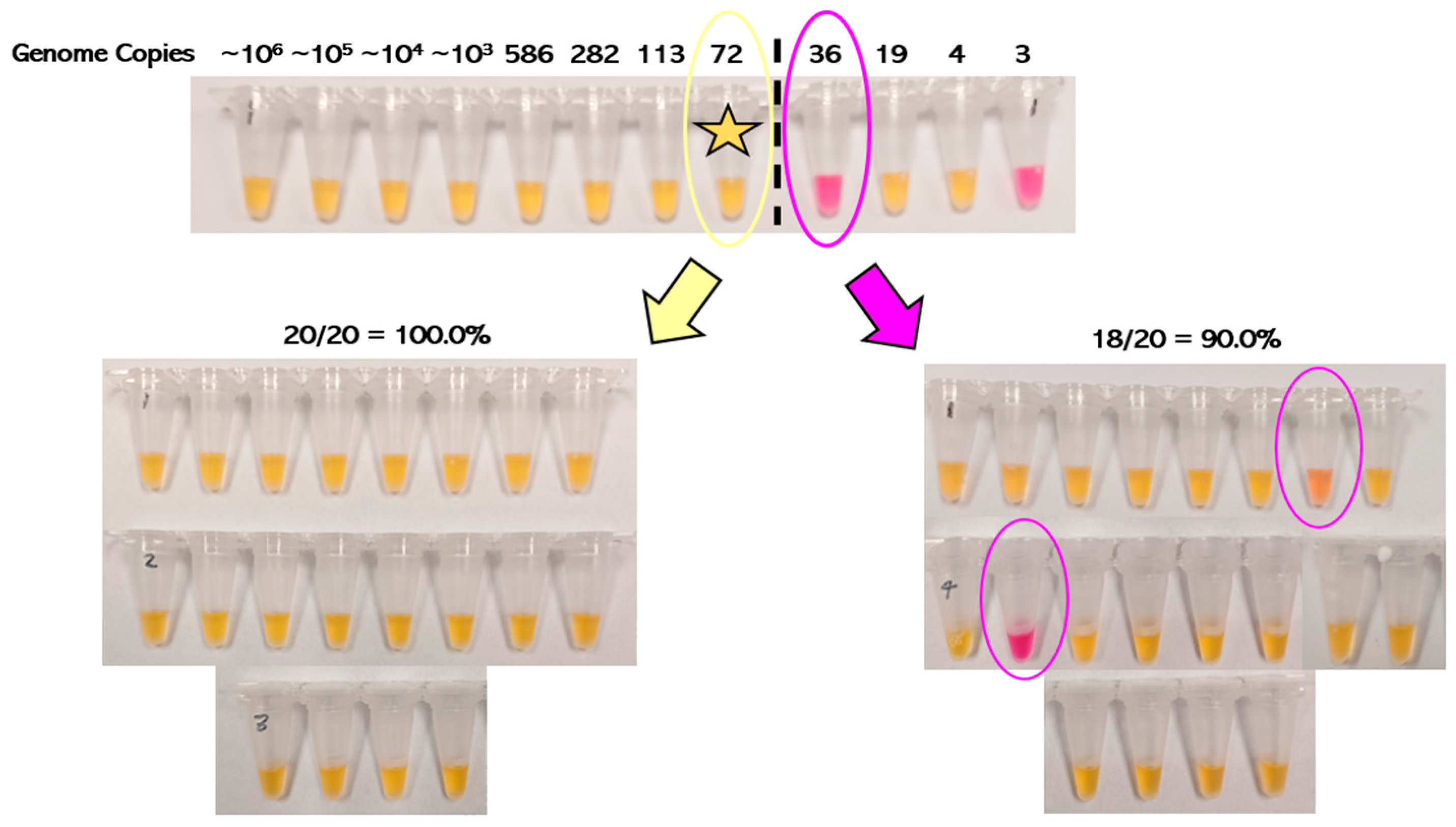
2.5. Analytical Sensitivity
2.6. Analytical Specificity
2.7. Assay Robustness
2.8. Banked Clinical Fecal Samples
2.9. Diagnostic Sensitivity and Specificity
2.10. Blinded Method Testing
3. Results
3.1. Analytical Sensitivity of RT-LAMP
3.2. Analytical Specificity of RT-LAMP
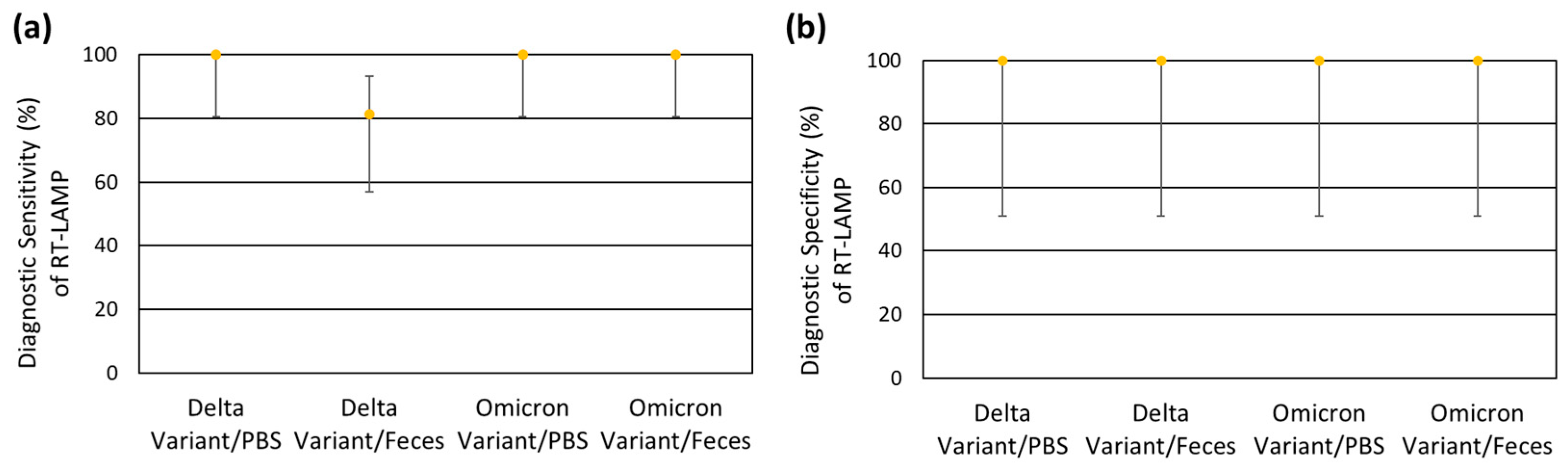
3.3. Robustness of RT-LAMP
3.3.1. Incubation Temperature
3.3.2. Incubation Length
3.3.3. Reaction Volume
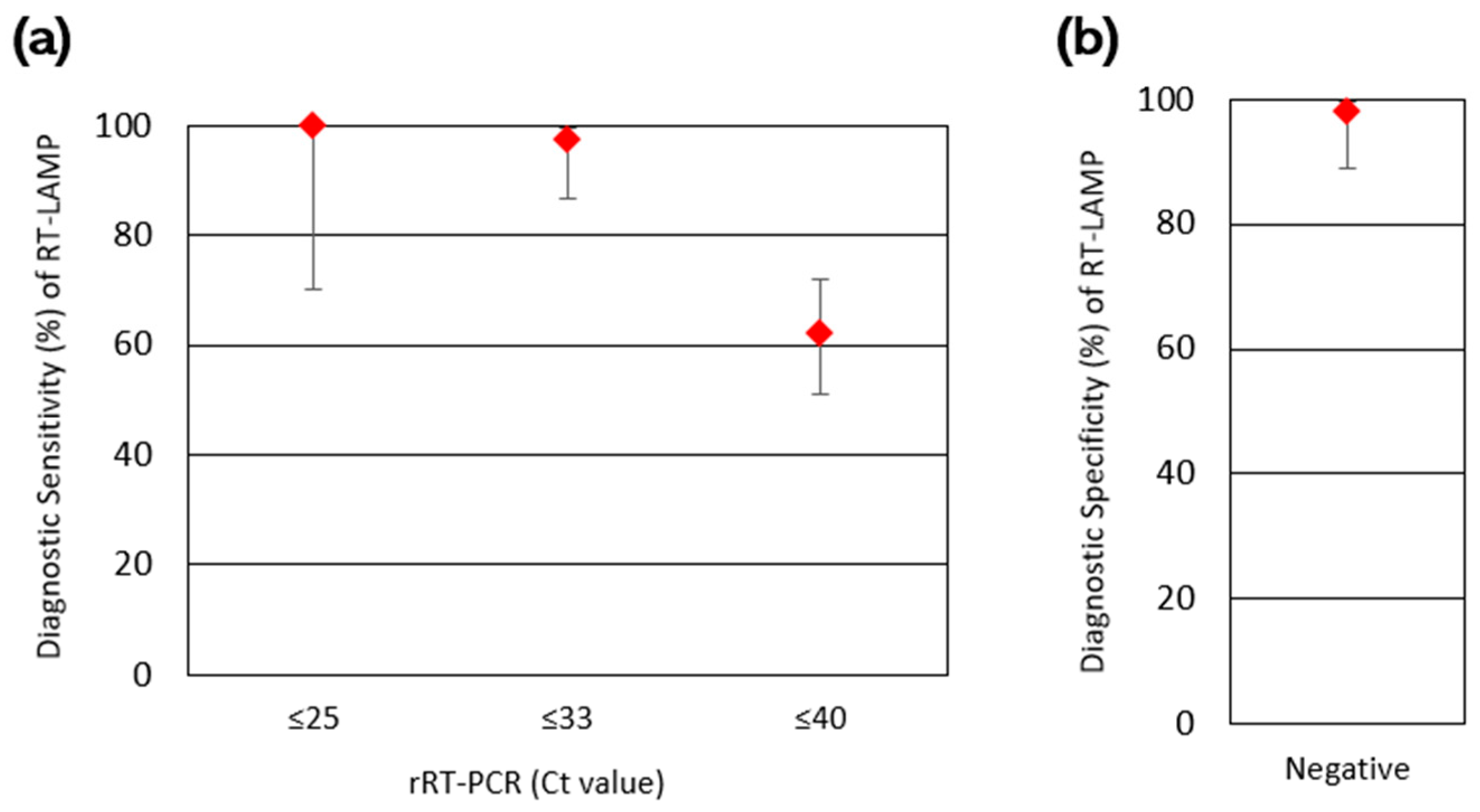
3.4. Diagnostic Sensitivity and Specificity of RT-LAMP
3.5. Blinded Method Testing of RT-LAMP
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| COVID-19 | Coronavirus disease 19 |
| LAMP | Loop-mediated isothermal amplification |
| RT-LAMP | Reverse transcription loop-mediated isothermal amplification |
| rRT-PCR | Real-time reverse-transcription polymerase chain reaction |
| dNTP | Deoxynucleoside triphosphate |
| PBS | Phosphate-buffered saline |
| N | Nucleocapsid |
| E | Envelope |
| NTC | No-template control |
| Ct | Cycle threshold |
| LoD | Limit of detection |
| HCoV-NL63 | Human coronavirus NL63 |
| PHEV | Porcine hemagglutinating encephalomyelitis virus |
| PEDV | Porcine epidermic diarrhea virus |
| PDCoV | Porcine deltacoronavirus |
| TGEV | Transmissible gastroenteritis virus |
| BdCoV | Bottlenose dolphin coronavirus |
| FCoV | Feline coronavirus |
| BCoV | Bovine coronavirus |
| BRSV | Bovine respiratory syncytial virus |
| BMT | Blinded method testing |
| Vet-LIRN | FDA’s Veterinary Laboratory Investigation and Response Network |
| CI | Confidence interval |
| DSe | Diagnostic sensitivity |
| DSp | Diagnostic specificity |
| dUTP | Deoxyuridine triphosphate |
| UDG | Uracil DNA glycosylase |
Appendix A
| Primer | Sequence |
|---|---|
| E1-F3 | TGAGTACGAACTTATGTACTCAT |
| E1-B3 | TTCAGATTTTTAACACGAGAGT |
| E1-FIP | ACCACGAAAGCAAGAAAAAGAAGTTCGTTTCGGAAGAGACAG |
| E1-BIP | TTGCTAGTTACACTAGCCATCCTTAGGTTTTACAAGACTCACGT |
| E1-LF | CGCTATTAACTATTAACG |
| E1-LB | GCGCTTCGATTGTGTGCGT |
| N2-F3 | ACCAGGAACTAATCAGACAAG |
| N2-B3 | GACTTGATCTTTGAAATTTGGATCT |
| N2-FIP | TTCCGAAGAACGCTGAAGCGGAACTGATTACAAACATTGGCC |
| N2-BIP | CGCATTGGCATGGAAGTCACAATTTGATGGCACCTGTGTA |
| N2-LF | GGGGGCAAATTGTGCAATTTG |
| N2-LB | CTTCGGGAACGTGGTTGACC |
References
- Woo, P.C.Y.; de Groot, R.J.; Haagmans, B.; Lau, S.K.P.; Neuman, B.W.; Perlman, S.; Sola, I.; van der Hoek, L.; Wong, A.C.P.; Yeh, S.-H. ICTV Virus Taxonomy Profile: Coronaviridae 2023. J. Gen. Virol. 2023, 104, 001843. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Statement on the Update of WHO’s Working Definitions and Tracking System for SARS-CoV-2 Variants of Concern and Variants of Interest. Available online: https://www.who.int/news/item/16-03-2023-statement-on-the-update-of-who-s-working-definitions-and-tracking-system-for-sars-cov-2-variants-of-concern-and-variants-of-interest (accessed on 8 January 2025).
- WHO COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/cases?n=o (accessed on 2 January 2025).
- SARS-ANI VIS. Available online: https://vis.csh.ac.at/sars-ani/ (accessed on 7 January 2025).
- Qui, X.; Liu, Y.; Sha, A. SARS-CoV-2 and natural infection in animals. J. Med. Virol. 2023, 95, e28147. [Google Scholar] [CrossRef]
- Jan Molenaar, R.J.; Vreman, S.; Hakze-van der Honing, R.W.; Zwart, R.; de Rond, J.; Weesendorp, E.; Smit, L.A.M.; Koopmans, M.; Bouwstra, R.; Stegeman, A.; et al. Clinical and Pathological Findings in SARS-CoV-2 Disease Outbreaks in Farmed Mink (Neovison vison). Vet. Pathol. 2022, 57, 653–657. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 Pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Drozd, M.; Ritter, J.M.; Samuelson, J.P.; Parker, M.; Wang, L.; Sander, S.J.; Yoshicedo, J.; Wright, L.; Odani, J.; Shrader, T.; et al. Mortality associated with SARS-CoV-2 in nondomestic felids. Vet. Pathol. 2024, 61, 609–620. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Fonager, J.; Rasmussen, M.; Mundbjerg, K.; Lohse, L.; Strandbygaard, B.; Jørgensen, C.S.; Alfaro-Núñez, A.; et al. SARS-CoV-2 Transmission between Mink (Neovison vison) and Humans, Denmark. Emerg. Infect. Dis. 2021, 27, 547–551. [Google Scholar] [CrossRef]
- Yen, H.-L.; Sit, T.H.C.; Brackman, C.J.; Chuk, S.S.Y.; Gu, H.; Tam, K.W.S.; Leung, G.M.L.; Peiris, M.P.; Poon, L.L.M. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: A case study. Lancet 2022, 399, 1070–1078. [Google Scholar] [CrossRef]
- Sila, T.; Sunghan, J.; Laochareonsuk, W.; Surasombatpattana, S.; Kongkamol, C.; Ingviya, T.; Siripaitoon, P.; Kositpantawong, N.; Kanchanaswuan, S.; Hortiwakul, T.; et al. Suspected Cat-to-Human Transmission of SARS-CoV-2, Thailand, July–September 2021. Emerg. Infect. Dis. 2022, 28, 1485–1488. [Google Scholar] [CrossRef]
- Siegrist, A.A.; Richardson, K.L.; Ghai, R.R.; Pope, B.; Yeadon, J.; Culp, B.; Behravesh, B.; Liu, L.; Brown, J.A.; Boyer, L.V. Probable Transmission of SARS-CoV-2 from African Lion to Zoo Employees, Indiana, USA, 2021. Emerg. Infect. Dis. 2023, 29, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.; Lung, O.; Maguire, F.; Kruczkiewicz, P.; Kotwa, J.D.; Buchanan, T.; Gagnier, M.; Guthrie, J.L.; Jardine, C.M.; Marchand-Austin, A.; et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat. Microbiol. 2022, 7, 2011–2024. [Google Scholar] [CrossRef]
- Feng, A.; Bevins, S.; Chandler, J.; DeLiberto, T.J.; Ghai, R.; Lantz, K.; Lenoch, J.; Retchless, A.; Shriner, S.; Tang, C.Y.; et al. Transmission of SARS-CoV-2 in free-ranging white-tailed deer in the United States. Nat. Commun. 2023, 14, 4078. [Google Scholar] [CrossRef]
- Van Aart, A.E.; Velkers, F.C.; Fischer, E.A.J.; Broens, E.M.; Egberink, H.; Zhao, S.; Engelsma, M.; Hakze-van der Honing, R.W.; Harders, F.; de Rooij, M.M.T.; et al. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound. Emerg. Dis. 2022, 69, 3001–3007. [Google Scholar] [CrossRef]
- Pu, R.; Liu, S.; Ren, X.; Shi, D.; Ba, Y.; Huo, Y.; Zhang, W.; Ma, L.; Liu, Y.; Yang, Y.; et al. The screening value of RT-LAMP and RT-PCR in the diagnosis of COVID-19: Systematic review and meta-analysis. J. Virol. Methods 2022, 300, 114392. [Google Scholar] [CrossRef] [PubMed]
- Musso, N.; Constantino, A.; La Spina, S.; Finocchiaro, A.; Andronico, F.; Stracquadanio, S.; Liotta, L.; Visalli, R.; Emmanuele, G. New SARS-CoV-2 Infection Detected in an Italian Pet Cat by RT-qPCR from Deep Pharyngeal Swab. Pathogens 2020, 9, 746. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220-20. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Munnink, B.B.O.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Bartlett, S.L.; Diel, D.G.; Wang, L.; Zec, S.; Laverack, S.; Martins, M.; Cardia Caserta, L.; Killian, M.L.; Terio, K.; Olmstead, C.; et al. SARS-COV-2 infection and longitudinal fecal screening in malayan tigers (Panthera tigris jacksoni), amur tigers (Panthera Tigris altaica), and African lions (Panthera leo krugeri) at the Bronx Zoo, New York, USA. J. Zoo Wildl. Med. 2021, 51, 733–744. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.J.M.; Cahill, J.; Aidelberg, G.; Aronoff, R.; Bektaş, A.; Bezdan, D.; Butler, D.J.; Chittur, S.V.; Codyre, M.; Federici, F.; et al. Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications. J. Biomol. Tech. 2021, 32, 228–275. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Moehling, T.J.; Choi, G.; Dugan, L.C.; Salit, M.; Meagher, R.J. LAMP Diagnostics at the Point-of-Care: Emerging Trends and Perspectives for the Developer Community. Expert Rev. Mol. Diagn. 2021, 1, 43–61. [Google Scholar] [CrossRef]
- Kundrod, K.A.; Natoli, M.E.; Chang, M.M.; Smith, C.A.; Paul, S.; Ogoe, D.; Goh, C.; Santhanaraj, A.; Price, A.; Eldin, K.W.; et al. Sample-to-answer, extraction-free, real-time RT-LAMP test for SARS-CoV-2 in nasopharyngeal, nasal, and saliva samples: Implications and use for surveillance testing. PLoS ONE 2022, 17, e0264130. [Google Scholar] [CrossRef] [PubMed]
- Saharan, P.; Khatri, P.; Dingolia, S.; Duhan, J.S.; Gahlawat, S.K. Rapid Detection of Viruses Using Loop-Mediated Isothermal Amplification (LAMP): A Review. In Biotechnology: Prospects and Applications, 1st ed.; Salar, R.K., Gahlawat, S.K., Siwach, P., Duhan, J.S., Eds.; Springer: New Delhi, India, 2013; pp. 287–306. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Template for Developers of Molecular Diagnostic Tests. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised (accessed on 23 January 2025).
- Fowler, V.L.; Armson, B.; Gonzales, J.L.; Wise, E.M.; Howson, E.L.A.; Vincent-Mistiaen, Z.; Fouch, S.; Maltby, C.J.; Grippon, S.; Munro, S.; et al. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J. Infect. 2021, 82, 117–125. [Google Scholar] [CrossRef]
- Binom: Binomial Confidence Intervals for Several Parameterizations. Available online: https://cran.r-project.org/package=binom (accessed on 26 January 2025).
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- United States Food and Drug Administration (U.S. FDA). Official Website of the Veterinary Laboratory Investigation and Response Network (Vet-LIRN). Available online: https://www.fda.gov/animal-veterinary/science-research/veterinary-laboratory-investigation-and-response-network (accessed on 13 February 2025).
- Leonardi-Cattolica, A.; Kayastha, S.; Miller, M.; Guag, J.; Tkachenko, A.; Lowe, J.; Allender, M.; Terio, K.; Wang, L. Evaluation of Fecal Sample Pooling for Real-Time RT-PCR Testing SARS-CoV-2 in Animals. Viruses 2024, 16, 1651. [Google Scholar] [CrossRef]
- Amaral, C.; Antunes, W.; Moe, E.; Duarte, A.G.; Lima, L.M.P.; Santos, C.; Gomes, I.L.; Afonso, G.S.; Vieira, R.; Teles, H.S.S.; et al. A molecular test based on RT-LAMP for rapid, sensitive and inexpensive colorimetric detection of SARS-CoV-2 in clinical samples. Sci. Rep. 2021, 11, 16430. [Google Scholar] [CrossRef]
- SARS-CoV-2 Rapid Colorimetric LAMP Assay Kit Instruction Manual. Available online: https://www.neb.com/en-us/-/media/nebus/files/manuals/manuale2019.pdf?rev=640485747af74e879b323c9464380faf&hash=7519FE15B9C15DF9626B1D42050F942F (accessed on 25 January 2025).
- Choi, G.; Moehling, T.J.; Meagher, R.J. Advances in RT-LAMP for COVID-19 testing and diagnosis. Expert Rev. Mol. Diagn. 2023, 23, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Zoelzer, F.; Burger, A.L.; Dierkes, P.W. Unraveling differences in fecal microbiota stability in mammals: From high variable carnivores and consistently stable herbivores. Anim. Microbiome 2021, 3, 77. [Google Scholar] [CrossRef]
- La Scola, B.; Le Bideau, M.; Andreani, J.; Thuan Hoang, V.; Grimaldier, C.; Colson, P.; Gautret, P.; Raoult, D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1059–1061. [Google Scholar] [CrossRef]
- Walsh, K.A.; Jordan, K.; Clyne, B.; Rohde, D.; Drummond, L.; Byrne, P.; Ahern, S.; Carty, P.G.; O’Brien, K.K.; O’Murchu, E.; et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020, 81, 357–371. [Google Scholar] [CrossRef]
- Larremore, D.B.; Wilder, B.; Lester, E.; Shehata, S.; Burke, J.M.; Hay, J.A.; Tambe, M.; Mina, M.J.; Parker, R. Test sensitivity is secondary to frequency and turn-around time for COVID-19 screening. Sci. Adv. 2021, 7, eabd5393. [Google Scholar] [CrossRef]
- Paltiel, A.D.; Zheng, A.; Walensky, R.P. Assessment of SARS-CoV-2 Screening Strategies to Permit the Safe Reopening of College Campuses in the United States. JAMA Netw. Open 2020, 3, e2016818. [Google Scholar] [CrossRef] [PubMed]
- Chin, E.T.; Huynh, B.Q.; Chapman, L.A.C.; Murrill, M.; Basu, S.; Lo, N.C. Frequency of Routine Testing for Coronavirus Disease 2019 (COVID-19) in High-risk Healthcare Environments to Reduce Outbreaks. Clin. Infect. Dis. 2020, 73, e3127–e3129. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.G.; Kang, J.S.; Hwang, S.Y.; Song, J.; Jeong, S.H. Magnetic bead-based nucleic acid purification kit: Clinical application and performance evaluation in stool specimens. J. Microbiol. Methods 2016, 124, 62–68. [Google Scholar] [CrossRef]
- Tsengel, U.; Wu, T.-Y.; Chen, Y.-N. Rapid detection of bat coronaviruses from fecal samples using loop-mediated isothermal amplification assay in the field. J. Virol. Methods 2024, 330, 115035. [Google Scholar] [CrossRef]
- Hedman, J.; Rådström, P. Overcoming Inhibition in Real-Time Diagnostic PCR. In PCR Detection of Microbial Pathogens, 2nd ed.; Wilks, M.K., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 17–48. [Google Scholar] [CrossRef]
- Thakur, S.; Sasi, S.; Pillai, S.G.; Nag, A.; Shukla, D.; Singhal, R.; Phalke, S.; Velu, G.S.K. SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics, and Vaccines. Front. Med. 2022, 9, 815389. [Google Scholar] [CrossRef]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, M.; Moore, T. Emerging SARS-CoV-2 Variants: A Review of Its Mutations, Its Implications and Vaccine Efficacy. Vaccines 2021, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Hoque, M.N.; Islam, M.R.; Islam, I.; Mishu, I.D.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 2020, 22, 100997. [Google Scholar] [CrossRef] [PubMed]
- New England Biolabs Primer Monitor Tool. Available online: https://primer-monitor.neb.com/ (accessed on 2 February 2025).
- Tamanaha, E.; Zhang, Y.; Tanner, N.A. Profiling RT-LAMP tolerance of sequence variation for SARS-CoV-2 RNA detection. PLoS ONE 2022, 17, e0259610. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, G.T.; Stracke, M.C.; Coelho, B.D.O.; Sanchuki, H.B.S.; de Oliveira, V.K.; Marchini, F.K.; Zanette, D.L.; Aoki, M.N.; Viana, E.R.; Blanes, L. A new RT-LAMP-on-a-Chip Instrument for SARS-CoV-2 diagnostics. Microchem. J. 2022, 180, 107600. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, G.; Buss, J.; Barry, A.J.; Patton, G.C.; Tanner, N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine hydrochloride. Biotechniques 2020, 69, 178–185. [Google Scholar] [CrossRef]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2020, 67, 415–424. [Google Scholar] [CrossRef]
- Ongerth, J.E.; Danielson, R.E. RT qLAMP-Direct Detection of SARS-CoV-2 in Raw Sewage. J. Biomol. Tech. 2021, 32, 206–213. [Google Scholar] [CrossRef]
- Bivins, A.; Lott, M.; Shaffer, M.; Wu, Z.; North, D.; Lipp, E.K.; Bibby, K. Building-level wastewater surveillance using tampon swabs and RT-LAMP for rapid SARS-CoV-2 RNA detection. Environ. Sci. Water Res. Technol. 2022, 8, 173–183. [Google Scholar] [CrossRef]
- Cui, J.; Tian, A.; Wang, H.; Yu, Y.; Hao, J.; Wang, L.; Shi, C.; Ma, C. Hydrogel loop-mediated isothermal amplification for ultra-fast diagnosis of Helicobacter pylori in stool samples without nucleic acid extraction. Anal. Chim. Acta 2025, 1333, 343384. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, X.; Gu, A.; Dobelle, L.; Cid, C.A.; Li, J.; Hoffman, M.R. Membrane-Based In-Gel Loop-Mediated Isothermal Amplification (mgLAMP) System for SARS-CoV-2 Quantification in Environmental Waters. Environ. Sci. Technol. 2022, 56, 862–873. [Google Scholar] [CrossRef]
- Gibson, K.E.; Schwab, K.J.; Spencer, S.K.; Borchardt, M.A. Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 2012, 46, 4281–4291. [Google Scholar] [CrossRef] [PubMed]
- Ooi, K.H.; Liu, M.M.; Moo, J.R.; Nimsamer, P.; Payungpom, S.; Kaewsapsek, P.; Tan, M.H. A Sensitive and Specific Fluorescent RT-LAMP Assay for SARS-CoV-2 Detection in Clinical Samples. ACS Synth. Biol. 2022, 11, 448–463. [Google Scholar] [CrossRef] [PubMed]
- Natoli, M.; Kundrod, K.; Chang, M.; Smith, C.; Paul, S.; Eldin, K.; Patel, K.; Baker, E.; Schmeler, K.; Richards-Kortum, R. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) for point-of-care detection of SARS-CoV-2: A clinical study to evaluate agreement with RT-qPCR. Lancet Glob. Health 2021, 9, 53. [Google Scholar] [CrossRef]
- Rabe, B.A.; Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA 2020, 117, 24450–24458. [Google Scholar] [CrossRef]
- Bokelmann, L.; Nickel, O.; Maricic, T.; Pääbo, S.; Meyer, M.; Borte, S.; Riesenberg, S. Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nat. Commun. 2021, 12, 1467. [Google Scholar] [CrossRef]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Bakar, W.A.; Abas, W.; et al. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip 2016, 16, 611–621. [Google Scholar] [CrossRef]
- Lu, X.; Du, J.; You, M.; Chen, L.; Xu, F.; Chen, F. From crisis to innovation in point-of-care testing: Lessons from the COVID-19 pandemic and future directions. Trends Anal. Chem. 2025, 184, 118131. [Google Scholar] [CrossRef]



| Viruses Tested | Animal Species | Type of Sample |
|---|---|---|
| Human coronavirus NL63 (HCoV-NL63) | Human | Isolate 1 |
| Porcine hemagglutinating encephalomyelitis virus (PHEV) | Pig | Isolate |
| Porcine epidemic diarrhea virus (PEDV) | Pig | Feces |
| PEDV/porcine deltacoronavirus (PDCoV)/transmissible gastroenteritis virus (TGEV) 2 | Pig | Isolate |
| Bottlenose dolphin coronavirus (BdCoV) | Bottlenose dolphin | Feces |
| Feline coronavirus (FCoV) | Domestic cat | Lung |
| FCoV | Domestic cat | Liver and Kidney |
| Bovine coronavirus (BCoV) | Bovine | Feces |
| BCoV/bovine respiratory syncytial virus (BRSV) 2 | Bovine | Isolate 3 |
| SARS-CoV-2 | Felidae | Feces |
| Animal Species | Number of Fecal Samples | Number of Animals |
|---|---|---|
| Sumatran tiger | 79 | 7 |
| African lion | 15 | 8 |
| Amur tiger | 8 | 3 |
| Malayan tiger | 7 | 2 |
| Cheetah | 7 | 1 |
| Asian small-clawed otter | 5 | 1 |
| White-cheeked gibbon | 3 | 3 |
| Bearded emperor tamarin | 1 | 2 |
| Giant anteater | 1 | 1 |
| Total | 126 | 28 |
| RT-LAMP | |||||||
|---|---|---|---|---|---|---|---|
| Ct | Positive | Negative | Total | DSe | DSp | ||
| rRT-PCR | Positive | ≤25 | 9 | 0 | 9 | 100.0% | - |
| ≤33 | 38 | 1 | 39 | 97.4% | - | ||
| ≤40 | 49 | 30 | 79 | 62.0% | - | ||
| Negative | Negative | 1 | 46 | 47 | - | 97.9% | |
| Total | 50 | 76 | 126 | ||||
| (A) Delta Variant/PBS Matrix | RT-LAMP | |||||
| Target Concentration/ Reaction | Average Ct | Positive | Negative | Total | ||
| rRT-PCR | Positive | 250 | 33.2 | 6 | 0 | 6 |
| 500 | 31.9 | 2 | 0 | 2 | ||
| 1000 | 31.1 | 6 | 0 | 6 | ||
| 2500 | 29.7 | 2 | 0 | 2 | ||
| Negative | 0 | 0.0 | 0 | 4 | 4 | |
| Total | 16 | 4 | 20 | |||
| (B) Delta Variant/Fecal Matrix * | RT-LAMP | |||||
| Target Concentration/ Reaction | Average Ct | Positive | Negative | Total | ||
| rRT-PCR | Positive | 250 | 33.7 | 3 | 3 | 6 |
| 500 | 32.9 | 2 | 0 | 2 | ||
| 1000 | 32.4 | 6 | 0 | 6 | ||
| 2500 | 30.4 | 2 | 0 | 2 | ||
| Negative | 0 | 0.0 | 0 | 4 | 4 | |
| Total | 13 | 7 | 20 | |||
| (C) Omicron Variant/PBS Matrix | RT-LAMP | |||||
| Target Concentration/ Reaction | Average Ct | Positive | Negative | Total | ||
| rRT-PCR | Positive | 250 | 33.6 | 6 | 0 | 6 |
| 500 | 32.9 | 2 | 0 | 2 | ||
| 1000 | 32.0 | 6 | 0 | 6 | ||
| 2500 | 30.6 | 2 | 0 | 2 | ||
| Negative | 0 | 0.0 | 0 | 4 | 4 | |
| Total | 16 | 4 | 20 | |||
| (D) Omicron Variant/Fecal Matrix * | RT-LAMP | |||||
| Target Concentration/ Reaction | Average Ct | Positive | Negative | Total | ||
| rRT-PCR | Positive | 250 | 34.8 | 6 | 0 | 6 |
| 500 | 34.0 | 2 | 0 | 2 | ||
| 1000 | 33.5 | 6 | 0 | 6 | ||
| 2500 | 31.4 | 2 | 0 | 2 | ||
| Negative | 0 | 0.0 | 0 | 4 | 4 | |
| Total | 16 | 4 | 20 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pepper, A.; Kayastha, S.; Miller, M.; Guag, J.; Tkachenko, A.; Allender, M.; Terio, K.; Wang, L. Evaluation of RT-LAMP for SARS-CoV-2 Detection in Animal Feces. Viruses 2025, 17, 783. https://doi.org/10.3390/v17060783
Pepper A, Kayastha S, Miller M, Guag J, Tkachenko A, Allender M, Terio K, Wang L. Evaluation of RT-LAMP for SARS-CoV-2 Detection in Animal Feces. Viruses. 2025; 17(6):783. https://doi.org/10.3390/v17060783
Chicago/Turabian StylePepper, Aimee, Sandipty Kayastha, Megan Miller, Jake Guag, Andriy Tkachenko, Matthew Allender, Karen Terio, and Leyi Wang. 2025. "Evaluation of RT-LAMP for SARS-CoV-2 Detection in Animal Feces" Viruses 17, no. 6: 783. https://doi.org/10.3390/v17060783
APA StylePepper, A., Kayastha, S., Miller, M., Guag, J., Tkachenko, A., Allender, M., Terio, K., & Wang, L. (2025). Evaluation of RT-LAMP for SARS-CoV-2 Detection in Animal Feces. Viruses, 17(6), 783. https://doi.org/10.3390/v17060783







