How HIV-1 Uses the Metabolite Inositol Hexakisphosphate to Build Its Capsid
Abstract
1. Introduction
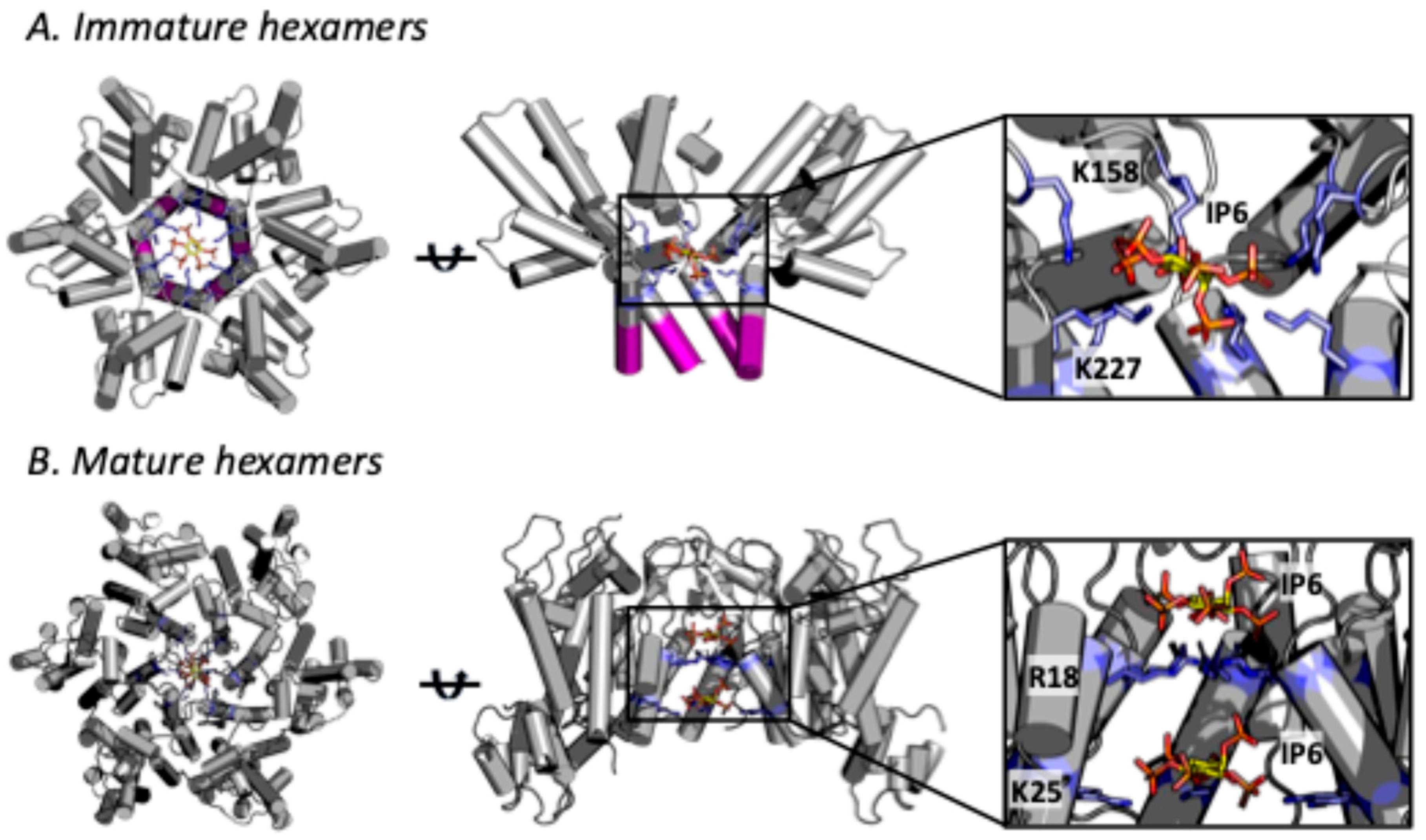
2. Role of IP6 in Immature Lattice Formation and HIV-1 Production
3. Role of IP6 in Mature Capsid Formation and HIV-1 Infectivity
4. IP6 Stabilises the HIV-1 Capsid
5. HIV-1 Can Build Its Immature Lattice but Not Its Capsid Without IP6
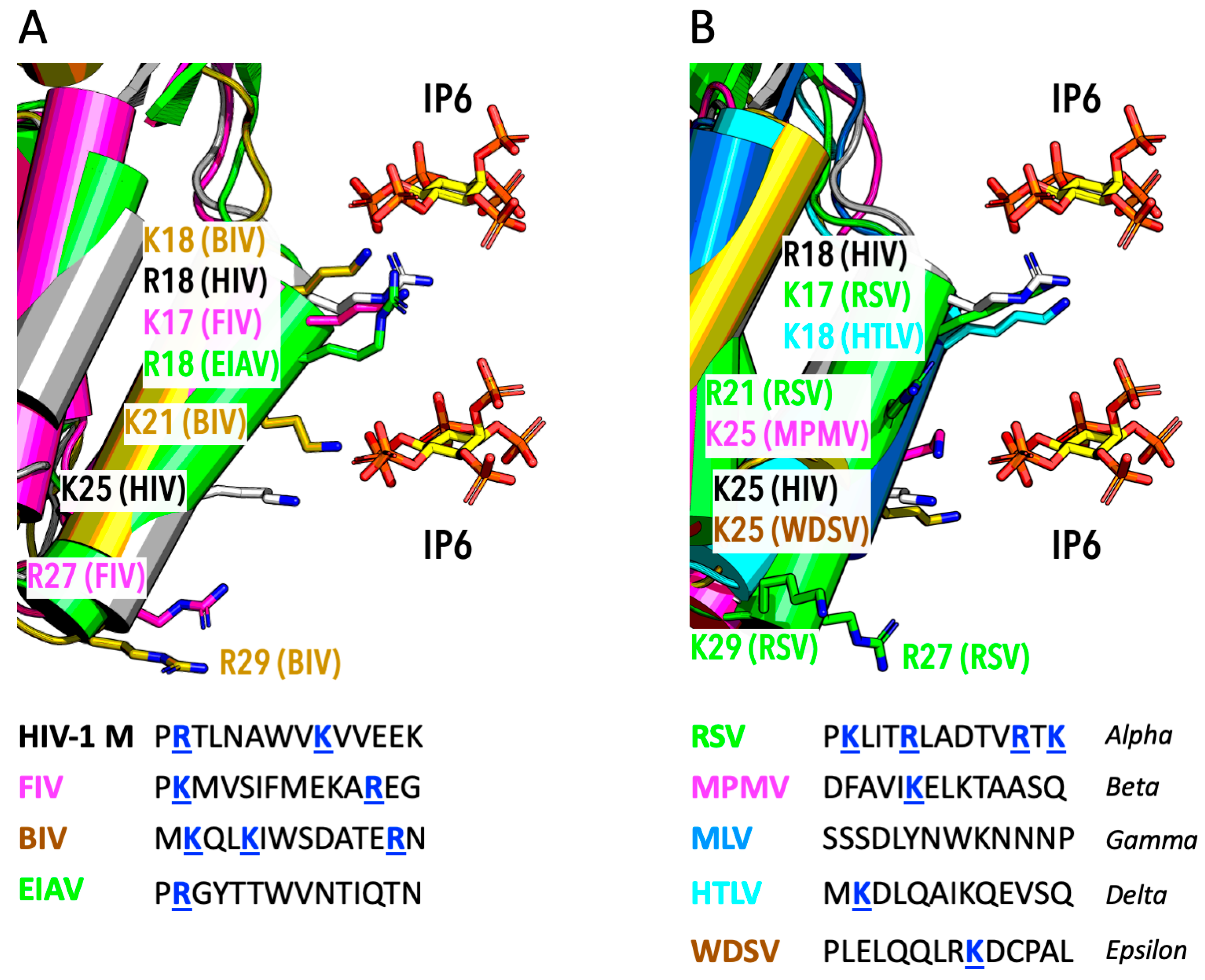
6. IP6 Usage by Other Lentiviruses
7. IP6 Usage by Other Diverse Retroviruses
8. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jang, S.; Engelman, A.N. Capsid-host interactions for HIV-1 ingress. Microbiol. Mol. Biol. Rev. 2023, 87, e0004822. [Google Scholar] [CrossRef]
- Christensen, D.E.; Ganser-Pornillos, B.K.; Johnson, J.S.; Pornillos, O.; Sundquist, W.I. Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science 2020, 370, eabc8420. [Google Scholar] [CrossRef] [PubMed]
- Jacques, D.A.; McEwan, W.A.; Hilditch, L.; Price, A.J.; Towers, G.J.; James, L.C. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 2016, 536, 349–353. [Google Scholar] [CrossRef]
- Papa, G.; Albecka, A.; Mallery, D.; Vaysburd, M.; Renner, N.; James, L.C. IP6-stabilised HIV capsids evade cGAS/STING-mediated host immune sensing. EMBO Rep. 2023, 24, e56275. [Google Scholar] [CrossRef]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.A.; Selwood, D.L.; James, L.C.; Noursadeghi, M.; et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 2013, 503, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Naghavi, M.H. The host cytoskeleton: A key regulator of early HIV-1 infection. FEBS J. 2024, 291, 1835–1848. [Google Scholar] [CrossRef]
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493. [Google Scholar] [CrossRef] [PubMed]
- Dharan, A.; Bachmann, N.; Talley, S.; Zwikelmaier, V.; Campbell, E.M. Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 2020, 5, 1088–1095. [Google Scholar] [CrossRef]
- Zila, V.; Margiotta, E.; Turonova, B.; Muller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Borner, K.; Rada, J.; Muller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e18. [Google Scholar] [CrossRef]
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 2018, 24, 392–404.e8. [Google Scholar] [CrossRef]
- Singh, P.K.; Bedwell, G.J.; Engelman, A.N. Spatial and Genomic Correlates of HIV-1 Integration Site Targeting. Cells 2022, 11, 655. [Google Scholar] [CrossRef] [PubMed]
- Ay, S.; Burlaud-Gaillard, J.; Gazi, A.; Tatirovsky, Y.; Cuche, C.; Diana, J.S.; Scoca, V.; Di Santo, J.P.; Roingeard, P.; Mammano, F.; et al. In vivo HIV-1 nuclear condensates safeguard against cGAS and license reverse transcription. EMBO J. 2024, 44, 166–199. [Google Scholar] [CrossRef] [PubMed]
- Burdick, R.C.; Morse, M.; Rouzina, I.; Williams, M.C.; Hu, W.S.; Pathak, V.K. HIV-1 uncoating requires long double-stranded reverse transcription products. Sci. Adv. 2024, 10, eadn7033. [Google Scholar] [CrossRef]
- Francis, A.C.; Marin, M.; Prellberg, M.J.; Palermino-Rowland, K.; Melikyan, G.B. HIV-1 Uncoating and Nuclear Import Precede the Completion of Reverse Transcription in Cell Lines and in Primary Macrophages. Viruses 2020, 12, 1234. [Google Scholar] [CrossRef]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef]
- Dostalkova, A.; Kaufman, F.; Krizova, I.; Vokata, B.; Ruml, T.; Rumlova, M. In Vitro Quantification of the Effects of IP6 and Other Small Polyanions on Immature HIV-1 Particle Assembly and Core Stability. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]
- Kucharska, I.; Ding, P.; Zadrozny, K.K.; Dick, R.A.; Summers, M.F.; Ganser-Pornillos, B.K.; Pornillos, O. Biochemical Reconstitution of HIV-1 Assembly and Maturation. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, L.; Sun, D.; Ning, J.; Liu, J.; Kotecha, A.; Olek, M.; Frosio, T.; Fu, X.; Himes, B.A.; Kleinpeter, A.B.; et al. CryoET structures of immature HIV Gag reveal six-helix bundle. Commun. Biol. 2021, 4, 481. [Google Scholar] [CrossRef]
- Schur, F.K.; Hagen, W.J.; Rumlova, M.; Ruml, T.; Muller, B.; Krausslich, H.G.; Briggs, J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef]
- Mallery, D.L.; Faysal, K.M.R.; Kleinpeter, A.; Wilson, M.S.C.; Vaysburd, M.; Fletcher, A.J.; Novikova, M.; Bocking, T.; Freed, E.O.; Saiardi, A.; et al. Cellular IP6 Levels Limit HIV Production while Viruses that Cannot Efficiently Package IP6 Are Attenuated for Infection and Replication. Cell Rep. 2019, 29, 3983–3996.e4. [Google Scholar] [CrossRef]
- Mallery, D.L.; Kleinpeter, A.B.; Renner, N.; Faysal, K.M.R.; Novikova, M.; Kiss, L.; Wilson, M.S.C.; Ahsan, B.; Ke, Z.; Briggs, J.A.G.; et al. A stable immature lattice packages IP6 for HIV capsid maturation. Sci. Adv. 2021, 7, eabe4716. [Google Scholar] [CrossRef]
- Renner, N.; Kleinpeter, A.; Mallery, D.L.; Albecka, A.; Rifat Faysal, K.M.; Bocking, T.; Saiardi, A.; Freed, E.O.; James, L.C. HIV-1 is dependent on its immature lattice to recruit IP6 for mature capsid assembly. Nat. Struct. Mol. Biol. 2023, 30, 370–382. [Google Scholar] [CrossRef]
- Ricana, C.L.; Lyddon, T.D.; Dick, R.A.; Johnson, M.C. Primate lentiviruses require Inositol hexakisphosphate (IP6) or inositol pentakisphosphate (IP5) for the production of viral particles. PLoS Pathog. 2020, 16, e1008646. [Google Scholar] [CrossRef]
- Sowd, G.A.; Aiken, C. Inositol phosphates promote HIV-1 assembly and maturation to facilitate viral spread in human CD4+ T cells. PLoS Pathog. 2021, 17, e1009190. [Google Scholar] [CrossRef]
- Renner, N.; Mallery, D.L.; Faysal, K.M.R.; Peng, W.; Jacques, D.A.; Bocking, T.; James, L.C. A lysine ring in HIV capsid pores coordinates IP6 to drive mature capsid assembly. PLoS Pathog. 2021, 17, e1009164. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Glass, B.; Hagen, W.J.; Krausslich, H.G.; Briggs, J.A. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Zhu, Y.; Yang, Z.; Xu, C.; Chaban, Y.; Nesterova, T.; Ning, J.; Bocking, T.; Parker, M.W.; Monnie, C.; et al. Structure of native HIV-1 cores and their interactions with IP6 and CypA. Sci. Adv. 2021, 7, eabj5715. [Google Scholar] [CrossRef] [PubMed]
- Stacey, J.C.V.; Tan, A.; Lu, J.M.; James, L.C.; Dick, R.A.; Briggs, J.A.G. Two structural switches in HIV-1 capsid regulate capsid curvature and host factor binding. Proc. Natl. Acad. Sci. USA 2023, 120, e2220557120. [Google Scholar] [CrossRef]
- Schirra, R.T.; Dos Santos, N.F.B.; Zadrozny, K.K.; Kucharska, I.; Ganser-Pornillos, B.K.; Pornillos, O. A molecular switch modulates assembly and host factor binding of the HIV-1 capsid. Nat. Struct. Mol. Biol. 2023, 30, 383–390. [Google Scholar] [CrossRef]
- Mallery, D.L.; Marquez, C.L.; McEwan, W.A.; Dickson, C.F.; Jacques, D.A.; Anandapadamanaban, M.; Bichel, K.; Towers, G.J.; Saiardi, A.; Bocking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 2018, 7, e35335. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Cheng, A.; Yeager, M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell 2007, 131, 70–79. [Google Scholar] [CrossRef]
- Kleinpeter, A.; Mallery, D.L.; Renner, N.; Albecka, A.; Klarhof, J.O.; Freed, E.O.; James, L.C. HIV-1 adapts to lost IP6 coordination through second-site mutations that restore conical capsid assembly. Nat. Commun. 2024, 15, 8017. [Google Scholar] [CrossRef]
- Rankovic, S.; Varadarajan, J.; Ramalho, R.; Aiken, C.; Rousso, I. Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Yu, A.; Lee, E.M.Y.; Briggs, J.A.G.; Ganser-Pornillos, B.K.; Pornillos, O.; Voth, G.A. Strain and rupture of HIV-1 capsids during uncoating. Proc. Natl. Acad. Sci. USA 2022, 119, e2117781119. [Google Scholar] [CrossRef]
- Jennings, J.; Shi, J.; Varadarajan, J.; Jamieson, P.J.; Aiken, C. The Host Cell Metabolite Inositol Hexakisphosphate Promotes Efficient Endogenous HIV-1 Reverse Transcription by Stabilizing the Viral Capsid. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Burdick, R.C.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc. Natl. Acad. Sci. USA 2021, 118, e2019467118. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.; Bryer, A.J.; Andino-Moncada, J.R.; Shi, J.; Hong, J.; Torres, C.; Harel, S.; Francis, A.C.; Perilla, J.R.; Aiken, C.; et al. Elasticity of the HIV-1 core facilitates nuclear entry and infection. PLoS Pathog. 2024, 20, e1012537. [Google Scholar] [CrossRef] [PubMed]
- Rankovic, S.; Deshpande, A.; Harel, S.; Aiken, C.; Rousso, I. HIV-1 uncoating occurs via a series of rapid biomechanical changes in the core related to individual stages of reverse transcription. J. Virol. 2021, 95, 10–1128. [Google Scholar] [CrossRef]
- Sowd, G.A.; Shi, J.; Fulmer, A.; Aiken, C. HIV-1 capsid stability enables inositol phosphate-independent infection of target cells and promotes integration into genes. PLoS Pathog. 2023, 19, e1011423. [Google Scholar] [CrossRef]
- Talledge, N.; Yang, H.; Shi, K.; Coray, R.; Yu, G.; Arndt, W.G.; Meng, S.; Baxter, G.C.; Mendonca, L.M.; Castano-Diez, D.; et al. HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation. J. Mol. Biol. 2023, 435, 168143. [Google Scholar] [CrossRef]
- Dick, R.A.; Xu, C.; Morado, D.R.; Kravchuk, V.; Ricana, C.L.; Lyddon, T.D.; Broad, A.M.; Feathers, J.R.; Johnson, M.C.; Vogt, V.M.; et al. Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly. PLoS Pathog. 2020, 16, e1008277. [Google Scholar] [CrossRef] [PubMed]
- Krebs, A.S.; Liu, H.F.; Zhou, Y.; Rey, J.S.; Levintov, L.; Shen, J.; Howe, A.; Perilla, J.R.; Bartesaghi, A.; Zhang, P. Molecular architecture and conservation of an immature human endogenous retrovirus. Nat. Commun. 2023, 14, 5149. [Google Scholar] [CrossRef]
- Qu, K.; Glass, B.; Dolezal, M.; Schur, F.K.M.; Murciano, B.; Rein, A.; Rumlova, M.; Ruml, T.; Krausslich, H.G.; Briggs, J.A.G. Structure and architecture of immature and mature murine leukemia virus capsids. Proc. Natl. Acad. Sci. USA 2018, 115, E11751–E11760. [Google Scholar] [CrossRef] [PubMed]
- Obr, M.; Ricana, C.L.; Nikulin, N.; Feathers, J.R.; Klanschnig, M.; Thader, A.; Johnson, M.C.; Vogt, V.M.; Schur, F.K.M.; Dick, R.A. Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer. Nat. Commun. 2021, 12, 3226. [Google Scholar] [CrossRef]
- Obr, M.; Percipalle, M.; Chernikova, D.; Yang, H.; Thader, A.; Pinke, G.; Porley, D.; Mansky, L.M.; Dick, R.A.; Schur, F.K.M. Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice. Nat. Struct. Mol. Biol. 2024, 32, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Dostalkova, A.; Vokata, B.; Kaufman, F.; Ulbrich, P.; Ruml, T.; Rumlova, M. Effect of Small Polyanions on In Vitro Assembly of Selected Members of Alpha-, Beta- and Gammaretroviruses. Viruses 2021, 13, 129. [Google Scholar] [CrossRef]
- Biswas, B.; Lai, K.K.; Bracey, H.; Datta, S.A.K.; Harvin, D.; Sowd, G.A.; Aiken, C.; Rein, A. Essential functions of inositol hexakisphosphate (IP6) in murine leukemia virus replication. mBio 2024, 15, e0115824. [Google Scholar] [CrossRef]
- Yu, R.; Phalora, P.; Li, N.; Böcking, T.; Jacques, D.A. The Human T-cell Leukemia Virus capsid protein is a potential drug target. bioRxiv 2024, 32, 268–276. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
James, L.C. How HIV-1 Uses the Metabolite Inositol Hexakisphosphate to Build Its Capsid. Viruses 2025, 17, 689. https://doi.org/10.3390/v17050689
James LC. How HIV-1 Uses the Metabolite Inositol Hexakisphosphate to Build Its Capsid. Viruses. 2025; 17(5):689. https://doi.org/10.3390/v17050689
Chicago/Turabian StyleJames, Leo C. 2025. "How HIV-1 Uses the Metabolite Inositol Hexakisphosphate to Build Its Capsid" Viruses 17, no. 5: 689. https://doi.org/10.3390/v17050689
APA StyleJames, L. C. (2025). How HIV-1 Uses the Metabolite Inositol Hexakisphosphate to Build Its Capsid. Viruses, 17(5), 689. https://doi.org/10.3390/v17050689



