Text Correction
The authors wish to make the following correction to this paper [1]:
There was an error in the original publication. In Section 3.2. “Rodent Chaphamaparvoviruses from Zambia”, the names ‘Mwangazi virus’ and ‘Nyamadzi virus’ have been switched in the following two sentences: “The region of D1 shows indel and sequence variation between the viruses, and two deduced D1 sites appear possible in the Mwangazi virus, the second one with better conservation (Supplementary Table S4). In the Nyamadzi virus, the analogous site does not conform with the canonical consensus, and another canonical site is located 5′ of the p10 termination codon so that a p10/p15 fusion protein would be generated that is not observed in the other viruses”.
A correction has been made to Section 3.2. in the first paragraph:
“We identified two 4 kb contigs with BLASTn homology of approximately 80% to murine chaphamaparvoviruses that branched in phylogenetic analyses separate from classified species (Figure 2A,B). Genomic features were analogous to those of mouse kidney parvovirus (MKPV; Chaphamaparvovirus rodent1), capuchin kidney parvovirus (CKPV, Chaphamaparvovirus primate1), and Tasmanian devil-associated chapparvovirus 2 (TdChPV2, Chaphamaparvovirus dasyurid2), which are species in the genus Chaphamaparvovirus (subfamily Hamaparvovirinae, family Parvoviridae). A common feature of these viruses is a 5′ p10 ORF that is also present in the viruses from Zambia. Based on NS1 and VP1 analyses, these viruses are also close to Ursus americanus parvovirus (UaPV; Chaphamaparvovirus carnivoran3) and Ursus thibetanus ussuricus chapparvovirus (UtPV; not classified) (Figure 2A,B), but detailed analysis is hindered by the 5′ truncated sequence for these viruses (GenBank Accession NC_077031 and OR779981). Both Zambian viruses, named Mwangazi virus and Nyamadzi virus, include the SF3 helicase family signature motifs Walker A, B, B’, and C in NS1 [43,44] and a domain of unknown function (DUF) 3648 described for NS1 of Brazilian bat chaphamaparvoviruses (Figure 2C,D) [45]. Like other chaphamaviruses, they lack a PLA2 domain in VP that is found in other parvovirus genera [46]. Two major splice donor sites (D1, D2) and three acceptor sites (A1–A3) have been experimentally mapped for MKPV [22]. In Mwangazi and Nyamadzi viruses, A1–A3 and D2 appear largely conserved. The region of D1 shows indel and sequence variation between the viruses, and two deduced D1 sites appear possible in the Nyamadzi virus, the second one with better conservation (Supplementary Table S4). In the Mwangazi virus, the analogous site does not conform with the canonical consensus, and another canonical site is located 5′ of the p10 termination codon so that a p10/p15 fusion protein would be generated that is not observed in the other viruses (D1a, Supplementary Table S4). The region between D1 and A1 differs in additional aspects between the viruses. NS2-P and p15 in Mwangazi and Nyamadzi viruses constitute one continuous ORF, and expression of p15 functionality may not require efficient D1/A1 splicing, whereas splicing may be essential in the other viruses where NS2-P and p15 are in different frames (MKPV, CKPV) or separated by a stop codon (MuCPV, TdChPV2). Based on the NS1 amino acid identity of 74% between each other and less than 85% with the existing species, both viruses qualify as novel species in the genus Chaphamaparvovirus (Supplementary Table S5) [47].”
Error in Figure/Table
In the original publication [1], there was a mistake in Figure 2 as published. In panels C and D, the names ’Mwangazi virus’ and ’Nyamadzi virus’ have been switched and associated with the opposite schematic. The corrected version of Figure 2 appears below.
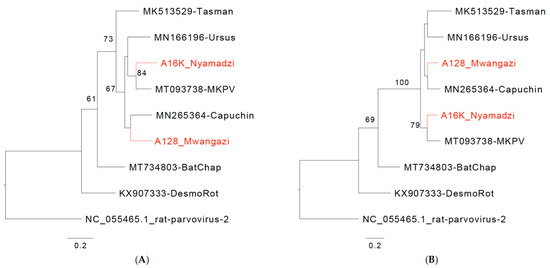
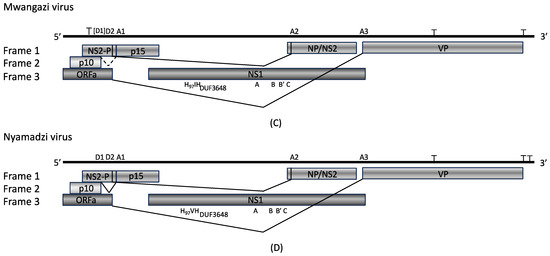
Figure 2.
Chaphamaparvoviruses from Zambia. Phylogenetic relationship of Mwangazi and Nyamadzi viruses to other viruses in the genus Chaphamaparvovirus based on NS1 (A) and VP (B) amino acid sequences. Phylogeny was reconstructed with the maximum likelihood method by applying a JTT+G4 substitution model for NS1 and a Q.yeast+F+G4 substitution model for VP1, selected by Model Finder implemented in IQtree; bootstrap values (>60%) resulting from 1000 pseudoreplicates are indicated at the respective nodes; the scale bars indicate the number of amino acid substitutions per site, and GenBank accession numbers are given next to the branches. The red font indicates viruses described in this study. (C) Schematic of Mwangazi virus genome organization. (D) Schematic of Nyamadzi virus genome organization. Gray shading indicates the three possible reading frames. Predicted major splice sites (donor sites D1, D2, and acceptor sites A1–A3), polyadenylation signals (T), SF3 helicase (H97), Walker A, B, B’, C, and domain of unknown function (DUF) 3648 motifs are indicated.
In the original publication [1], there was a mistake in Supplementary Table S4 as published. The names ’Mwangazi virus’ and ’Nyamadzi virus’ have been switched. The corrected version of Supplementary Table S4 appears below.

Table S4.
Conservation of splice sites in Mwangazi and Nyamadzi virus.
Table S4.
Conservation of splice sites in Mwangazi and Nyamadzi virus.
| Canonical donor consensus | mAG GTr | mAG GTr | |
| MKPV | D1 GAAGGAG GTGAGTCAG | D2 GCCGAAG GTAATTAAA | |
| CKPV | D1 GAAAGAG GTGAGTCGC | D2 CCTGAAG GTACTTATC | |
| Nyamadzi virus | D1 GGTGGAG GTGAGGGAG | D2 GCGGAAG GTACTTATT | |
| Mwangazi virus | D1 GCAGAAG GAGAATCGG | D2 CCAGAAG GTACTTATT | |
| D1a CCACAAG GTGCGAAAA | |||
| Canonical acceptor consensus | cAG Gk | cAG Gk | cAG Gk |
| MKPV | A1 CTTCTTACAG ATGTCTAT | A2 TTATTTGCAG AGCTAGTG | A3 TTATTTACAG AAACACTA |
| CKPV | A1 ATGCATGCAG ATGTCTAT | A2 TCTTTTGCAG AACTAGTG | A3 TTATTTACAG CAACAATA |
| Nyamadzi virus | A1 TAATTTACAG ATGTCTCT | A2 TTATTTGCAG AGCTAGTG | A3 TCATTTACAG AAACAATA |
| Mwangazi virus | A1 TCATCTACAG ATGTCTAT | A2 TTGTTTGCAG AATTAGTG | A3 TTATTTGCAG AACAAATA |
The authors state that the scientific conclusions are unaffected. The corrections were approved by the Academic Editor. The original publication has also been updated. The authors apologize for any inconvenience caused to the readers by these changes.
Reference
- Moonga, L.C.; Chipinga, J.; Collins, J.P.; Kapoor, V.; Saasa, N.; Nalubamba, K.S.; Hang’ombe, B.M.; Namangala, B.; Lundu, T.; Lu, X.-J.; et al. Application of a Sensitive Capture Sequencing Approach to Reservoir Surveillance Detects Novel Viruses in Zambian Wild Rodents. Viruses 2024, 16, 1754. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).