Deletion of the Human Cytomegalovirus US2 to US11 Gene Family Members Impairs the Type-I Interferon Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, BAC-Cloning, Western Blots, and Viruses
2.2. Infection of HFF
2.3. RNA-Seq Analysis
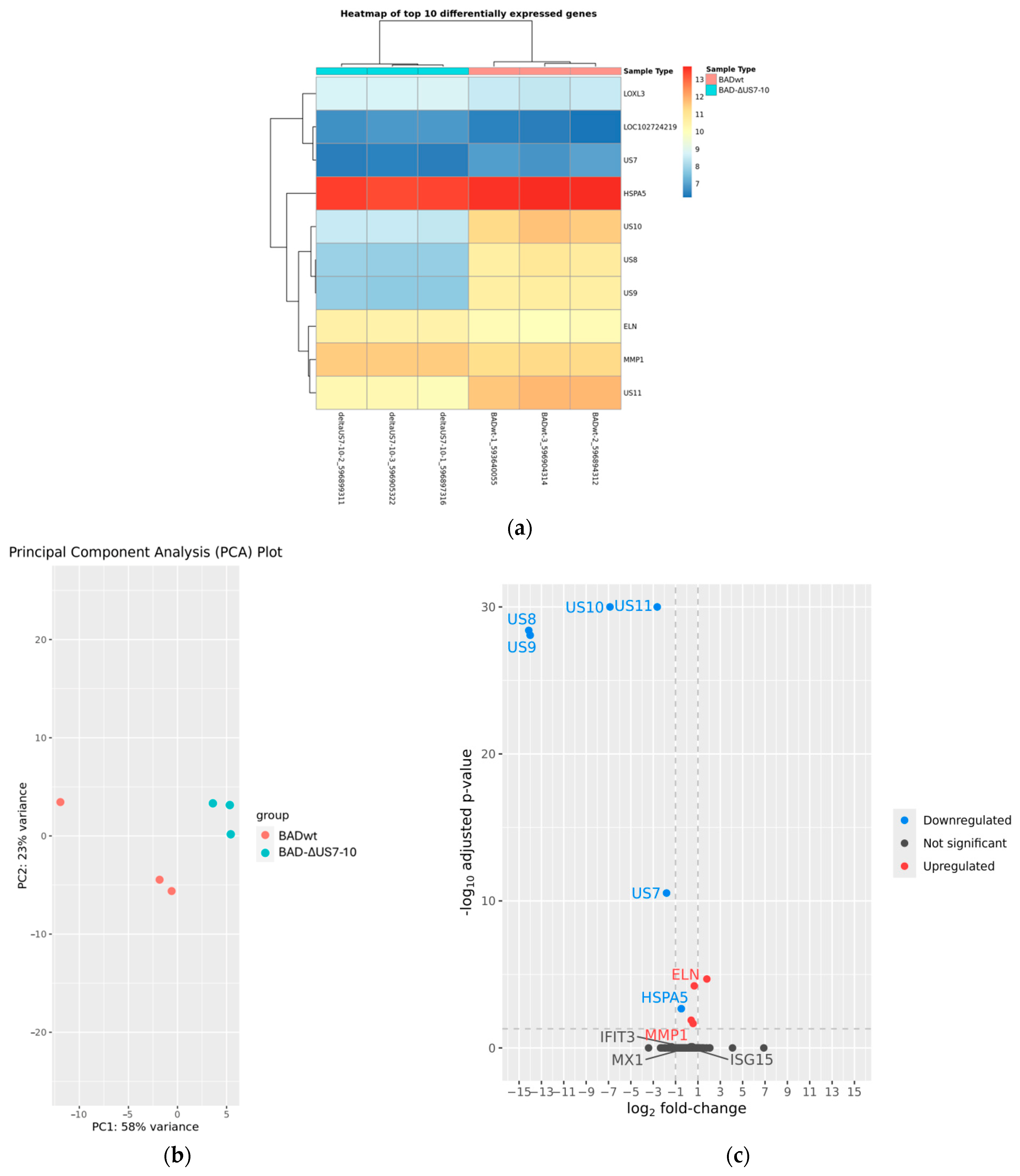
3. Results
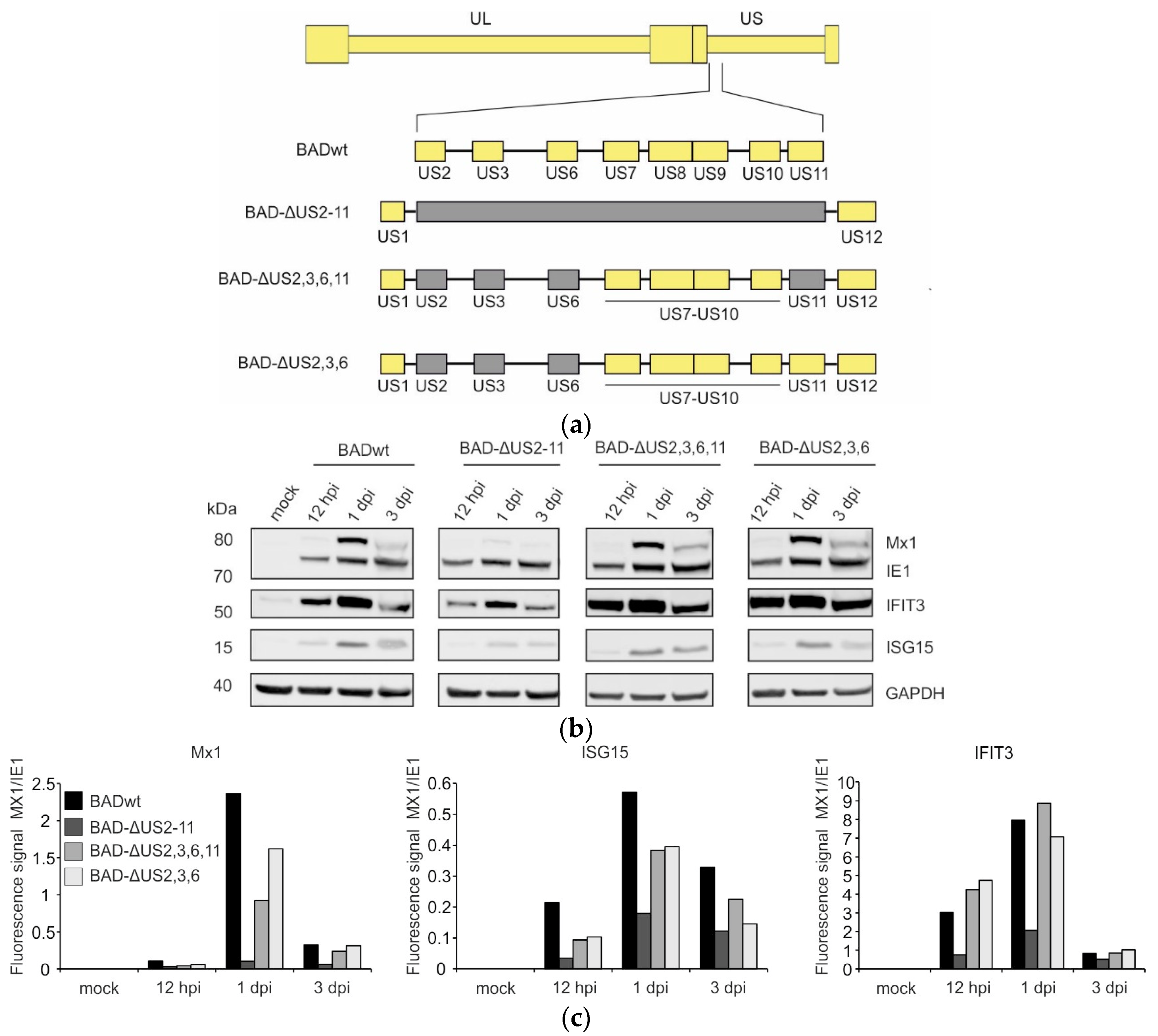
3.1. Deletion of Immune Evasion Genes US2–US11 Leads to Impairment in the Induction of ISGs
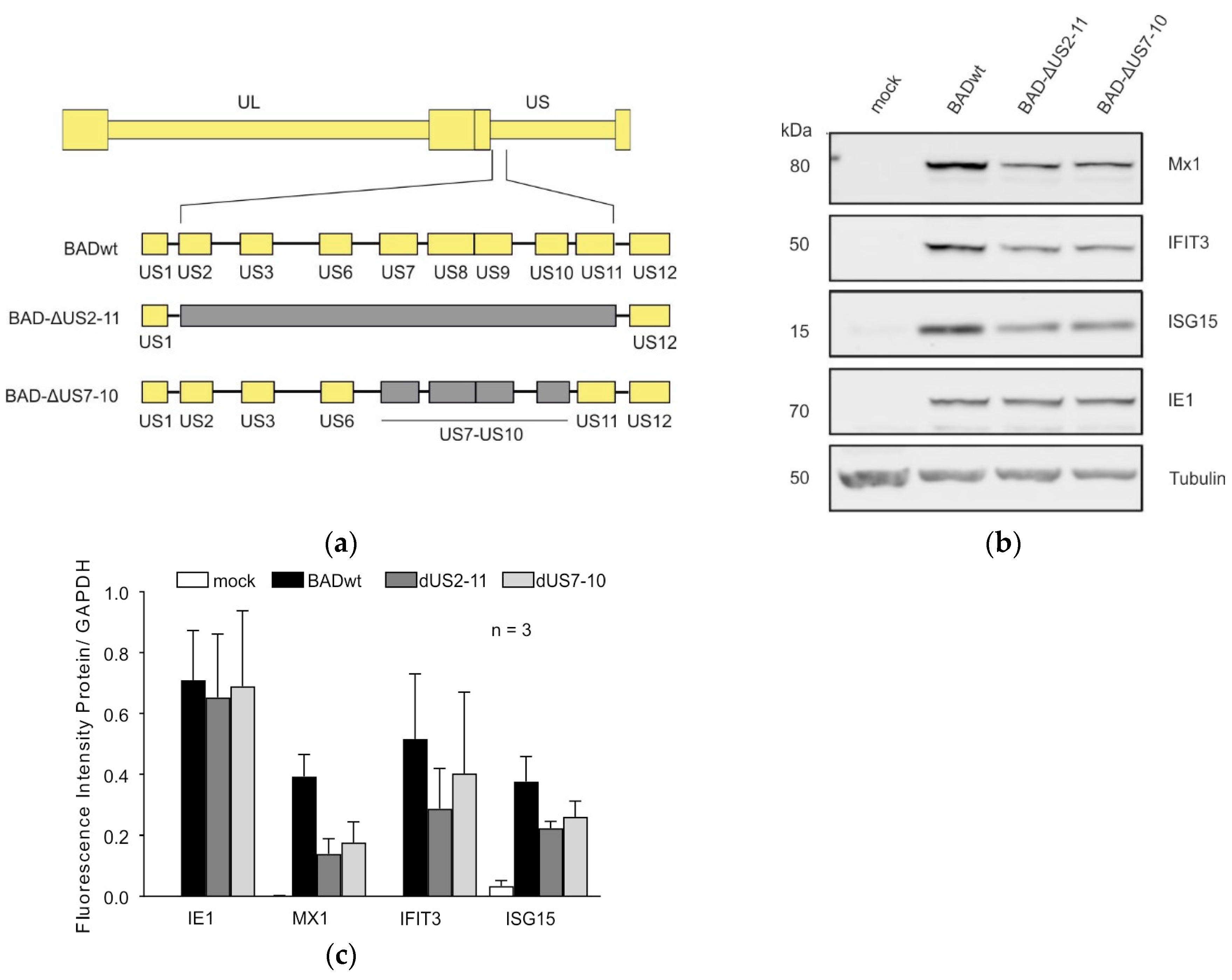
3.2. Deletion of Immune Evasion Genes US7–US10 Impairs the Induction of ISGs
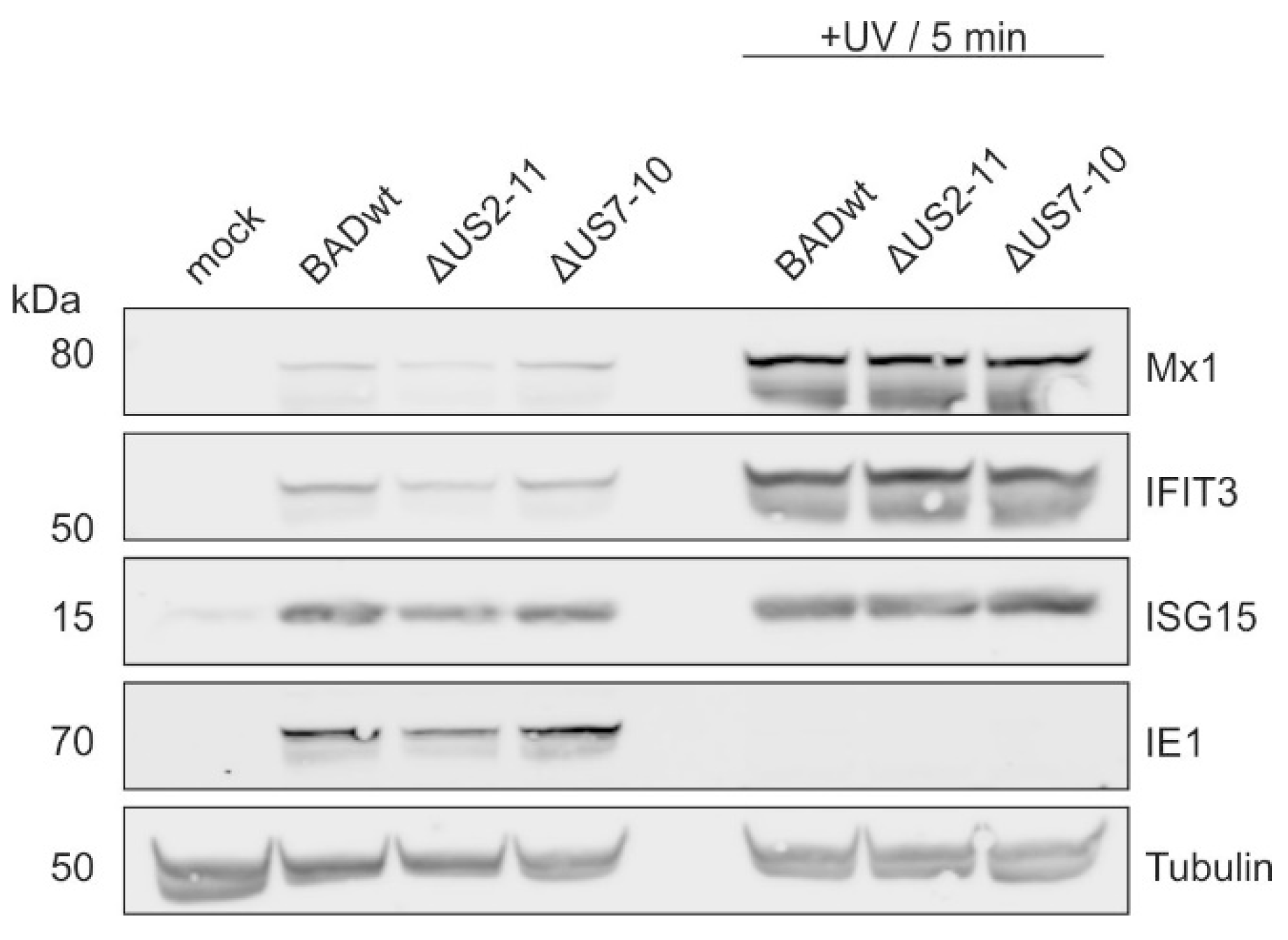
3.3. Induction of IFIT3 and ISG15 Does Not Require Interferon Signaling
3.4. Modulation of ISG-Levels by US2–US11 Requires Viral Gene Expression
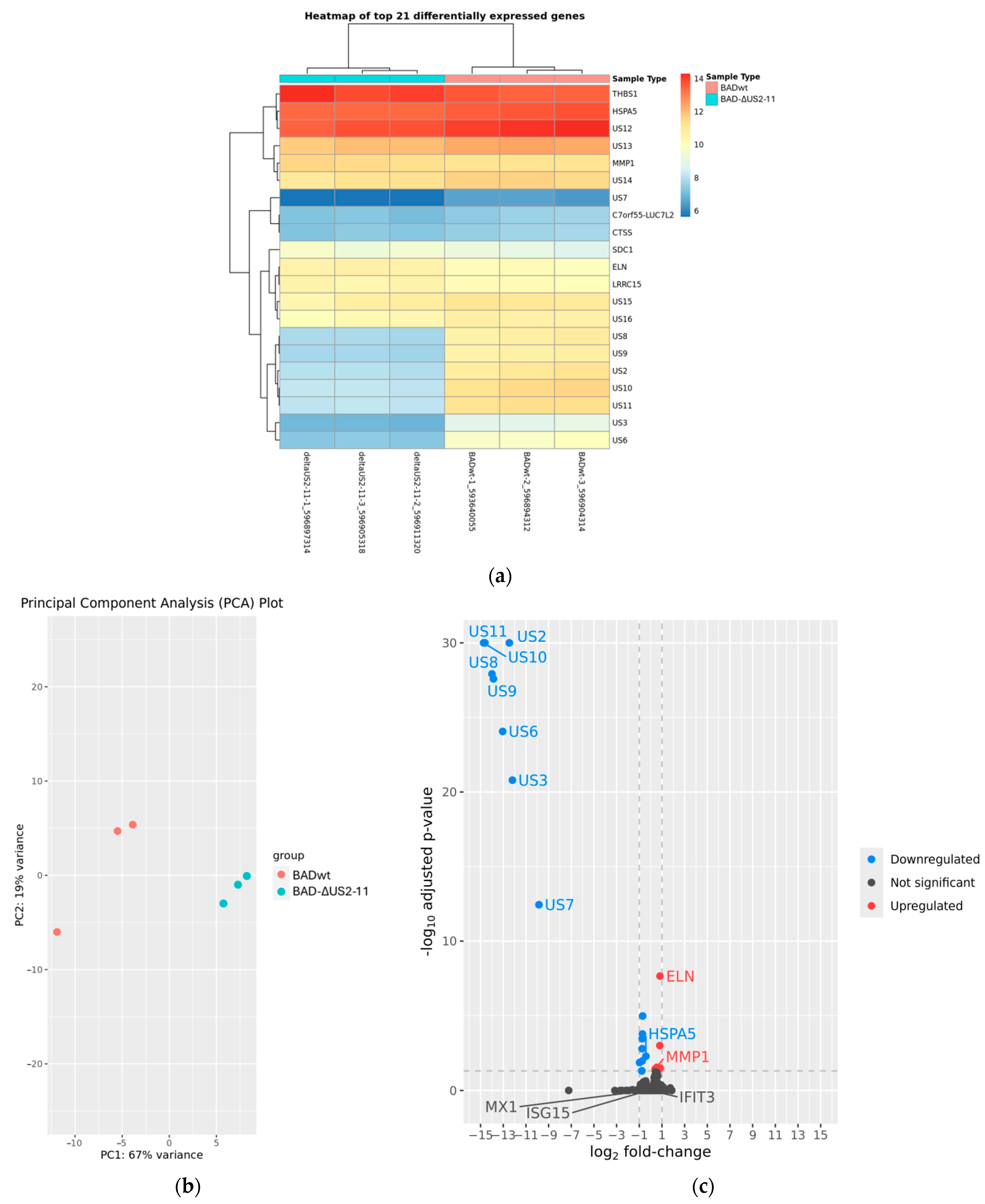
3.5. Modulation of ISG Induction by US2–US11 Is Not Caused by Differential Gene Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, Y.; Agueda-Pinto, A.; Brune, W. No Time to Die: How Cytomegaloviruses Suppress Apoptosis, Necroptosis, and Pyroptosis. Viruses 2024, 16, 1272. [Google Scholar] [CrossRef]
- Sabbaghian, M.; Gheitasi, H.; Fadaee, M.; Javadi Henafard, H.; Tavakoli, A.; Shekarchi, A.A.; Poortahmasebi, V. Human cytomegalovirus microRNAs: Strategies for immune evasion and viral latency. Arch. Virol. 2024, 169, 157. [Google Scholar] [CrossRef] [PubMed]
- Muscolino, E.; Luoto, L.M.; Brune, W. Viral Induced Protein Aggregation: A Mechanism of Immune Evasion. Int. J. Mol. Sci. 2021, 22, 9624. [Google Scholar] [CrossRef]
- Diggins, N.L.; Skalsky, R.L.; Hancock, M.H. Regulation of Latency and Reactivation by Human Cytomegalovirus miRNAs. Pathogens 2021, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Dell’Oste, V.; Biolatti, M.; Galitska, G.; Griffante, G.; Gugliesi, F.; Pasquero, S.; Zingoni, A.; Cerboni, C.; De Andrea, M. Tuning the Orchestra: HCMV vs. Innate Immunity. Front. Microbiol. 2020, 11, 661. [Google Scholar] [CrossRef]
- Lim, E.Y.; Jackson, S.E.; Wills, M.R. The CD4+ T Cell Response to Human Cytomegalovirus in Healthy and Immunocompromised People. Front. Cell Infect. Microbiol. 2020, 10, 202. [Google Scholar] [CrossRef]
- Li, S.; Xie, Y.; Yu, C.; Zheng, C.; Xu, Z. The battle between host antiviral innate immunity and immune evasion by cytomegalovirus. Cell Mol. Life Sci. 2024, 81, 341. [Google Scholar] [CrossRef]
- Noriega, V.; Redmann, V.; Gardner, T.; Tortorella, D. Diverse immune evasion strategies by human cytomegalovirus. Immunol. Res. 2012, 54, 140–151. [Google Scholar] [CrossRef]
- Jones, T.R.; Hanson, L.K.; Sun, L.; Slater, J.S.; Stenberg, R.M.; Campbell, A.E. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 1995, 69, 4830–4841. [Google Scholar] [CrossRef]
- Johnson, D.C.; Hill, A.B. Herpesvirus evasion of the immune system. Curr. Top. Microbiol. Immunol. 1998, 232, 149–177. [Google Scholar] [CrossRef]
- Ploegh, H.L. Viral strategies of immune evasion. Science 1998, 280, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Tomazin, R.; Boname, J.; Hegde, N.R.; Lewinsohn, D.M.; Altschuler, Y.; Jones, T.R.; Cresswell, P.; Nelson, J.A.; Riddell, S.R.; Johnson, D.C. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 1999, 5, 1039–1043. [Google Scholar] [CrossRef]
- Hegde, N.R.; Tomazin, R.A.; Wisner, T.W.; Dunn, C.; Boname, J.M.; Lewinsohn, D.M.; Johnson, D.C. Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: A novel mechanism for evading major histocompatibility complex class II antigen presentation. J. Virol. 2002, 76, 10929–10941. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Ra, E.A.; Lee, T.A.; Choi, H.J.; Lee, E.; Kang, S.; Seo, J.Y.; Lee, S.; Park, B. HCMV-encoded US7 and US8 act as antagonists of innate immunity by distinctively targeting TLR-signaling pathways. Nat. Commun. 2019, 10, 4670. [Google Scholar] [CrossRef]
- Choi, H.J.; Park, A.; Kang, S.; Lee, E.; Lee, T.A.; Ra, E.A.; Lee, J.; Lee, S.; Park, B. Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat. Commun. 2018, 9, 125. [Google Scholar] [CrossRef]
- Kollert-Jöns, A.; Bogner, E.; Radsak, K. A 15-kilobase-pair region of the human cytomegalovirus genome which includes US1 through US13 is dispensable for growth in cell culture. J. Virol. 1991, 65, 5184–5189. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.R.; Muzithras, V.P. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J. Virol. 1992, 66, 2541–2546. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.R.; Muzithras, V.P.; Gluzman, Y. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J. Virol. 1991, 65, 5860–5872. [Google Scholar] [CrossRef]
- Borst, E.M.; Hahn, G.; Koszinowski, U.H.; Messerle, M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: A new approach for construction of HCMV mutants. J. Virol. 1999, 73, 8320–8329. [Google Scholar] [CrossRef]
- Sinzger, C.; Hahn, G.; Digel, M.; Katona, R.; Sampaio, K.L.; Messerle, M.; Hengel, H.; Koszinowski, U.; Brune, W.; Adler, B. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. J. Gen. Virol. 2008, 89 Pt 2, 359–368. [Google Scholar] [CrossRef]
- Marchini, A.; Liu, H.; Zhu, H. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 2001, 75, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Schilling, E.M.; Scherer, M.; Stamminger, T. Intrinsic Immune Mechanisms Restricting Human Cytomegalovirus Replication. Viruses 2021, 13, 179. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.L.; van den Boomen, D.J.; Tomasec, P.; Weekes, M.P.; Antrobus, R.; Stanton, R.J.; Ruckova, E.; Sugrue, D.; Wilkie, G.S.; Davison, A.J.; et al. Plasma membrane profiling defines an expanded class of cell surface proteins selectively targeted for degradation by HCMV US2 in cooperation with UL141. PLoS Pathog. 2015, 11, e1004811. [Google Scholar] [CrossRef] [PubMed]
- Besold, K.; Wills, M.; Plachter, B. Immune evasion proteins gpUS2 and gpUS11 of human cytomegalovirus incompletely protect infected cells from CD8 T cell recognition. Virology 2009, 391, 5–19. [Google Scholar] [CrossRef]
- Yu, D.; Smith, G.A.; Enquist, L.W.; Shenk, T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 2002, 76, 2316–2328. [Google Scholar] [CrossRef]
- Zimmermann, C.; Krämer, N.; Krauter, S.; Strand, D.; Sehn, E.; Wolfrum, U.; Freiwald, A.; Butter, F.; Plachter, B. Autophagy interferes with human cytomegalovirus genome replication, morphogenesis, and progeny release. Autophagy 2020, 17, 779–795. [Google Scholar] [CrossRef]
- Noriega, V.M.; Hesse, J.; Gardner, T.J.; Besold, K.; Plachter, B.; Tortorella, D. Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol. Immunol. 2012, 51, 245–253. [Google Scholar] [CrossRef]
- Warming, S.; Costantino, N.; DL, C.; Jenkins, N.A.; Copeland, N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005, 33, e36. [Google Scholar] [CrossRef]
- Penner, I.; Büscher, N.; Krauter, S.; Plachter, B. Subviral Dense Bodies of Human Cytomegalovirus Enhance Interferon-Beta Responses in Infected Cells and Impair Progeny Production. Viruses 2023, 15, 1333. [Google Scholar] [CrossRef]
- Fleckenstein, B.; Muller, I.; Collins, J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene 1982, 18, 39–46. [Google Scholar] [CrossRef]
- Behera, S.; Catreux, S.; Rossi, M.; Truong, S.; Huang, Z.; Ruehle, M.; Visvanath, A.; Parnaby, G.; Roddey, C.; Onuchic, V.; et al. Comprehensive genome analysis and variant detection at scale using DRAGEN. Nat. Biotechnol. 2024. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC10802302/ (accessed on 1 February 2025). [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Hesse, J.; Ameres, S.; Besold, K.; Krauter, S.; Moosmann, A.; Plachter, B. Suppression of CD8+ T-cell recognition in the immediate-early phase of human cytomegalovirus infection. J. Gen. Virol. 2013, 94, 376–386. [Google Scholar] [CrossRef]
- Zhu, H.; Cong, J.P.; Shenk, T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: Induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 1997, 94, 13985–13990. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.A.; Pietropaolo, R.L.; Compton, T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell Biol. 1999, 19, 3607–3613. [Google Scholar] [CrossRef]
- Gonzalez-Perez, A.C.; Stempel, M.; Wyler, E.; Urban, C.; Piras, A.; Hennig, T.; Ganskih, S.; Wei, Y.; Heim, A.; Landthaler, M.; et al. The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral UL4/UL5 Transcripts. mBio 2021, 12, e02683-20. [Google Scholar] [CrossRef] [PubMed]
- Biolatti, M.; Gugliesi, F.; Dell’Oste, V.; Landolfo, S. Modulation of the innate immune response by human cytomegalovirus. Infect. Genet. Evol. 2018, 64, 105–114. [Google Scholar] [CrossRef]
- Hunter, L.M.; Kite, J.; Fletcher-Etherington, A.; Nightingale, K.; Nobre, L.; Antrobus, R.; Fielding, C.A.; Stanton, R.J.; Weekes, M.P. HCMV US2 co-opts TRC8 to degrade the endoplasmic reticulum-resident protein LMAN2L. J. Gen. Virol. 2024, 105, 001980. [Google Scholar] [CrossRef]
- Machold, R.P.; Wiertz, E.J.; Jones, T.R.; Ploegh, H.L. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J. Exp. Med. 1997, 185, 363–366. [Google Scholar] [CrossRef]
- Jones, T.R.; Sun, L. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 1997, 71, 2970–2979. [Google Scholar] [CrossRef]
- Erhard, F.; Halenius, A.; Zimmermann, C.; L’Hernault, A.; Kowalewski, D.J.; Weekes, M.P.; Stevanovic, S.; Zimmer, R.; Dölken, L. Improved Ribo-seq enables identification of cryptic translation events. Nat. Methods 2018, 15, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Weisburd, B.; Michalski, A.; Le, V.T.; Hein, M.Y.; Huang, S.X.; Ma, M.; Shen, B.; Qian, S.B.; Hengel, H.; et al. Decoding human cytomegalovirus. Science 2012, 338, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Shin, J.; Kim, Y.; Evnouchidou, I.; Kim, D.; Kim, Y.K.; Kim, Y.E.; Ahn, J.H.; Riddell, S.R.; et al. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat. Immunol. 2011, 12, 984–991. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Sampaio, K.L.; Weyell, A.; Subramanian, N.; Wu, Z.; Sinzger, C. A TB40/E-derived human cytomegalovirus genome with an intact US-gene region and a self-excisable BAC cassette for immunological research. Biotechniques 2017, 63, 205–214. [Google Scholar] [CrossRef]
- Nobre, L.V.; Nightingale, K.; Ravenhill, B.J.; Antrobus, R.; Soday, L.; Nichols, J.; Davies, J.A.; Seirafian, S.; Wang, E.C.; Davison, A.J.; et al. Human cytomegalovirus interactome analysis identifies degradation hubs, domain associations and viral protein functions. Elife 2019, 8, e49894. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penner, I.; Krämer, N.; Hirsch, J.; Büscher, N.; Schmidt, H.; Plachter, B. Deletion of the Human Cytomegalovirus US2 to US11 Gene Family Members Impairs the Type-I Interferon Response. Viruses 2025, 17, 426. https://doi.org/10.3390/v17030426
Penner I, Krämer N, Hirsch J, Büscher N, Schmidt H, Plachter B. Deletion of the Human Cytomegalovirus US2 to US11 Gene Family Members Impairs the Type-I Interferon Response. Viruses. 2025; 17(3):426. https://doi.org/10.3390/v17030426
Chicago/Turabian StylePenner, Inessa, Nadine Krämer, Julia Hirsch, Nicole Büscher, Hanno Schmidt, and Bodo Plachter. 2025. "Deletion of the Human Cytomegalovirus US2 to US11 Gene Family Members Impairs the Type-I Interferon Response" Viruses 17, no. 3: 426. https://doi.org/10.3390/v17030426
APA StylePenner, I., Krämer, N., Hirsch, J., Büscher, N., Schmidt, H., & Plachter, B. (2025). Deletion of the Human Cytomegalovirus US2 to US11 Gene Family Members Impairs the Type-I Interferon Response. Viruses, 17(3), 426. https://doi.org/10.3390/v17030426





