Development and Validation of a Differentiating Infected from Vaccinated Animals (DIVA) Enzyme-Linked Immunosorbent Assay (ELISA) Strategy for Distinguishing Between Hendra-Infected and Vaccinated Horses
Abstract
1. Introduction
2. Materials and Methods
2.1. Serum Samples
2.2. HeV DIVA ELISA
3. Results
3.1. Analytical Sensitivity (ASe)
3.2. Analytical Specificity (ASp)
| Antisera | G Protein (% Inhibition) | N Protein (% Inhibition) |
|---|---|---|
| Anti-PPR (Caprine) | 32 | −7 |
| Anti-Rinderpest (Rabbit) | 18 | −29 |
| Anti-Measles (Human) | −2 | −37 |
| Anti-Canine Distemper Virus (Horse) | −3 | −25 |
| Anti-NDV V4 (Rabbit) | 26 | −14 |
| Anti-J Virus (Rabbit) | 15 | 4 |
| Anti-Parainfluenza Type 1 (Horse) | 10 | −1 |
| Anti-Parainfluenza Type 2 (Horse) | −10 | −46 |
| Anti-Parainfluenza Type 3 (Horse) | 4 | −24 |
| Anti-Parainfluenza Type 1 (Guinea pig) | 24 | −28 |
| Anti-Parainfluenza Type 4A (Guinea pig) | 5 | −14 |
| Anti-Parainfluenza Type 4B (Guinea pig) | 23 | −30 |
| Anti-Mumps Enders Strain (Horse) | 3 | −11 |
| Anti-Nariva (Rabbit) | 11 | −32 |
| Anti-Tioman Virus (Pig) | 35 | −19 |
| Anti-Menangle Virus (Rabbit) | 10 | −34 |
| Anti-Menangle Virus (Pig) | 29 | −14 |
| Anti-Blue Eye Rubulavirus (Pig) | −8 | −10 |
| Anti-Mossman Virus (Rabbit) | 16 | −18 |
| Anti-Japanese Encephalitis (Pig) | 3 | −22 |
| Anti-Murray Valley Encephalitis Virus (Pig) | 8 | 26 |
| Anti-West Nile Virus (Horse) | 10 | 2 |
| Anti-African Horse Sickness Virus (Horse) | 13 | −9 |
| Anti-Canine Distemper Virus (Horse) | 8 | −27 |
| Anti-Hendra Virus (Horse) | 99 | 94 |
3.3. Diagnostic Specificity (DSp)
3.4. Diagnostic Sensitivity (DSe)
3.5. Repeatability
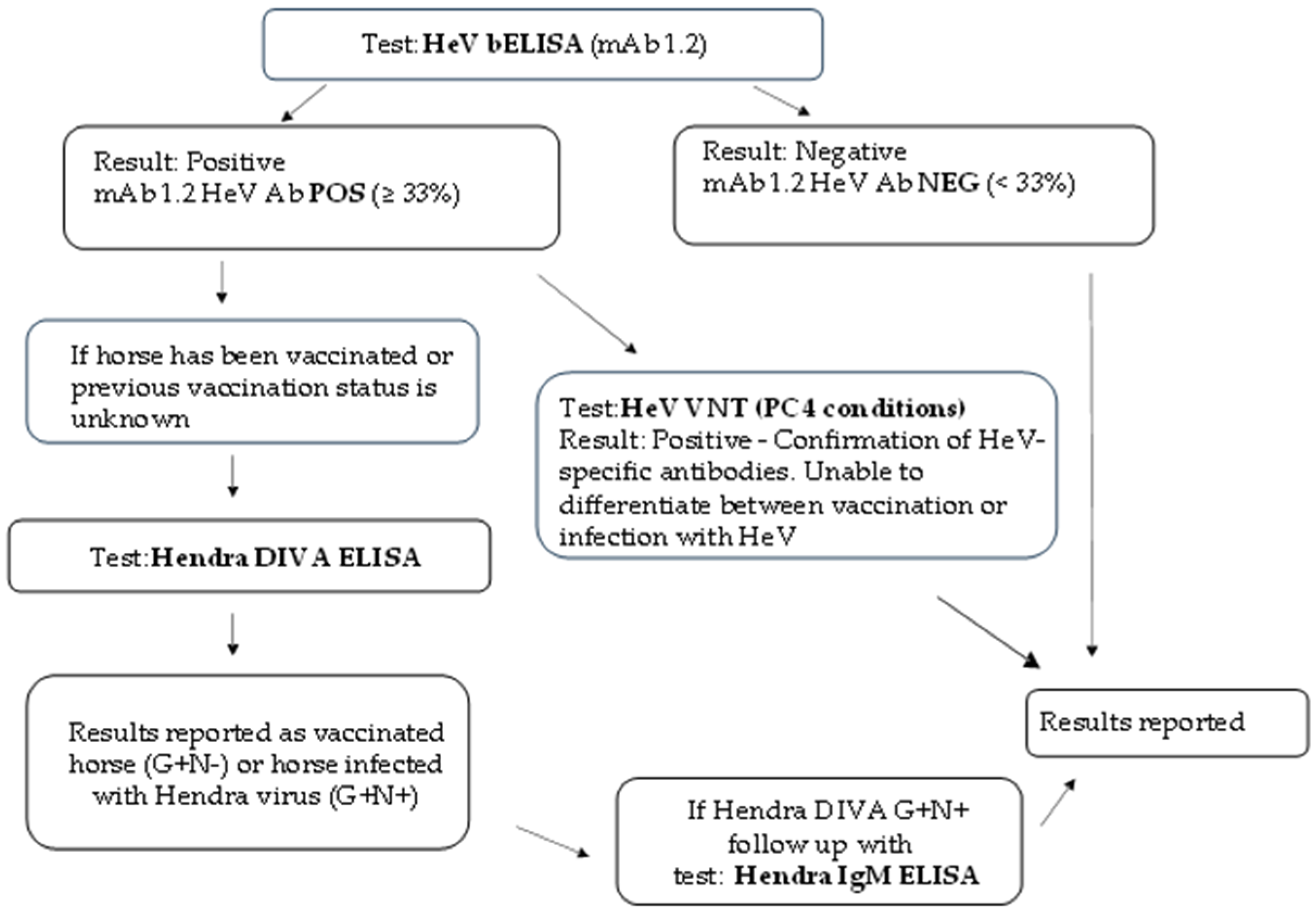
3.6. New Algorithm for HeV Serology Testing at ACDP
4. Discussion
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSe | Diagnostic sensitivity |
| DSp | Diagnostic specificity |
| ASe | Analytical sensitivity |
| ASp | Analytical specificity |
| DIVA | Differentiating infected from vaccinated animals |
| ELISA | Enzyme-linked immunosorbent assay |
| VNT | Virus neutralization test |
| HeV | Hendra virus |
References
- Eaton, B.T.; Broder, C.C.; Middleton, D.; Wang, L.F. Hendra and Nipah viruses: Different and dangerous. Nat. Rev. Microbiol. 2006, 4, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef] [PubMed]
- Queensland Business. Available online: https://www.business.qld.gov.au/industries/service-industries-professionals/service-industries/veterinary-surgeons/guidelines-hendra/incident-summary (accessed on 16 December 2024).
- OIE. Nipah and Hendra virus Diseases. In OIE Terrestrial Manual 2018; OIE: Paris, France, 2018; Chapter 3.1.14. [Google Scholar]
- Crameri, G.; Wang, L.F.; Morrissy, C.; White, J.; Eaton, B.T. A rapid immune plaque assay for the detection of Hendra and Nipah viruses and anti-virus antibodies. J. Virol. Methods 2002, 99, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Amaya, M.; Yin, R.; Yan, L.; Borisevich, V.; Adhikari, B.N.; Bennett, A.; Malagon, F.; Cer, R.Z.; Bishop-Lilly, K.A.; Dimitrov, A.S.; et al. A Recombinant Chimeric Cedar Virus-Based Surrogate Neutralization Assay Platform for Pathogenic Henipaviruses. Viruses 2023, 15, 1077. [Google Scholar] [CrossRef]
- Bossart, K.N.; Crameri, G.; Dimitrov, A.S.; Mungall, B.A.; Feng, Y.R.; Patch, J.R.; Choudhary, A.; Wang, L.F.; Eaton, B.T.; Broder, C.C. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 2005, 79, 6690–6702. [Google Scholar] [CrossRef]
- Colling, A.; Lunt, R.; Bergfeld, J.; McNabb, L.; Halpin, K.; Juzva, S.; Newberry, K.; Morrissy, C.; Loomes, C.; Warner, S.; et al. A network approach for provisional assay recognition of a Hendra virus antibody ELISA: Test validation with low sample numbers from infected horses. J. Vet. Diagn. Investig. 2018, 30, 362–369. [Google Scholar] [CrossRef]
- McNabb, L.; Barr, J.; Crameri, G.; Juzva, S.; Riddell, S.; Colling, A.; Boyd, V.; Broder, C.; Wang, L.F.; Lunt, R. Henipavirus microsphere immuno-assays for detection of antibodies against Hendra virus. J. Virol. Methods 2014, 200, 22–28. [Google Scholar] [CrossRef]
- White, J.R.; Boyd, V.; Crameri, G.S.; Duch, C.J.; van Laar, R.K.; Wang, L.F.; Eaton, B.T. Location of, immunogenicity of and relationships between neutralization epitopes on the attachment protein (G) of Hendra virus. J. Gen. Virol. 2005, 86, 2839–2848. [Google Scholar] [CrossRef]
- Di Rubbo, A.; McNabb, L.; Klein, R.; White, J.R.; Colling, A.; Dimitrov, D.S.; Broder, C.C.; Middleton, D.; Lunt, R.A. Optimization and diagnostic evaluation of monoclonal antibody-based blocking ELISA formats for detection of neutralizing antibodies to Hendra virus in mammalian sera. J. Virol. Methods 2019, 274, 113731. [Google Scholar] [CrossRef]
- Tan, R.; Hodge, A.; Klein, R.; Edwards, N.; Huang, J.A.; Middleton, D.; Watts, S.P. Virus-neutralising antibody responses in horses following vaccination with Equivac(R) HeV: A field study. Aust. Vet. J. 2018, 96, 161–166. [Google Scholar] [CrossRef]
- Wang, X.; Wise, J.C.; Stewart, A.J. Hendra Virus: An Update on Diagnosis, Vaccination, and Biosecurity Protocols for Horses. Vet. Clin. N. Am. Equine Pract. 2023, 39, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.; Graham, K.; Durr, P.A. Sero-Monitoring of Horses Demonstrates the Equivac((R)) HeV Hendra Virus Vaccine to Be Highly Effective in Inducing Neutralising Antibody Titres. Vaccines 2021, 9, 731. [Google Scholar] [CrossRef]
- Juozapaitis, M.; Serva, A.; Zvirbliene, A.; Slibinskas, R.; Staniulis, J.; Sasnauskas, K.; Shiell, B.J.; Wang, L.F.; Michalski, W.P. Generation of henipavirus nucleocapsid proteins in yeast Saccharomyces cerevisiae. Virus Res. 2007, 124, 95–102. [Google Scholar] [CrossRef] [PubMed]
- McNabb, L.; Andiani, A.; Bulavaite, A.; Zvirbliene, A.; Sasnauskas, K.; Lunt, R. Development and validation of an IgM antibody capture ELISA for early detection of Hendra virus. J. Virol. Methods 2021, 298, 114296. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Wang, L.F.; Lam, S.K.; Crameri, G.; Yu, M.; Wise, T.; Boyle, D.; Hyatt, A.D.; Eaton, B.T. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 2001, 283, 215–229. [Google Scholar] [CrossRef]
- Pasick, J. Application of DIVA vaccines and their companion diagnostic tests to foreign animal disease eradication. Anim. Health Res. Rev. 2004, 5, 257–262. [Google Scholar] [CrossRef]
- Browning, C.F.J.; Di Nardo, A.; Henry, L.; Pollard, T.; Hendry, L.; Romey, A.; Relmy, A.; Eble, P.; Brocchi, E.; Grazioli, S.; et al. Inter-laboratory comparison of 2 ELISA kits used for foot-and-mouth disease virus nonstructural protein serology. J. Vet. Diagn. Investig. 2020, 32, 933–937. [Google Scholar] [CrossRef]
- Sorensen, K.J.; Madsen, K.G.; Madsen, E.S.; Salt, J.S.; Nqindi, J.; Mackay, D.K. Differentiation of infection from vaccination in foot-and-mouth disease by the detection of antibodies to the non-structural proteins 3D, 3AB and 3ABC in ELISA using antigens expressed in baculovirus. Arch. Virol. 1998, 143, 1461–1476. [Google Scholar] [CrossRef]
- Carey, K.J.; Smith, I.; Barr, J.; Caruso, S.; Au, G.G.; Hartley, C.A.; Bailey, K.E.; Perriam, W.; Broder, C.C.; Gilkerson, J.R. Foals of mares vaccinated for Hendra virus have a suboptimal response to HeV vaccination. Vet. Microbiol. 2024, 295, 110167. [Google Scholar] [CrossRef]
- Zhu, W.; Pickering, B.; Smith, G.; Pinette, M.; Truong, T.; Babiuk, S.; Kobasa, D.; Banadyga, L.; Yang, M. Development and laboratory evaluation of a competitive ELISA for serodiagnosis of Nipah and Hendra virus infection using recombinant Nipah glycoproteins and a monoclonal antibody. Front. Vet. Sci. 2023, 10, 1120367. [Google Scholar] [CrossRef]
- Daniels, P.; Ksiazek, T.; Eaton, B.T. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes Infect. 2001, 3, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Balkema-Buschmann, A.; Fischer, K.; McNabb, L.; Diederich, S.; Singanallur, N.B.; Ziegler, U.; Keil, G.M.; Kirkland, P.D.; Penning, M.; Sadeghi, B.; et al. Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins. Microorganisms 2022, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
| Variable | N Assay | G Assay |
|---|---|---|
| Infected | 19 | 19 |
| Vaccinated | 502 | 502 |
| Negative | 1138 | 1138 |
| Cut-off | <49 | <36 |
| DSe | 100% | 100% |
| DSp | 100% | 99.91% |
| Sample IDs | G Protein | Result | N Protein | Result | HeV VNT | Result |
|---|---|---|---|---|---|---|
| 08-02438-0027 | 97.29 | Positive | 95.81 | Positive | 2048 | Positive |
| 08-02480-0001 | 97.87 | Positive | 95.46 | Positive | 4096 | Positive |
| 08-02667-0002 | 98.61 | Positive | 95.16 | Positive | 2048 | Positive |
| 08-02669-0001 | 98.73 | Positive | 92.98 | Positive | 2048 | Positive |
| 08-02668-0001 | 98.56 | Positive | 94.68 | Positive | 2048 | Positive |
| 08-02844-0001 | 41 | Positive | 94.57 | Positive | 16 | Positive |
| 08-02813-0001 | 98.49 | Positive | 92.33 | Positive | 512 | Positive |
| 06-03803-0004 | 39 | Positive | 93.12 | Positive | 20 | Positive |
| 09-02723-0001 | 92.89 | Positive | 93.1 | Positive | 1024 | Positive |
| 09-02723-0002 | 96.08 | Positive | 92.72 | Positive | 16 | Positive |
| 09-02844-0007 | 98.9 | Positive | 94.89 | Positive | >16 | Positive |
| 09-02844-0008 | 98.74 | Positive | 94.67 | Positive | 640 | Positive |
| 163910/1 | 99.36 | Positive | 95.74 | Positive | 640 | Positive |
| 163910/2 | 98.99 | Positive | 95.79 | Positive | 640 | Positive |
| 163910/3 | 98.2 | Positive | 97.82 | Positive | 1280 | Positive |
| 163910/4 | 81.78 | Positive | 93.05 | Positive | 20 | Positive |
| 163910/5 | 99.22 | Positive | 94.71 | Positive | 640 | Positive |
| 163910/6 | 99.12 | Positive | 94.98 | Positive | 640 | Positive |
| 163910/7 | 99.48 | Positive | 95.8 | Positive | 640 | Positive |
| Positive (1) | Vaccinated (2) | Negative (3) | ||||
|---|---|---|---|---|---|---|
| G | N | G | N | G | N | |
| Count | 12 | 12 | 12 | 12 | 12 | 12 |
| Average PI | 98.9 | 98.6 | 99.9 | −23.6 | 12.7 | −18.7 |
| Standard deviation | 0.5 | 0.8 | 1.1 | 14.9 | 14.4 | 14.8 |
| Maximum PI | 99.7 | 99.6 | 101.3 | −1.7 | 32.1 | −1.8 |
| Minimum PI | 97.9 | 97.3 | 97 | −63.2 | −12.9 | −54.6 |
| Median PI | 98.9 | 98.7 | 100.1 | −19.5 | 10.6 | −15 |
| Results | G+ | N+ | G+ | N- | G- | N- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McNabb, L.; McMahon, A.; Woube, E.G.; Agnihotri, K.; Colling, A.; Broder, C.C.; Kucinskaite-Kodze, I.; Petraityte-Burneikiene, R.; Bowden, T.R.; Halpin, K. Development and Validation of a Differentiating Infected from Vaccinated Animals (DIVA) Enzyme-Linked Immunosorbent Assay (ELISA) Strategy for Distinguishing Between Hendra-Infected and Vaccinated Horses. Viruses 2025, 17, 354. https://doi.org/10.3390/v17030354
McNabb L, McMahon A, Woube EG, Agnihotri K, Colling A, Broder CC, Kucinskaite-Kodze I, Petraityte-Burneikiene R, Bowden TR, Halpin K. Development and Validation of a Differentiating Infected from Vaccinated Animals (DIVA) Enzyme-Linked Immunosorbent Assay (ELISA) Strategy for Distinguishing Between Hendra-Infected and Vaccinated Horses. Viruses. 2025; 17(3):354. https://doi.org/10.3390/v17030354
Chicago/Turabian StyleMcNabb, Leanne, Amy McMahon, Ezana Getachew Woube, Kalpana Agnihotri, Axel Colling, Christopher C. Broder, Indre Kucinskaite-Kodze, Rasa Petraityte-Burneikiene, Timothy R. Bowden, and Kim Halpin. 2025. "Development and Validation of a Differentiating Infected from Vaccinated Animals (DIVA) Enzyme-Linked Immunosorbent Assay (ELISA) Strategy for Distinguishing Between Hendra-Infected and Vaccinated Horses" Viruses 17, no. 3: 354. https://doi.org/10.3390/v17030354
APA StyleMcNabb, L., McMahon, A., Woube, E. G., Agnihotri, K., Colling, A., Broder, C. C., Kucinskaite-Kodze, I., Petraityte-Burneikiene, R., Bowden, T. R., & Halpin, K. (2025). Development and Validation of a Differentiating Infected from Vaccinated Animals (DIVA) Enzyme-Linked Immunosorbent Assay (ELISA) Strategy for Distinguishing Between Hendra-Infected and Vaccinated Horses. Viruses, 17(3), 354. https://doi.org/10.3390/v17030354







