The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Specimen Collection
2.2. Virus Detection and Subtype Identification
2.3. Genome Sequencing and Assembly
2.4. Phylogenetic Analysis, Estimation of Evolution Rate, and Positive Selection Analysis
2.5. Vaccine Efficacy and Antiviral Resistance
3. Results
3.1. The Prevalence of Influenza A(H3N2) Viruses in Hangzhou from 2010 to 2022
3.2. Phylogenetic Analysis and Clade Identification of Influenza A(H3N2) Viruses
3.3. Shannon Entropy Diversity
3.4. Evolutionary Rates and Selection Pressure Analysis
3.5. Antigenic Characteristics and Vaccine Strain Match Analysis
4. Discussion
5. Limitations and Strengths
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HPD | Highest probability density |
| dN/dS | Non-synonymous/synonymous rate ratio |
| SLAC | Single-likelihood ancestor counting |
| MEME | Mixed effects model of evolution |
| FUBAR | Fast, unconstrained Bayesian approximation |
References
- Lei, R.; Tan, T.J.C.; Garcia, A.H.; Wang, Y.; Diefenbacher, M.; Teo, C.; Gopan, G.; Dargani, Z.T.; Teo, Q.W.; Graham, C.S.; et al. Prevalence and mechanisms of evolutionary contingency in human influenza H3N2 neuraminidase. Nat. Commun. 2022, 13, 6443. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Ke, C.; Fang, S.; Li, J.; Song, H.; Li, X.; Hu, T.; Wu, J.; Chen, T.; Yi, L.; et al. Co-circulation and persistence of multiple A/H3N2 influenza variants in China. Emerg. Microbes Infect. 2019, 8, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Uyeki, T.M. High-risk Groups for Influenza Complications. JAMA 2020, 324, 2334. [Google Scholar] [CrossRef] [PubMed]
- Jallow, M.M.; Barry, M.A.; Ndiaye, N.K.; Touré, C.T.; Kiori, D.; Sagne, S.N.; Sy, S.; Goudiaby, D.; Niang, M.N.; Diagne, M.M.; et al. Genetic and antigenic characterization of influenza A(H3N2) virus after 13 consecutive years of influenza surveillance in Senegal, 2010–2022. J. Med. Virol. 2024, 96, e70010. [Google Scholar] [CrossRef]
- Dorji, T.; Dorji, K.; Gyeltshen, S. Evolution of Influenza A(H3N2) Viruses in Bhutan for Two Consecutive Years, 2022 and 2023. Influ. Other Respir. Viruses 2024, 18, e70028. [Google Scholar] [CrossRef]
- Choi, Y.J.; Song, J.Y.; Wie, S.-H.; Choi, W.S.; Lee, J.; Lee, J.-S.; Kim, Y.K.; Kim, S.W.; Lee, S.H.; Park, K.-H.; et al. Real-world effectiveness of influenza vaccine over a decade during the 2011–2021 seasons—Implications of vaccine mismatch. Vaccine 2024, 42, 126381. [Google Scholar] [CrossRef]
- Shao, T.; Li, J.; Yu, X.; Kou, Y.; Zhou, Y.; Qian, X. Progressive antigenic drift and phylogeny of human influenza A (H3N2) virus over five consecutive seasons (2009–2013) in Hangzhou, China. Int. J. Infect. Dis. 2014, 29, 190–193. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Zhou, Y.; Kou, Y.; Yu, X.; Zheng, Z.; Yang, X.; Wang, H. Interim estimates of divergence date and vaccine strain match of human influenza A (H3N2) virus from systematic influenza surveillance (2010–2015) in Hangzhou, southeast of China. Int. J. Infect. Dis. 2015, 40, 17–24. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Phyu, W.W.; Saito, R.; Kyaw, Y.; Lin, N.; Win, S.M.K.; Win, N.C.; Di Ja, L.; Htwe, K.T.Z.; Aung, T.Z.; Tin, H.H.; et al. Evolutionary Dynamics of Whole-Genome Influenza A/H3N2 Viruses Isolated in Myanmar from 2015 to 2019. Viruses 2022, 14, 2414. [Google Scholar] [CrossRef]
- Pan, K.; Deem, M.W. Quantifying selection and diversity in viruses by entropy methods, with application to the haemagglutinin of H3N2 influenza. J. R. Soc. Interface 2011, 8, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Suptawiwat, O.; Ninpan, K.; Boonarkart, C.; Ruangrung, K.; Auewarakul, P. Evolutionary dynamic of antigenic residues on influenza B hemagglutinin. Virology 2017, 502, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Muñoz, E.T.; Deem, M.W. Epitope analysis for influenza vaccine design. Vaccine 2005, 23, 1144–1148. [Google Scholar] [CrossRef]
- E Bonomo, M.; Deem, M.W. Predicting Influenza H3N2 Vaccine Efficacy From Evolution of the Dominant Epitope. Clin. Infect. Dis. 2018, 67, 1129–1131. [Google Scholar] [CrossRef]
- Tamerius, J.D.; Shaman, J.; Alonso, W.J.; Bloom-Feshbach, K.; Uejio, C.K.; Comrie, A.; Viboud, C. Environmental predictors of seasonal influenza epidemics across temperate and tropical climates. PLoS Pathog. 2013, 9, e1003194. [Google Scholar] [CrossRef]
- Zhao, X.; Gu, Y.; Tang, X.; Jiang, C.; Fang, F.; Chu, W.; Tao, L.; Zhang, X.; Chen, M.; Wu, H.; et al. Whole-genome analysis of circulating influenza A virus (H3N2) strains in Shanghai, China from 2005 to 2023. Emerg. Microbes Infect. 2024, 13, 2396867. [Google Scholar] [CrossRef]
- Zhang, B.; Huang, W.; Pei, S.; Zeng, J.; Shen, W.; Wang, D.; Wang, G.; Chen, T.; Yang, L.; Cheng, P.; et al. Mechanisms for the circulation of influenza A (H3N2) in China: A spatiotemporal modelling study. PLoS Pathog. 2022, 18, e1011046. [Google Scholar] [CrossRef]
- Li, H.; Prevention, B.C.C.F.D.C.A.; Gao, G.F. Highly Effective Stratified Hub-and-Spoke Non-Pharmaceutical Intervention Provides New Insight into the Prevention of COVID-19 Transmission Comment. China CDC Wkly. 2020, 2, 985–986. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Prevention, B.C.C.F.D.C.A.; Gu, Y.; Liu, H.; Zhou, L. Epidemiology Features and Effectiveness of Vaccination and Non-Pharmaceutical Interventions of Delta and Lambda SARS-CoV-2 Variants. China CDC Wkly. 2021, 3, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Walker, G.; Kim, K.W.; Stelzer-Braid, S.; Scotch, M.; Rawlinson, W.D. The resurgence of influenza A/H3N2 virus in Australia after the relaxation of COVID-19 restrictions during the 2022 season. J. Med. Virol. 2024, 96, e29922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhou, J.; Ni, R.; Zhao, X.; Chen, Y.; Sun, Y.; Liu, Z.; Han, X.; Luo, C.; Fu, X.; et al. Genomic Analyses Uncover Evolutionary Features of Influenza A/H3N2 Viruses in Yunnan Province, China, from 2017 to 2022. Viruses 2024, 16, 138. [Google Scholar] [CrossRef]
- Shin, W.-J.; Seong, B.L. Novel antiviral drug discovery strategies to tackle drug-resistant mutants of influenza virus strains. Expert Opin. Drug Discov. 2018, 14, 153–168. [Google Scholar] [CrossRef]
- Xu, J.; Luo, Q.; Huang, Y.; Li, J.; Ye, W.; Yan, R.; Zhou, X.; He, Z.; Liu, G.; Zhu, Q. Influenza neuraminidase mutations and resistance to neuraminidase inhibitors. Emerg. Microbes Infect. 2024, 13, 2429627. [Google Scholar] [CrossRef]
- Wang, P.; Guo, J.; Zhou, Y.; Zhu, M.; Fang, S.; Sun, F.; Huang, C.; Zhu, Y.; Zhou, H.; Pan, B.; et al. The C-terminal amino acid motifs of NS1 protein affect the replication and virulence of naturally NS-truncated H1N1 canine influenza virus. Emerg. Microbes Infect. 2024, 13, 2400546. [Google Scholar] [CrossRef]
- Jiang, Y.; Dou, H.; Wang, X.; Song, T.; Jia, Y.; Yue, Y.; Li, L.; He, F.; Kong, L.; Wu, Z.; et al. Analysis of seasonal H3N2 influenza virus epidemic characteristics and whole genome features in Jining City from 2018 to 2023. J. Med. Virol. 2024, 96, e29846. [Google Scholar] [CrossRef]
- Fall, A.; Han, L.; Yunker, M.; Gong, Y.-N.; Li, T.-J.; Norton, J.M.; Abdullah, O.; E Rothman, R.; Fenstermacher, K.Z.J.; Morris, C.P.; et al. Evolution of Influenza A (H3N2) Viruses in 2 Consecutive Seasons of Genomic Surveillance, 2021–2023. Open Forum Infect. Dis. 2023, 10, ofad577. [Google Scholar] [CrossRef]
- Galli, C.; Pellegrinelli, L.; Giardina, F.; Ferrari, G.; Renteria, S.C.U.; Novazzi, F.; Masi, E.; Pagani, E.; Piccirilli, G.; Mauro, M.V.; et al. On the lookout for influenza viruses in Italy during the 2021–2022 season: Along came A (H3N2) viruses with a new phylogenetic makeup of their hemagglutinin. Virus Res. 2022, 324, 199033. [Google Scholar] [CrossRef]
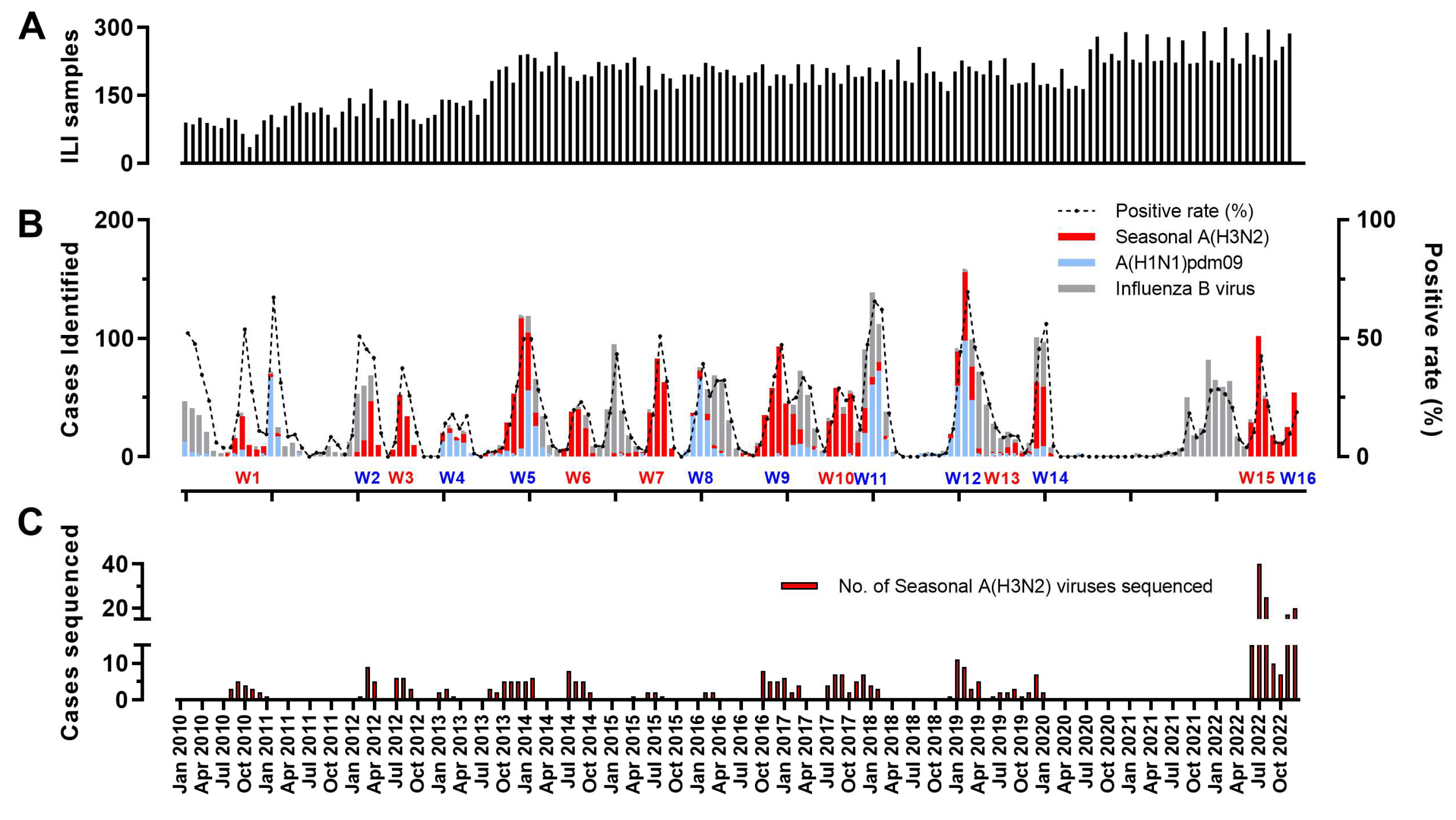
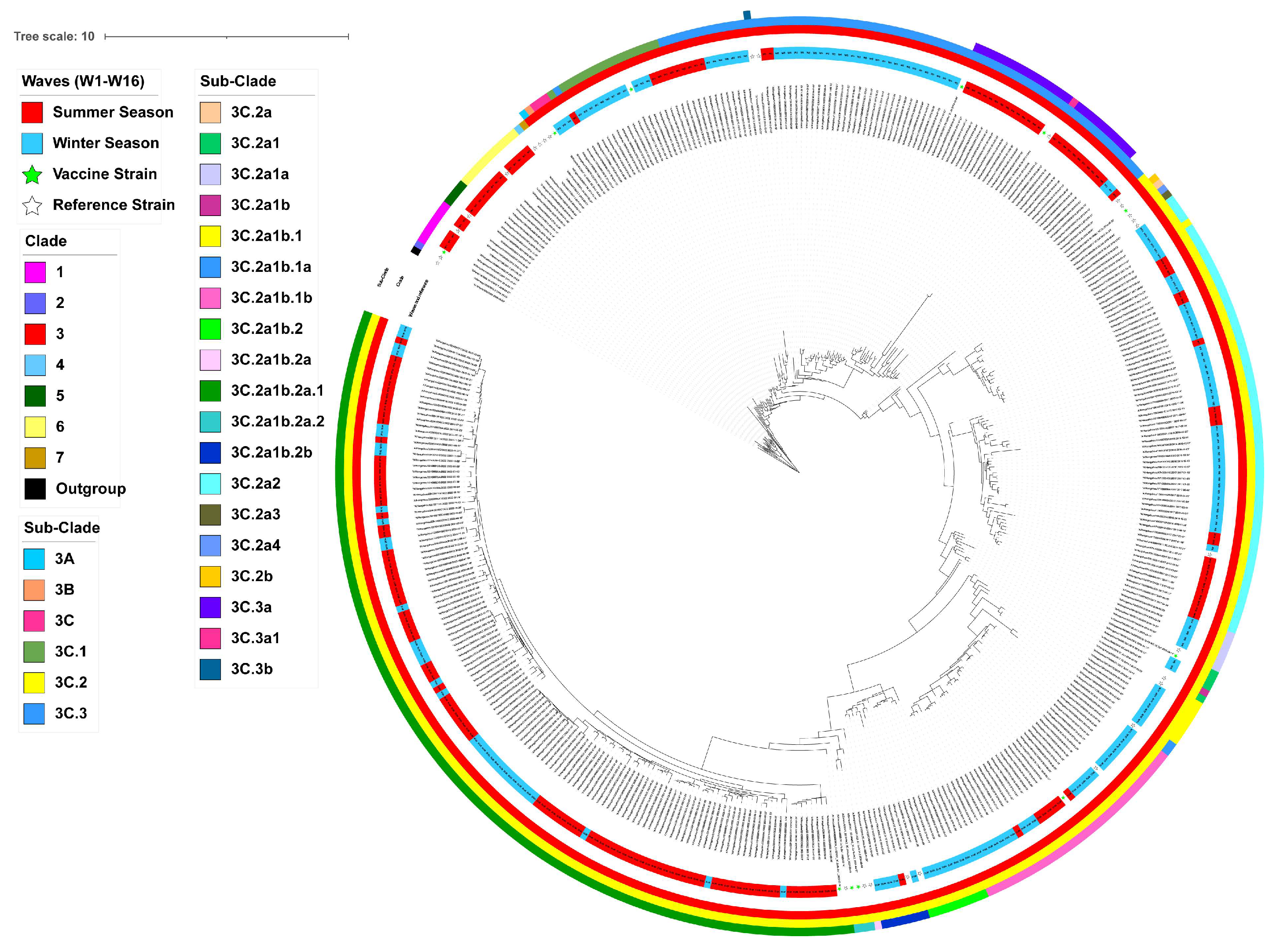
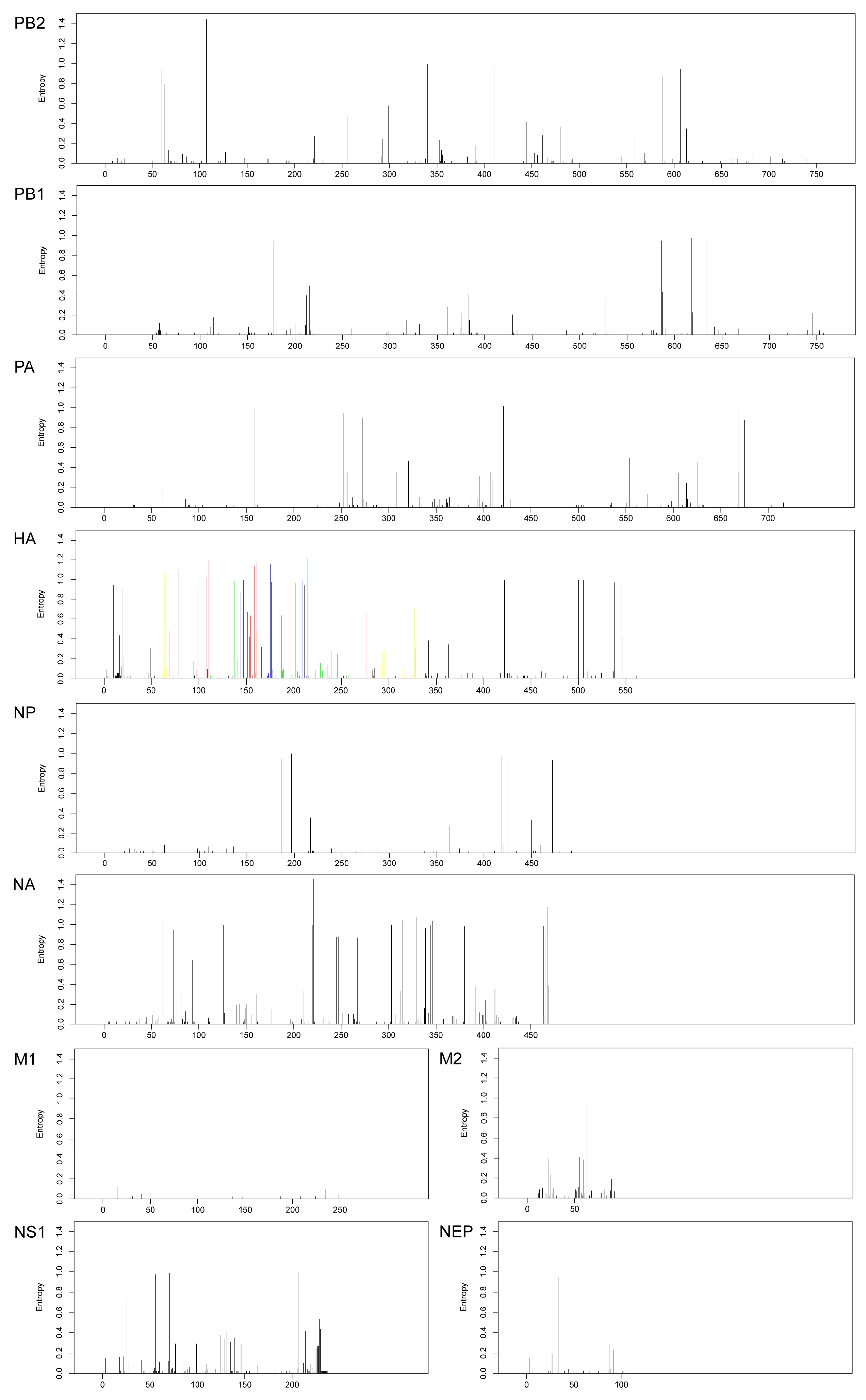
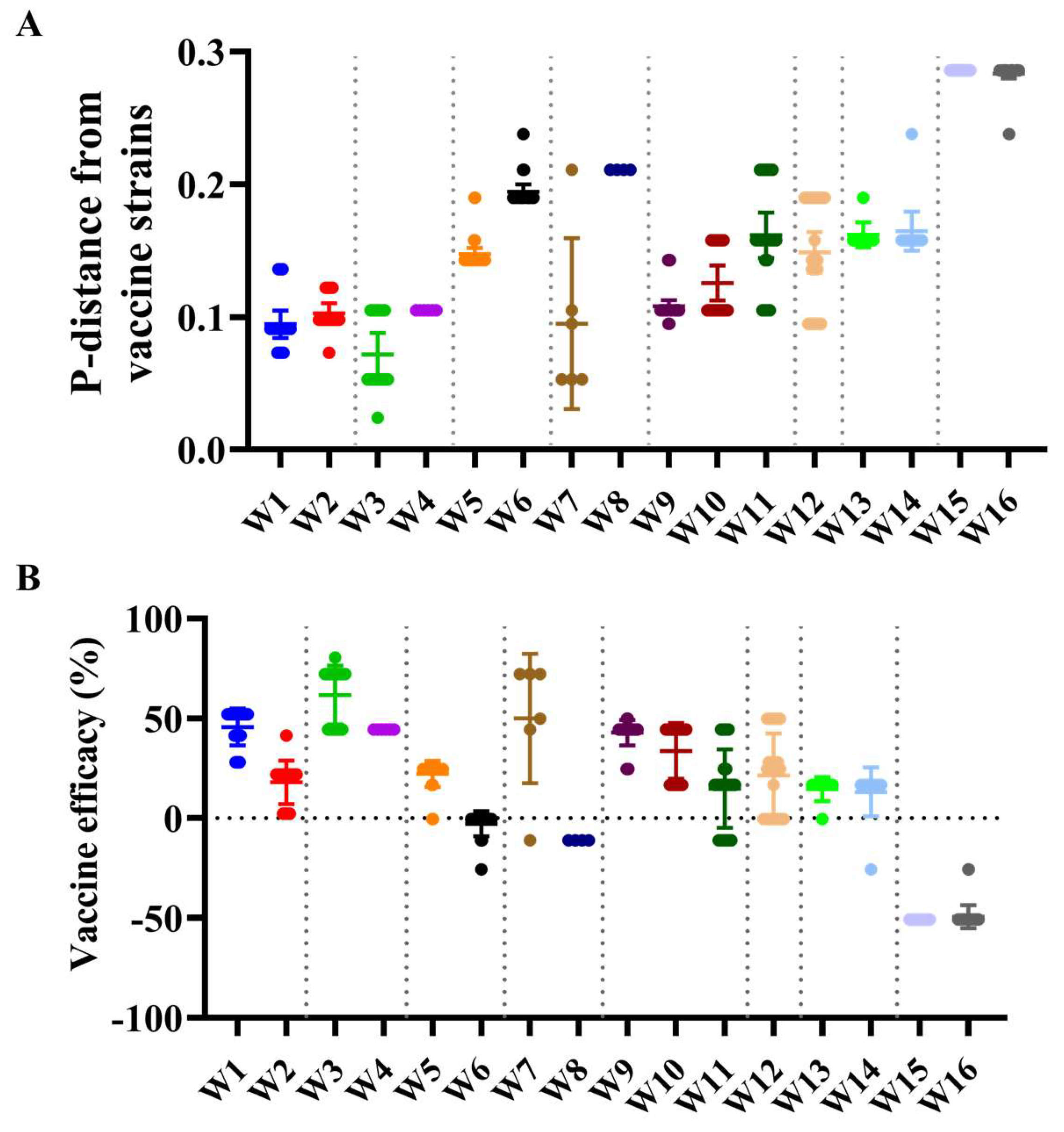
| Segments (n = 367) | Evolutionary Rate (95% HPD) | Protein | dN/dS | Sites Under Diversifying Positive Selection (Amino Acid Position) | ||
|---|---|---|---|---|---|---|
| SLAC a | MEME a | FUBAR b | ||||
| HA | 3.96 × 10−3 (3.53–4.38) | HA | 0.218 | 21, 176, 277 | 21, 26, 53, 110, 147, 176, 209 | 21, 69, 110, 176, 209, 277 |
| NA | 3.77 × 10−3 (3.31–4.24) | NA | 0.266 | 93, 380 | 45, 93, 370, 380 | 93, 380 |
| NS | 3.65 × 10−3 (2.92–4.42) | NS1 | 0.346 | None | None | 22, 71, 230 |
| NEP | 0.205 | None | None | None | ||
| PB1 | 3.09 × 10−3 (2.67–3.57) | PB1 | 0.072 | None | None | None |
| PB2 | 2.91 × 10−3 (2.50–3.30) | PB2 | 0.076 | 67 | 67, 513 | 67 |
| PA | 2.81 × 10−3 (2.47–3.16) | PA | 0.094 | None | 32, 109, 321, 500, 628 | 272, 321 |
| MP | 2.63 × 10−3 (2.16–3.10) | M1 | 0.037 | None | None | None |
| M2 | 0.464 | None | 28, 68 | 28, 59, 68 | ||
| NP | 2.62 × 10−3 (2.21–3.06) | NP | 0.060 | None | None | 270 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Cao, F.; Yu, Y.; Yu, X.; Zhou, Y.; Cheng, S.; Qiu, X.; Ao, L.; Yang, X.; Sun, Z.; et al. The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity. Viruses 2025, 17, 526. https://doi.org/10.3390/v17040526
Zheng X, Cao F, Yu Y, Yu X, Zhou Y, Cheng S, Qiu X, Ao L, Yang X, Sun Z, et al. The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity. Viruses. 2025; 17(4):526. https://doi.org/10.3390/v17040526
Chicago/Turabian StyleZheng, Xueling, Feifei Cao, Yue Yu, Xinfen Yu, Yinyan Zhou, Shi Cheng, Xiaofeng Qiu, Lijiao Ao, Xuhui Yang, Zhou Sun, and et al. 2025. "The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity" Viruses 17, no. 4: 526. https://doi.org/10.3390/v17040526
APA StyleZheng, X., Cao, F., Yu, Y., Yu, X., Zhou, Y., Cheng, S., Qiu, X., Ao, L., Yang, X., Sun, Z., & Li, J. (2025). The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity. Viruses, 17(4), 526. https://doi.org/10.3390/v17040526






