Development of New Probe-Based Real-Time RT-qPCR Assays for the Detection of All Known Strains of Bovine Ephemeral Fever Viruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Viral RNA Extraction
2.3. Real-Time RT-qPCR Assay Preparation
2.3.1. Probe–Primer Mix Preparation
2.3.2. Master Mix Preparation
2.4. Repeatability (Reproducible Quality)
2.5. Analytical Sensitivity
2.6. Analytical Specificity
2.7. Diagnostic Sensitivity and Specificity
3. Results and Discussion
3.1. Reproducible RNA Extraction from Cattle Field Samples
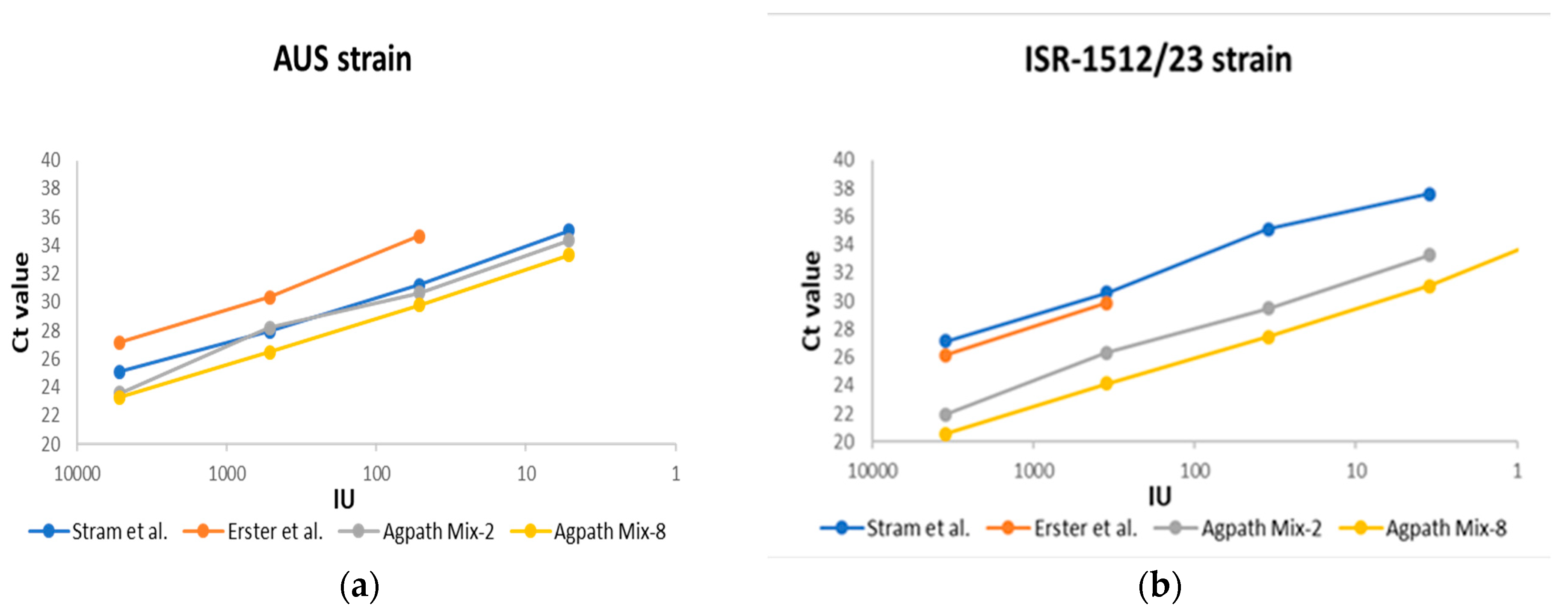
3.2. Analytical Sensitivity (The Limit of Detection (LOD))
3.3. Analytical Specificity
3.4. Diagnostic Sensitivity (DSe) and Diagnostic Specificity (SPe)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walker, P.J. Bovine ephemeral fever in Australia and the world. Curr. Top. Microbiol. Immunol. 2005, 292, 57–80. [Google Scholar] [PubMed]
- Walker, P.J.; Klement, E. Epidemiology and control of bovine ephemeral fever. Vet. Res. 2015, 46, 124. [Google Scholar] [CrossRef] [PubMed]
- MDS Veterinary Manual. Available online: https://www.msdvetmanual.com/generalized-conditions/bovine-ephemeral-fever/bovine-ephemeral-fever (accessed on 12 January 2024).
- Barigye, R.; Davis, S.; Hunt, R.; Hunt, N.; Walsh, S.; Elliott, N.; Burnup, C.; Aumann, S.; Day, C.; Dyrting, K.; et al. Viral neurotropism, peripheral neuropathy and other morphological abnormalities in bovine ephemeral fever virus-infected downer cattle. Aust. Vet. J. 2016, 94, 362–370. [Google Scholar] [CrossRef]
- Hill, M.W.; Schultz, K. Ataxia and paralysis associated with bovine ephemeral fever infection. Aust. Vet. J. 1977, 53, 217–221. [Google Scholar] [CrossRef]
- Pekmez, K.; Kaplan, M.; Çağırgan, A.; Arslan, F. The origin and molecular characterization of the BEF virus causing the small-scale epidemic in Western Turkey. Authorea 2024. [Google Scholar] [CrossRef]
- Rezatofighi, S.E.; Mirzadeh, K.; Mahmoodi, F. Molecular characterization and phylogenetic analysis of bovine ephemeral fever viruses in Khuzestan province of Iran in 2018 and 2020. BMC Vet. Res. 2022, 18, 19. [Google Scholar] [CrossRef]
- Hirashima, Y.; Nojiri, M.; Ohtsuka, Y.; Kato, T.; Shirafuji, H.; Kurazono, M.; Imafuji, T.; Yanase, T. Resurgence of bovine ephemeral fever in mainland Japan in 2015 after a 23-year absence. J. Vet. Med. Sci. 2017, 79, 904–911. [Google Scholar] [CrossRef]
- Chen, J.; Liu, M.; Li, Y.; Yang, L.; Tang, Y.; Dan, R.; Xie, M.; Fang, R.; Li, N.; Ye, C.; et al. Emergence and genomic analysis of a novel sublineage of bovine ephemeral fever virus in Southwest China. Front. Microbiol. 2023, 14, 1161287. [Google Scholar] [CrossRef]
- Golender, N.; Hoffmann, B.; Kenigswald, G.; Scheinin, S.; Kedmi, M.; Gleser, D.; Klement, E. Bovine Ephemeral Fever Viruses in Israel 2014–2023: Genetic Characterization of Local and Emerging Strains. Pathogens 2024, 13, 636. [Google Scholar] [CrossRef]
- Dacheux, L.; Dommergues, L.; Chouanibou, Y.; Doméon, L.; Schuler, C.; Bonas, S.; Luo, D.; Maufrais, C.; Cetre-Sossah, C.; Cardinale, E.; et al. Co-circulation and characterization of novel African arboviruses (genus Ephemerovirus) in cattle, Mayotte island, Indian Ocean, 2017. Transbound. Emerg. Dis. 2019, 66, 2601–2604. [Google Scholar] [CrossRef]
- Pyasi, S.; Gupta, A.; Hegde, N.R.; Nayak, D. Complete genome sequencing and assessment of mutation-associated protein dynamics of the first Indian bovine ephemeral fever virus (BEFV) isolate. Vet. Q. 2021, 41, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Erster, O.; Stram, R.; Menasherow, S.; Rubistein-Giuni, M.; Sharir, B.; Kchinich, E.; Stram, Y. High-resolution melting (HRM) for genotyping bovine ephemeral fever virus (BEFV). Virus Res. 2017, 229, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, S.M.; Kongsuwan, K.; Cowley, J.A.; Byrne, K.A.; Walker, P.J. Genome organization and transcription strategy in the complex GNS-L intergenic region of bovine ephemeral fever rhabdovirus. J. Gen. Virol. 1997, 6, 1309–1317. [Google Scholar] [CrossRef]
- Walker, P.J.; Byrne, K.A.; Cybinski, D.H.; Doolan, D.L.; Wang, Y. Proteins of bovine ephemeral fever virus. J. Gen. Virol. 1991, 72, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Blasdell, K.R.; Adams, M.M.; Davis, S.S.; Walsh, S.J.; Aziz-Boaron, O.; Klement, E.; Tesh, R.B.; Walker, P.J. A reverse-transcription PCR method for detecting all known ephemeroviruses in clinical samples. J. Virol. Methods. 2013, 191, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Golender, N.; Klement, E.; Ofer, L.; Hoffmann, B.; Wernike, K.; Beer, M.; Pfaff, F. Hefer valley virus: A novel ephemerovirus detected in the blood of a cow with severe clinical signs in Israel in 2022. Arch. Virol. 2023, 168, 234. [Google Scholar] [CrossRef]
- Niwa, T.; Shirafuji, H.; Ikemiyagi, K.; Nitta, Y.; Suzuki, M.; Kato, T.; Yanase, T. Occurrence of bovine ephemeral fever in Okinawa Prefecture, Japan, in 2012 and development of a reverse-transcription polymerase chain reaction assay to detect bovine ephemeral fever virus gene. J. Vet. Med. Sci. 2015, 77, 455–460. [Google Scholar] [CrossRef][Green Version]
- Gao, S.; Du, J.; Tian, Z.; Niu, Q.; Huang, D.; Wang, J.; Luo, J.; Liu, G.; Yin, H. A SYBR green I-based quantitative RT-PCR assay for bovine ephemeral fever virus and its utility for evaluating viral kinetics in cattle. J. Vet. Diagn. Investig. 2020, 32, 44–50. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, G.; Wang, H.; He, C.; Huan, Y.; He, H. Development of a recombinase polymerase amplification combined with lateral-flow dipstick assay for detection of bovine ephemeral fever virus. Mol. Cell. Probes 2018, 38, 31–37. [Google Scholar] [CrossRef]
- Stram, Y.; Kuznetzova, L.; Levin, A.; Yadin, H.; Rubinstein-Giuni, M. A real-time RT-quantative(q)PCR for the detection of bovine ephemeral fever virus. J. Virol. Methods 2005, 130, 1–6. [Google Scholar] [CrossRef]
- Zheng, F.; Lin, G.; Zhou, J.; Wang, G.; Cao, X.; Gong, X.; Qiu, C. A reverse-transcription, loop-mediated isothermal amplification assay for detection of bovine ephemeral fever virus in the blood of infected cattle. J. Virol. Methods 2011, 171, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Lew, A.; Corney, B.; Doogan, V.; Fordyce, G.; Bertram, J.; Holroyd, R.; McMillen, L.; Turner, L.; Smythe, L.; Fenwick, S.; et al. Improved Diagnosis of Reproductive Disease in Cattle; Final Report; Meat and Livestock Australia Limited: North Sydney, Australia, 2006; pp. 58–62. [Google Scholar]
- Toussaint, J.F.; Sailleau, C.; Breard, E.; Zientara, S.; De Clercq, K. Bluetongue Virus Detection by Two Real-Time RT-QPCRs Targeting Two Different Genomic Segments. J. Virol. Methods 2007, 140, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Hoffmann, B.; Kalthoff, D.; König, P.; Beer, M. Development and validation of a triplex real-time PCR assay for the rapid detection and differentiation of wild-type and glycoprotein E-deleted vaccine strains of Bovine herpesvirus type 1. J. Virol. Methods 2011, 174, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Cell Biology Protocols. MOI, pfu, and TCID50. Available online: https://www.sciencegateway.org/protocols/cellbio/cell/moipfu.htm (accessed on 6 February 2025).
- WOAH. Chapter 1.1.6. Principles and methods of validation of diagnostic assays for infectious diseases. In WOAH Terrestrial Manual 2023; WOAH: Paris, France, 2023; pp. 1–27. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/1.01.06_VALIDATION.pdf (accessed on 16 September 2024).
- Parikh, R.; Mathai, A.; Parikh, S.; Chandra Sekhar, G.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef]
- Akobeng, A.K. Understanding diagnostic tests 1: Sensitivity, specificity and predictive values. Acta Paediatr. 2007, 96, 338–341. [Google Scholar] [CrossRef]
| Sample | W.blood | B.coat | Serum/Plasma | Spleen | S+L | Brain | Inact.vac. | Total |
|---|---|---|---|---|---|---|---|---|
| № pos | 55 | 81 | 4 | 3 | 3 | 0 | 1 | 147 |
| № neg | 70 | 13 | 4 | 6 | 2 | 3 | 0 | 98 |
| Total | 125 | 94 | 8 | 9 | 5 | 3 | 1 | 245 |
| PCR Assay | Designation of Oligo | Sequence of Oligo 5′–3′ | Amplicon Length | Location | Source |
|---|---|---|---|---|---|
| BEFV- | BEFV-N-46-F | GTSTTTYAACAGGTCTCTTTCCT | 79 | N gene | Current |
| N-Mix-2- | BEFV-N-124-R | TCAGTTGGCTTAACAGCCTTG | study | ||
| BEFV-N-102-FAM | FAM-TCTCTTTCTTRTTCAATGTGCART ACAT-BHQ1 | ||||
| BEFV- | BEFV-N-311-F | GGAACTTTTATGARGTAATCATAGA | 85 | N gene | Current |
| N-Mix-8 | BEFV-N-395-R | CATACATCATCATTYTCATCYACATT | study | ||
| BEFV-N-363-FAM | FAM-CTTCTHCATCTATTGTCTGRTCATC-BHQ1 | ||||
| beta- | ACT-1030-F | AGCGCAAGTACTCCGTGTG | 106 | beta- | [25] |
| actin- | ACT-R-1135-R | CGGACTCATCGTACTCCTGCTT | actin- | [24] | |
| Mix5 | ACT-PROBE-HEX | HEX-TCGCTGTCCACCTTCCAGCAGATGT-BHQ2 | gene | [24] | |
| SYBR | BEFV-G-1140F | GAATCATTATGGGATMGGATC | G gene | [17] | |
| BEFV-G-1273R | CCTCCTGCTGGTGCTGTTTC | ||||
| BEFV- | BEFV-G-70F | GAGATCAAATGTCCACAACGTTTAA | 73 | G gene | [21] |
| G-Mix-1 | BEFV-G-142R | AATGTTCATCCTTTGCAAGATTATGA | |||
| BEFV-G-98FAM | FAM-AATTATCACTTCAAGCCC-MGB |
| BEFV-Mix-1/-2/-8 | Beta-Actin-Mix-5 | |
|---|---|---|
| Primer F | 10 μL | 2.5 μL |
| Primer R | 10 μL | 2.5 μL |
| Probe | 2.5 μL | 2.5 μL |
| Water | 77.5 μL | 92.5 μL |
| Total | 100 μL | 100 μL |
| Commercial Kit | |||||
|---|---|---|---|---|---|
| Reagents | Quanta | AgPath | Clara | Takara | Promega |
| Buffer | 6.25 μL | 6.25 μL | 3.12 μL | 6.25 μL | 6.25 μL |
| RT | - | 0.5 μL | - | 0.25 μL | 0.25 μL |
| EX Tag HS | - | - | - | 0.25 μL | - |
| BEFV-N-Mix-2/-8 * | 1 μL | 1 μL | 1 μL | 1 μL | 1 μL |
| Beta-actin-Mix-5 * | 1 μL | 1 μL | 1 μL | 1 μL | 1 μL |
| Water | 1.75 μL | 1.25 μL | 4.88 μL | 1.25 μL | 1.5 μL |
| Total | 10 μL | 10 μL | 10 μL | 10 μL | 10 μL |
| Commercial Kit | ||||||
|---|---|---|---|---|---|---|
| Cycle | Step | Quanta | AgPath | Clara | TaKaRa | Promega |
| 1x | RT | 10 min/50 °C | 10 min/45 °C | 10 min/52 °C | 5 min/45 °C | 15 min/45 °C |
| 1x | denaturation | 2 min/95 °C | 10 min/95 °C | 3 min/95 °C | 30 s/95 °C | 2 min/95 °C |
| denaturation | 10 s/95 °C | 15 s/95 °C | 15 s/95 °C | 5 s/95 °C | 15 s/95 °C | |
| 45x | annealing | 15 s/58 °C | 30 s/58 °C | 15 s/58 °C | 15 s/58 °C | 15 s/58 °C |
| elongation | 15 s/68 °C | 45 s/68 °C | 30 s/68 °C | 34 s/68 °C | 45 s/68 °C | |
| Assay Type/RT-qPCR Kit | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stram/Quanta | Erster/ Sensi | Quanta | Quanta | AgPath | AgPath | Clara | Clara | ||||||||||
| Sample (Replicate) | Source | Ct | β-ACT | Ct | Tm | MIX2 | β-ACT | MIX8 | β-ACT | MIX 2 | β-ACT | MIX 8 | β-ACT | MIX 2 | β-ACT | MIX 8 | β-ACT |
| A(1) | w.b. | NA | 28.97 | 27.66 | 78.36 | 34.12 | 30.42 | 32.63 | 31.25 | 31.28 | 27.38 | 30.87 | 28.06 | 32.57 | 29.07 | 30.81 | 29.33 |
| A(2) | w.b. | NA | 28.69 | 26.67 | 78.05 | 33.58 | 30.55 | 33.06 | 32.26 | 31.17 | 27.24 | 30.35 | 28.04 | 31.97 | 29.01 | 30.73 | 29.84 |
| A(3) | w.b. | NA | 29.51 | 26.15 | 78.28 | 33.1 | 30.84 | 31.76 | 31.81 | 30.63 | 27.98 | 29.85 | 27.95 | 31.82 | 29.93 | 30.28 | 30.5 |
| B(1) | w.b. | NA | 30.49 | 29.49 | 78.28 | 35.77 | 32.00 | 34.82 | 32.86 | 33.51 | 28.94 | 33.16 | 31.14 | 35.19 | 30.66 | 32.64 | 31.88 |
| B(2) | w.b. | NA | 30.31 | 29.99 | 78.36 | 36.76 | 31.6 | 35.66 | 32.49 | 32.95 | 29.07 | 32.97 | 29.79 | 36.02 | 31.99 | 32.62 | 32.22 |
| B(3) | w.b. | NA | 30.57 | 28.8 | 78.2 | 35.08 | 32.18 | 33.75 | 33.41 | 33.97 | 31.53 | 32.32 | 30.26 | 34.54 | 31.74 | 32.05 | 31.73 |
| C(1) | w.b. | 26.63 | 26.49 | 15.28 | 80.02 | 22.08 | 27.78 | 19.78 | 28.92 | 19.71 | 25.79 | 18.47 | 26.08 | 19.93 | 27.58 | 18.75 | 28.47 |
| C(2) | w.b. | 26.30 | 26.48 | 15.57 | 79.94 | 22.06 | 28.58 | 20.17 | 28.62 | 20.00 | 26.28 | 18.80 | 26.07 | 20.05 | 26.97 | 19.02 | 27.83 |
| C(3) | w.b. | 25.43 | 26.44 | 14.15 | 79.80 | 20.52 | 28.31 | 18.08 | 27.35 | 18.59 | 25.55 | 16.98 | 25.88 | 19.02 | 26.70 | 17.67 | 27.44 |
| D(1) | b.coat | NA | 26.23 | 22.18 | 78.05 | 26.57 | 27.82 | 24.71 | 28.44 | 23.96 | 24.78 | 23.20 | 25.17 | 25.03 | 26.63 | 23.07 | 27.31 |
| D(2) | b.coat | NA | 25.66 | 22.01 | 78.05 | 26.29 | 27.33 | 24.91 | 28.43 | 23.96 | 24.60 | 26.34 | 27.05 | 24.28 | 25.44 | 23.22 | 26.31 |
| D(3) | b.coat | NA | 26.06 | 22.27 | 78.05 | 27.03 | 28.28 | 25.64 | 28.95 | 24.33 | 24.74 | 23.47 | 24.44 | 24.94 | 26.26 | 23.44 | 26.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golender, N.; Klement, E.; Hoffmann, B. Development of New Probe-Based Real-Time RT-qPCR Assays for the Detection of All Known Strains of Bovine Ephemeral Fever Viruses. Viruses 2025, 17, 407. https://doi.org/10.3390/v17030407
Golender N, Klement E, Hoffmann B. Development of New Probe-Based Real-Time RT-qPCR Assays for the Detection of All Known Strains of Bovine Ephemeral Fever Viruses. Viruses. 2025; 17(3):407. https://doi.org/10.3390/v17030407
Chicago/Turabian StyleGolender, Natalia, Eyal Klement, and Bernd Hoffmann. 2025. "Development of New Probe-Based Real-Time RT-qPCR Assays for the Detection of All Known Strains of Bovine Ephemeral Fever Viruses" Viruses 17, no. 3: 407. https://doi.org/10.3390/v17030407
APA StyleGolender, N., Klement, E., & Hoffmann, B. (2025). Development of New Probe-Based Real-Time RT-qPCR Assays for the Detection of All Known Strains of Bovine Ephemeral Fever Viruses. Viruses, 17(3), 407. https://doi.org/10.3390/v17030407







