Detection and Molecular Characterization of Novel Porcine Parvovirus 8 Strains in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Sample Processing
2.3. PCR Detection of PPV8 and Sequencing
2.4. Sequence Alignment and Phylogenetic Analysis
2.5. Nucleotide Sequence Accession Numbers
3. Results
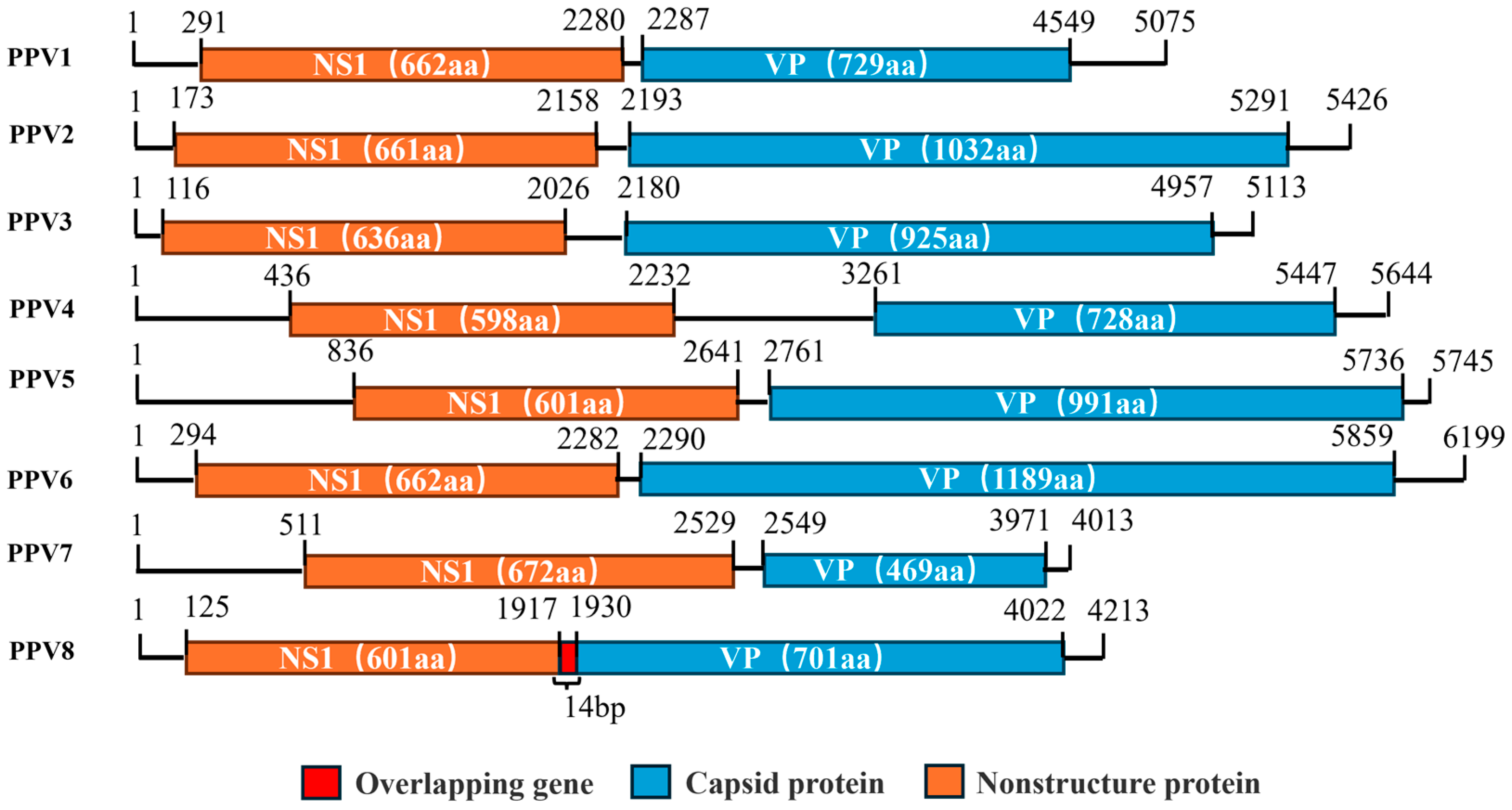
3.1. Genomic and Amino Acid Structure Analysis of PPV8
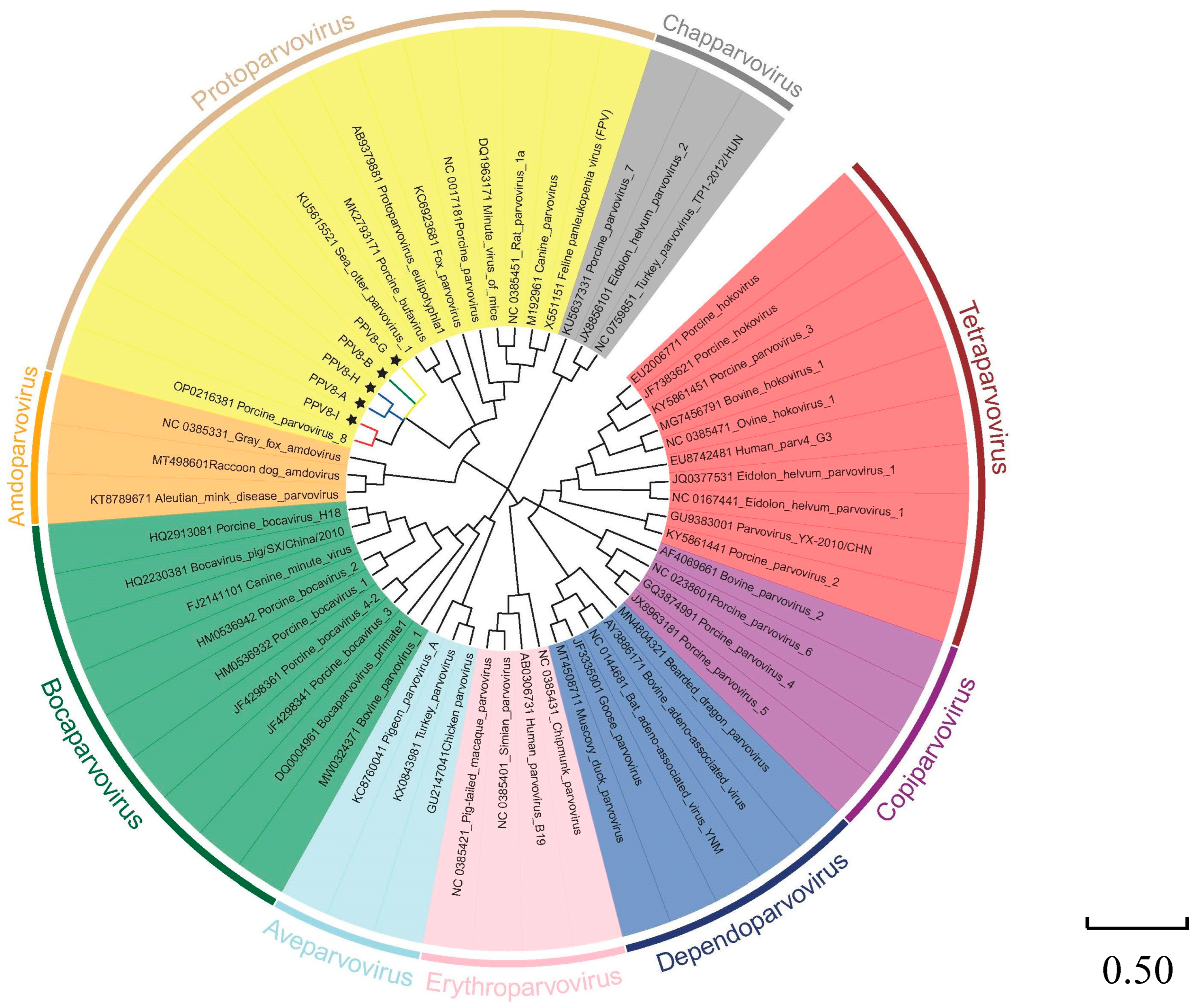
3.2. Phylogenetic Analysis of PPV8
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| PPV8-A | PPV8-B | PPV8-G | PPV8-H | PPV8-I | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome (nt) | NS (nt/aa) | VP (nt/aa) | Genome (nt) | NS (nt/aa) | VP (nt/aa) | Genome (nt) | NS (nt/aa) | VP (nt/aa) | Genome (nt) | NS (nt/aa) | VP (nt/aa) | Genome (nt) | NS (nt/aa) | VP (nt/aa) | ||
| Protoparvovirus | PPV1 | 44.44 | 47.89/36.67 | 52.15/34.33 | 44.54 | 47.40/31.80 | 52.56/34.70 | 44.50 | 47.59/31.66 | 52.69/34.43 | 44.54 | 47.59/31.66 | 52.88/34.15 | 44.64 | 47.54/31.66 | 52.92/34.56 |
| PPV8 | 98.36 | 98.67/99.17 | 97.69/98.57 | 98.69 | 98.84/99.00 | 98.53/99.43 | 98.53 | 98.73/99.00 | 98.44/98.86 | 97.77 | 99.00/99.67 | 98.20/99.26 | 98.84 | 98.89/99.67 | 98.48/99.14 | |
| Copiparvovirus | PPV4 | 29.91 | 36.98/19.14 | 40.05/13.93 | 30.26 | 37.29/19.44 | 39.72/13.91 | 30.22 | 37.24/20.64 | 40.04/14.19 | 30.35 | 37.05/19.29 | 40.24/14.04 | 30.29 | 37.93/19.29 | 39.99/13.91 |
| PPV5 | 32.37 | 39.61/16.92 | 29.37/12.70 | 32.90 | 40.38/19.46 | 30.28/12.75 | 31.59 | 39.50/19.91 | 30.27/12.30 | 32.41 | 39.97/19.31 | 30.78/12.40 | 31.93 | 39.9/19.31 | 30.84/12.40 | |
| PPV6 | 26.67 | 34.90/17.44 | 24.06/10.68 | 26.03 | 34.68/17.30 | 23.74/11.17 | 26.45 | 33.75/17.44 | 23.82/10.68 | 26.07 | 34.48/17.57 | 23.89/10.68 | 26.27 | 34.93/17.44 | 23.95/10.77 | |
| Tetraparvovirus | PPV2 | 30.48 | 34.84/28.22 | 26.67/11.75 | 30.55 | 35.23/14.34 | 26.63/12.94 | 30.59 | 34.97/14.48 | 27.51/11.80 | 30.30 | 34.58/14.48 | 26.89/11.70 | 30.40 | 35.01/14.48 | 28.41/12.09 |
| PPV3 | 31.26 | 34.45/14.57 | 29.55/12.09 | 31.02 | 34.50/14.71 | 29.47/12.18 | 31.30 | 34.50/14.71 | 29.56/12.29 | 31.23 | 34.98/14.57 | 30.88/12.39 | 31.52 | 35.16/14.57 | 29.77/12.18 | |
| Chapparvovirus | PPV7 | 37.62 | 37.36/8.74 | 29.69/9.86 | 38.24 | 37.66/8.89 | 29.72/8.26 | 38.80 | 37.41/8.70 | 29.68/9.69 | 37.43 | 37.28/8.89 | 30.00/10.26 | 38.05 | 37.46/9.79 | 29.76/10.11 |
| Virus Strain | Date | Origin | Accession No. |
|---|---|---|---|
| Muscovy duck parvovirus | 2020 | China | MT450871.1 |
| Bat adeno-associated virus YNM | 2010 | USA | NC_014468.1 |
| Bearded dragon parvovirus | 2019 | USA | MN480432.1 |
| Goose parvovirus | 2011 | China | JF333590.1 |
| Bovine adeno-associated virus | 2003 | USA | AY388617.1 |
| Bovine parvovirus 2 | 2001 | USA | AF406966.1 |
| Porcine parvovirus 4 | 2009 | USA | GQ387499.1 |
| Porcine parvovirus 5 | 2012 | USA | JX896318.1 |
| Porcine parvovirus 6 | 2013 | China | NC_023860.1 |
| Eidolon helvum parvovirus 1 | 2011 | The Netherlands | NC_016744.1 |
| Eidolon helvum parvovirus 1 | 2011 | The Netherlands | JQ037753.1 |
| Human parv4 G3 | 2008 | UK | EU874248.1 |
| Parvovirus YX-2010/CHN | 2010 | China | GU938300.1 |
| Porcine hokovirus | 2011 | Romania | JF738362.1 |
| Ovine hokovirus 1 | 2011 | China | NC_038547.1 |
| Bovine hokovirus 1 | 2018 | Brazil | MG745679.1 |
| Porcine hokovirus | 2007 | China | EU200677.1 |
| Porcine parvovirus 2 | 2017 | Brazil | KY586144.1 |
| Porcine parvovirus 3 | 2017 | Brazil | KY586145.1 |
| Human parvovirus B19 | 1999 | Japan | AB030673.1 |
| Chipmunk parvovirus | 2009 | USA | NC_038543.1 |
| Simian parvovirus | 2018 | USA | NC_038540.1 |
| Pig-tailed macaque parvovirus | 2018 | USA | NC_038542.1 |
| Raccoon dog amdovirus | 2020 | China | MT498601.1 |
| Gray fox amdovirus | 2011 | USA | NC_038533.1 |
| Aleutian mink disease parvovirus | 2015 | Canada | KT878967.1 |
| Minute virus of mice | 2005 | USA | DQ196317.1 |
| Canine parvovirus | 1988 | Swedeland | M19296.1 |
| Feline panleukopenia virus (FPV) | 1990 | Australia | X55115.1 |
| Sea otter parvovirus 1 | 2016 | USA | KU561552.1 |
| Porcine bufavirus | 2018 | China | MK279317.1 |
| Rat parvovirus 1a | 1997 | USA | NC_038545.1 |
| Protoparvovirus eulipotyphla1 | 2014 | Japan | AB937988.1 |
| Fox parvovirus | 2013 | The Netherlands | KC692368.1 |
| Porcine parvovirus | 1993 | USA | NC_001718. |
| Porcine parvovirus 8 | 2021 | China | OP021638.1 |
| Chicken parvovirus | 2009 | USA | GU214704.1 |
| Pigeon parvovirus A | 2013 | USA | KC876004.1 |
| Bovine parvovirus 1 | 2020 | China | MW032437.1 |
| Porcine bocavirus H18 | 2010 | China | HQ291308.1 |
| Bocaparvovirus primate1 | 2005 | Sweden | DQ000496.1 |
| Porcine bocavirus 2 | 2010 | China | HM053694.2 |
| Porcine bocavirus 1 | 2016 | China | HM053693.2 |
| Porcine bocavirus 3 | 2011 | China | JF429834.1 |
| Porcine bocavirus 4-2 | 2011 | China | JF429836.1 |
| Bocavirus pig/SX/China/2010 | 2010 | China | HQ223038.1 |
| Canine minute virus | 2008 | USA | FJ214110.1 |
| Turkey parvovirus | 2016 | China | KX084398.1 |
| Turkey parvovirus TP1-2012/HUN | 2018 | Hungary | KF880727.1 |
| Eidolon helvum parvovirus 2 | 2012 | UK | JX885610.1 |
| Porcine parvovirus 7 | 2016 | USA | KU563733.1 |
References
- Xie, C.; Tao, Y.; Zhang, Y.; Zhang, P.; Zhu, X.; Ha, Z.; Zhang, H.; Xie, Y.; Xia, X.; Jin, N.; et al. Codon Usage for Genetic Diversity, and Evolutionary Dynamics of Novel Porcine Parvoviruses 2 through 7 (PPV2–PPV7). Viruses 2022, 14, 170. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, X.; Ren, Y.; Cui, S.; Li, G.; Ren, X. Genome Sequence of Chinese Porcine Parvovirus Strain PPV2010. J. Virol. 2012, 86, 2379. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Chiorini, J.A.; Mukha, D.V.; Pintel, D.J.; Qiu, J.; Soderlund-Venermo, M.; Tattersall, P.; Tijssen, P.; Gatherer, D.; et al. The Family Parvoviridae. Arch. Virol. 2014, 159, 1239–1247. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef]
- Li, J.; Xiao, Y.; Qiu, M.; Li, X.; Li, S.; Lin, H.; Li, X.; Zhu, J.; Chen, N.; Jones, C.J. A Systematic Investigation Unveils High Coinfection Status of Porcine Parvovirus Types 1 through 7 in China from 2016 to 2020. Microbiol. Spectr. 2021, 9, e01294-21. [Google Scholar] [CrossRef]
- Wen, S.; Song, Y.; Lv, X.; Meng, X.; Liu, K.; Yang, J.; Diao, F.; He, J.; Huo, X.; Chen, Z.; et al. Detection and Molecular Characterization of Porcine Parvovirus 7 in Eastern Inner Mongolia Autonomous Region, China. Front. Vet. Sci. 2022, 9, 930123. [Google Scholar] [CrossRef]
- Xing, X.; Zhou, H.; Tong, L.; Chen, Y.; Sun, Y.; Wang, H.; Zhang, G. First Identification of Porcine Parvovirus 7 in China. Arch. Virol. 2018, 163, 209–213. [Google Scholar] [CrossRef]
- Streck, A.F.; Truyen, U. Porcine Parvovirus. Curr. Issues Mol. Biol. 2020, 37, 33–45. [Google Scholar] [CrossRef]
- Afolabi, K.O.; Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Prevalence of Porcine Parvoviruses in Some South African Swine Herds with Background of Porcine Circovirus Type 2 Infection. Acta Trop. 2019, 190, 37–44. [Google Scholar] [CrossRef]
- Streck, A.F.; Canal, C.W.; Truyen, U. Molecular Epidemiology Evolution of Porcine Parvoviruses. In Infection, Genetics and Evolution; Elsevier: Amsterdam, The Netherlands, 2015; pp. 300–306. [Google Scholar] [CrossRef]
- Zhao, K.; Hu, R.; Ni, J.; Liang, J.; He, X.; Du, Y.; Xu, Y.; Zhao, B.; Zhang, Q.; Li, C. Establishment of a Porcine Parvovirus (PPV) LAMP Visual Rapid Detection Method. J. Virol. Methods 2020, 284, 113924. [Google Scholar] [CrossRef]
- Oh, W.T.; Kim, R.Y.; Nguyen, V.G.; Chung, H.C.; Park, B.K. Perspectives on the Evolution of Porcine Parvovirus. Viruses 2017, 9, 196. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Coetzee, L.M.; Hemberger, M.Y.; Khaiseb, S.; Cattoli, G.; Dundon, W.G. Evidence Indicating Transmission of Porcine Parvovirus 1 between Warthogs and Domestic Pigs in Namibia. Vet. Res. Commun. 2023, 47, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Hijikata, M.; Abe, K.; Win, K.M.; Shimizu, Y.K.; Keicho, N.; Yoshikura, H. Identification of New Parvovirus DNA Sequence in Swine Sera from Myanmar. Jpn. J. Infect. Dis. 2001, 54, 244–245. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermudez, D.S.; Mogollon, J.D.; Franco-Rodriguez, C.; Jaime, J. The Novel Porcine Parvoviruses: Current State of Knowledge and Their Possible Implications in Clinical Syndromes in Pigs. Viruses 2023, 15, 2398. [Google Scholar] [CrossRef]
- Nelsen, A.; Lin, C.M.; Hause, B.M. Porcine Parvovirus 2 Is Predominantly Associated with Macrophages in Porcine Respiratory Disease Complex. Front. Vet. Sci. 2021, 8, 726884. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Tse, H.; Fu, C.T.Y.; Au, W.K.; Chen, X.C.; Tsoi, H.W.; Tsang, T.H.F.; Chan, J.S.Y.; Tsang, D.N.C.; et al. Identification of Novel Porcine and Bovine Parvoviruses Closely Related to Human Parvovirus 4. J. Gen. Virol. 2008, 89, 1840–1848. [Google Scholar] [CrossRef]
- Palinski, R.M.; Mitra, N.; Hause, B.M. Discovery of a Novel Parvovirinae Virus, Porcine Parvovirus 7, by Metagenomic Sequencing of Porcine Rectal Swabs. Virus Genes 2016, 52, 564–567. [Google Scholar] [CrossRef]
- Komina, A.; Anoyatbekova, A.; Krasnikov, N.; Yuzhakov, A. Identification and in Vitro Characterization of a Novel Porcine Parvovirus 6 in Russia. Vet. Res. Commun. 2024, 48, 417–425. [Google Scholar] [CrossRef]
- Kim, S.-C.; Kim, J.-H.; Kim, J.-Y.; Park, G.-S.; Jeong, C.-G.; Kim, W.-I. Prevalence of Porcine Parvovirus 1 through 7 (PPV1-PPV7) and Co-Factor Association with PCV2 and PRRSV in Korea. BMC Vet. Res. 2022, 18, 133. [Google Scholar] [CrossRef]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-Infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Q.; Shan, H.; Cao, Z.; Huang, J. Genome Characteristics of Atypical Porcine Pestivirus from Abortion Cases in Shandong Province, China. Virol. J. 2023, 20, 282. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Guzowska, M.; Stadejek, T. Detection Patterns of Porcine Parvovirus (PPV) and Novel Porcine Parvoviruses 2 through 6 (PPV2–PPV6) in Polish Swine Farms. Viruses 2019, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, K.K.; Wang, J.; Wang, X.P.; Zhao, L.; Sun, P.; Li, Y.D. Detection and Molecular Characterization of Novel Porcine Parvovirus 7 in Anhui Province from Central-Eastern China. Infect. Genet. Evol. 2019, 71, 31–35. [Google Scholar] [CrossRef]
- Zhao, W.; Sun, Y.; Guo, L.; Zhang, Y.; Huang, B.; Tian, K. Genome Sequence of Chinese Porcine Parvovirus Strain BJ2. Microbiol. Resour. Announc. 2023, 12, e00094-23. [Google Scholar] [CrossRef]
- Lyu, Z.; Zhang, X.; Xue, S.; Yang, X.; Liu, J.; Fan, K.; Dai, A. Detection and Genetic Evolution Analysis of Porcine Parvovirus Type 7 (PPV7) in Fujian Province. Infect. Genet. Evol. 2023, 115, 105515. [Google Scholar] [CrossRef]
- Deng, S.; Zhiyong, H.; Mengjiao, Z.; Shuangqi, F.; Jingyuan, Z.; Yunzhen, H.; Hailuan, X.; Jinding, C. Isolation and Phylogenetic Analysis of a New Porcine Parvovirus Strain GD2013 in China. J. Virol. Methods 2020, 275, 113748. [Google Scholar] [CrossRef]
- Parthiban, S.; Sowndhraya, R.K.V.; Raja, P.; Parthiban, M.; Ramesh, A.; Raj, G.D.; Senthilkumar, K.; Balasubramanyam, D.; Hemalatha, S.; Bharathi, R.; et al. Molecular Detection of Porcine Parvovirus 1–Associated Reproductive Failure in Southern India. Trop. Anim. Health Prod. 2022, 54, 195. [Google Scholar] [CrossRef]
- Thuy, N.T.D.; Trung, N.T.; Dung, T.Q.; Khoa, D.V.A.; Thuy, D.T.N.; Opriessnig, T. First Investigation of the Prevalence of Parvoviruses in Slaughterhouse Pigs and Genomic Characterization of Ungulate Copiparvovirus 2 in Vietnam. Arch. Virol. 2021, 166, 779–788. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Ni, J.; Qiao, C.; Han, X.; Han, T.; Kang, W.; Zi, Z.; Cao, Z.; Zhai, X.; Cai, X. Identification and Genomic Characterization of a Novel Porcine Parvovirus (PPV6) in China. Virol. J. 2014, 11, 203. [Google Scholar] [CrossRef]
- Li, L.-F.; Chen, N.; Zhang, X.; Huang, L.; Guo, Y.; Yan, G.; Chen, S.; Han, H.; Li, J.; Zhang, H.; et al. Identification and Genomic Characterization of a Novel Porcine Parvovirus in China. Front. Vet. Sci. 2022, 9, 1009103. [Google Scholar]
- Hargitai, R.; Boros, Á.; Pankovics, P.; Mátics, R.; Altan, E.; Delwart, E.; Reuter, G. Detection and Genetic Characterization of a Novel Parvovirus (Family Parvoviridae) in Barn Owls (Tyto Alba) in Hungary. Arch. Virol. 2021, 166, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Zádori, Z.; Szelei, J.; Lacoste, M.-C.; Li, Y.; Gariépy, S.; Raymond, P.; Allaire, M.; Nabi, I.R.; Tijssen, P. A Viral Phospholipase A 2 Is Required for Parvovirus Infectivity (Takahashi et al., 1998b) Remain to Be Confirmed (Moore, 2000). Dev. Cell 2001, 1, 291–302. [Google Scholar] [PubMed]
- Zhang, X.; Zheng, C.; Lv, Z.; Xue, S.; Chen, Y.; Liu, Y.; Huang, X.; Luo, G.; Yang, X.; Dai, A. Genetic and Epidemic Characteristics of Porcine Parvovirus 7 in the Fujian and Guangdong Regions of Southern China. Front. Vet. Sci. 2022, 9, 949764. [Google Scholar] [CrossRef]
- Park, G.N.; Song, S.; Cha, R.M.; Choe, S.E.; Shin, J.; Kim, S.Y.; Hyun, B.H.; Park, B.K.; An, D.J. Genetic Analysis of Porcine Parvoviruses Detected in South Korean Wild Boars. Arch. Virol. 2021, 166, 2249–2254. [Google Scholar] [CrossRef]



| Primer | Sequence (5′→3′) | PCR Conditions | |||
|---|---|---|---|---|---|
| Predenaturation | Denaturation, Annealing, Extension | Final Extension | |||
| PPV8 | OutF | TGTTGGTTTGCACCTAGCG | 98 °C 30 s | 98 °C 10 s, 56 °C 15 s, 72 °C 15 s | 72 °C 2 min |
| OutR | TGATGAGATGGTGGAACGC | ||||
| PPV8 | InF | TCCAAGTTGCCCTAGACAGC | 98 °C 30 s | 98 °C 10 s, 56 °C 15 s, 72 °C 15 s | 72 °C 2 min |
| InR | GCCTCGTACATGTGGACCTC | ||||
| ① | 119F | GAAGAAGAATCTGATTAAGGTAAGCC | 94 °C 5 min | 94 °C 30 s, 56 °C 30 s, 72 °C 30 s | 72 °C 7 min |
| 712R | GGATAGTTAATAGTGATAGAAGGAGC | ||||
| ② | 684F | TTACTAACTCAATGGTCAATGCTCCTTC | 94 °C 5 min | 94 °C 30 s, 52 °C 30 s, 72 °C 30 s | 72 °C 7 min |
| 2037R | GATTGTCTTCTAAGGACTGGC | ||||
| ③ | 1960F | CCAATACAGACTCACATCTTCTCTTGA | 94 °C 5 min | 94 °C 30 s, 60 °C 30 s, 72 °C 30 s | 72 °C 7 min |
| 3541R | TGGTTTGTTGTGACATCTCTGCTTCTAA | ||||
| ④ | 3488F | CTCATCATCCAAGAGAAGCTC | 94 °C 5 min | 94 °C 30 s, 55 °C 30 s, 72 °C 30 s | 72 °C 7 min |
| 4311R | ACCCAAGAGCGTTTTCAAAGA | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Hu, Y.; Qin, Y.; Li, Y.; Zhang, X.; Huang, H.; Liu, M.; Zheng, Y.; Lu, X.; Wang, Q.; et al. Detection and Molecular Characterization of Novel Porcine Parvovirus 8 Strains in China. Viruses 2025, 17, 543. https://doi.org/10.3390/v17040543
Chen W, Hu Y, Qin Y, Li Y, Zhang X, Huang H, Liu M, Zheng Y, Lu X, Wang Q, et al. Detection and Molecular Characterization of Novel Porcine Parvovirus 8 Strains in China. Viruses. 2025; 17(4):543. https://doi.org/10.3390/v17040543
Chicago/Turabian StyleChen, Wei, Yanqing Hu, Yan Qin, Yuying Li, Xinyu Zhang, Haixin Huang, Mengjia Liu, Yuping Zheng, Xuelian Lu, Qiaoqiong Wang, and et al. 2025. "Detection and Molecular Characterization of Novel Porcine Parvovirus 8 Strains in China" Viruses 17, no. 4: 543. https://doi.org/10.3390/v17040543
APA StyleChen, W., Hu, Y., Qin, Y., Li, Y., Zhang, X., Huang, H., Liu, M., Zheng, Y., Lu, X., Wang, Q., Yang, J., Kang, L., Xie, L., Zhao, B., Lan, T., & Sun, W. (2025). Detection and Molecular Characterization of Novel Porcine Parvovirus 8 Strains in China. Viruses, 17(4), 543. https://doi.org/10.3390/v17040543





