Immunogenic Potential of Selected Peptides from SARS-CoV-2 Proteins and Their Ability to Block S1/ACE-2 Binding
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptides Selection
2.2. Antigen Preparation
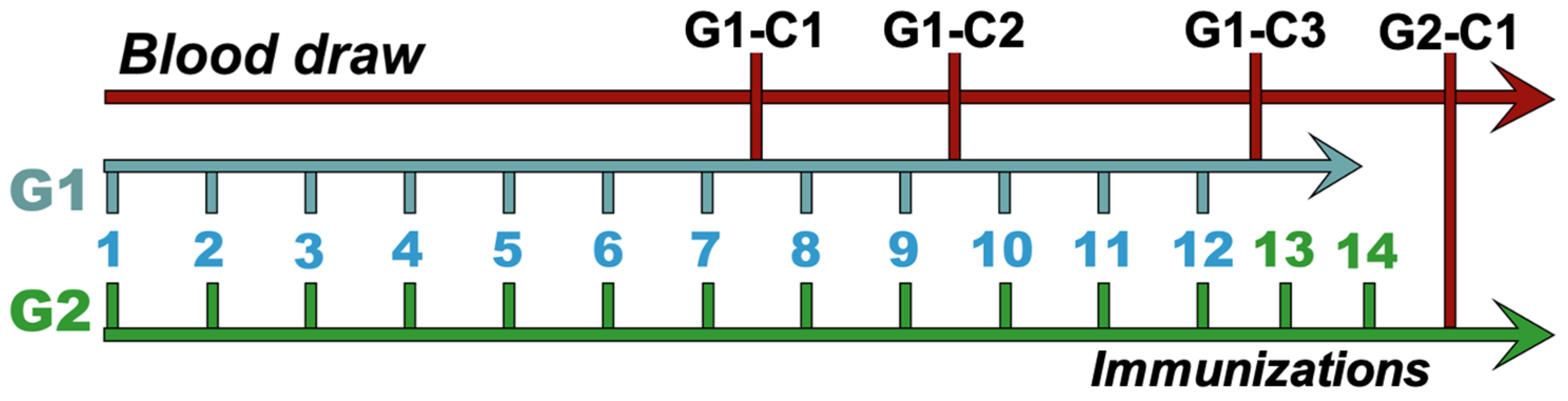
2.3. Animal Immunization
2.4. F(ab’)2 Fragment Purification
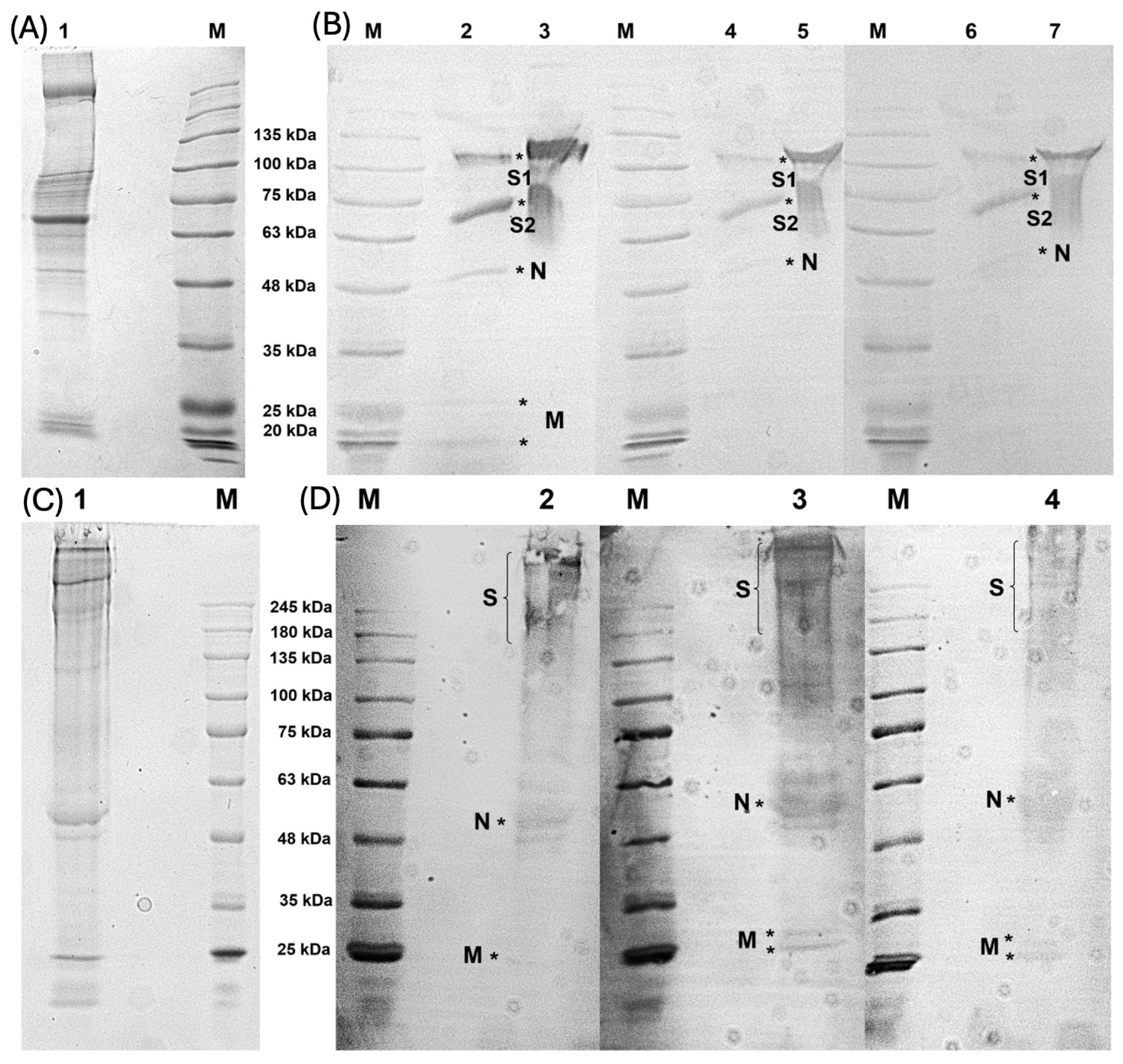
2.5. Polyacrylamide Gel Electrophoresis
2.6. Western Blot Assay
2.7. Immune Response and Antigen Recognition
2.8. ACE-2/S1 Blocking Assay
2.9. Statistical Analyses
3. Results
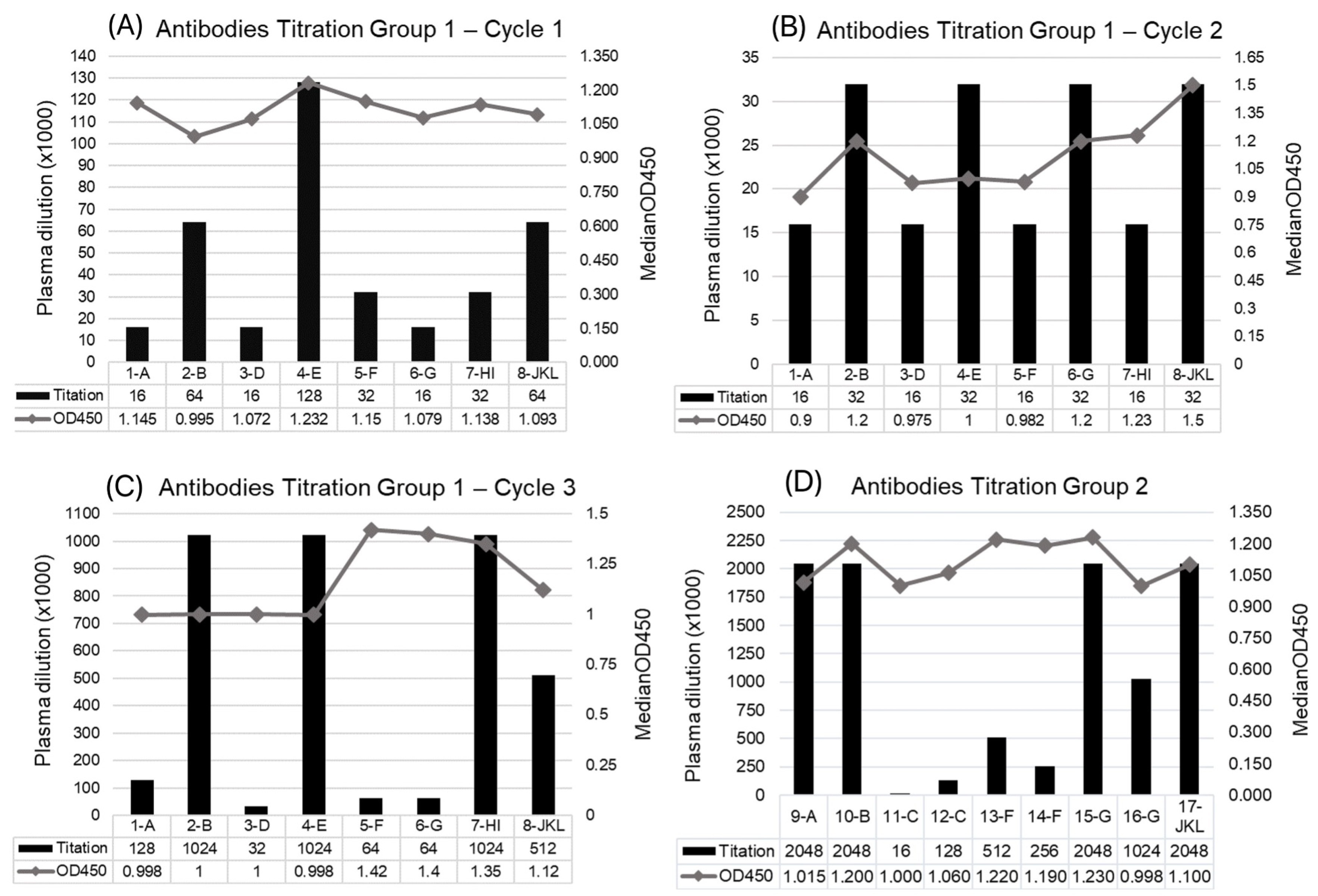
3.1. Immune Response of Horses to Peptides
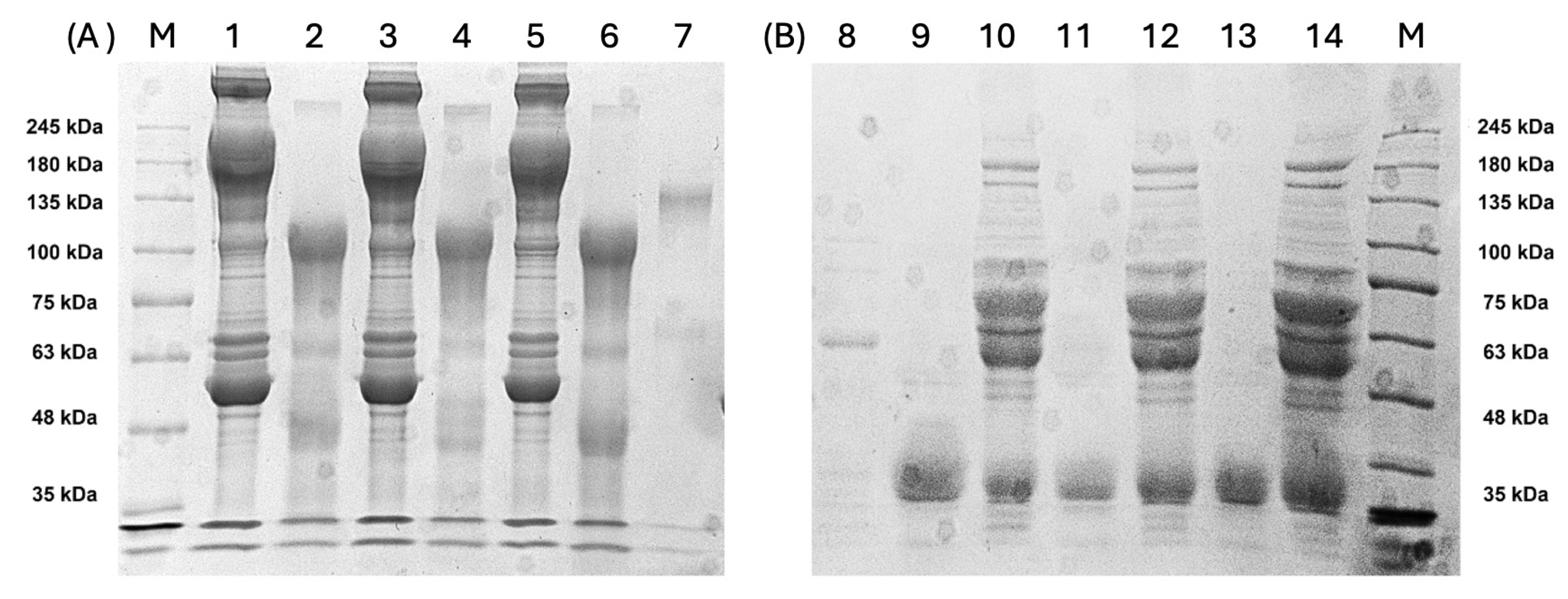
3.2. Characterization of Antibodies Produced and F(ab’)2 Enrichment
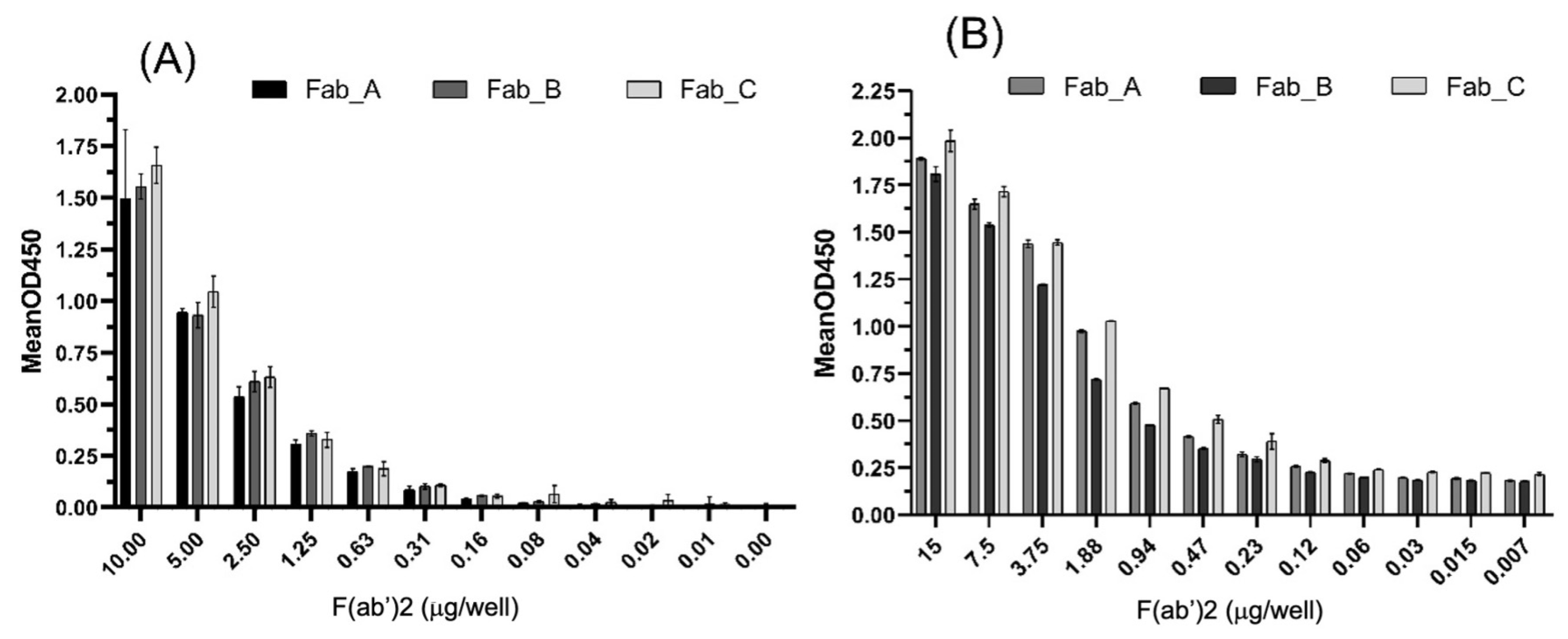
3.3. Ability of F(ab’)2 to Recognize Whole Viral Proteins
3.4. F(ab’)2 Sample Bound to Predicted N and M Proteins
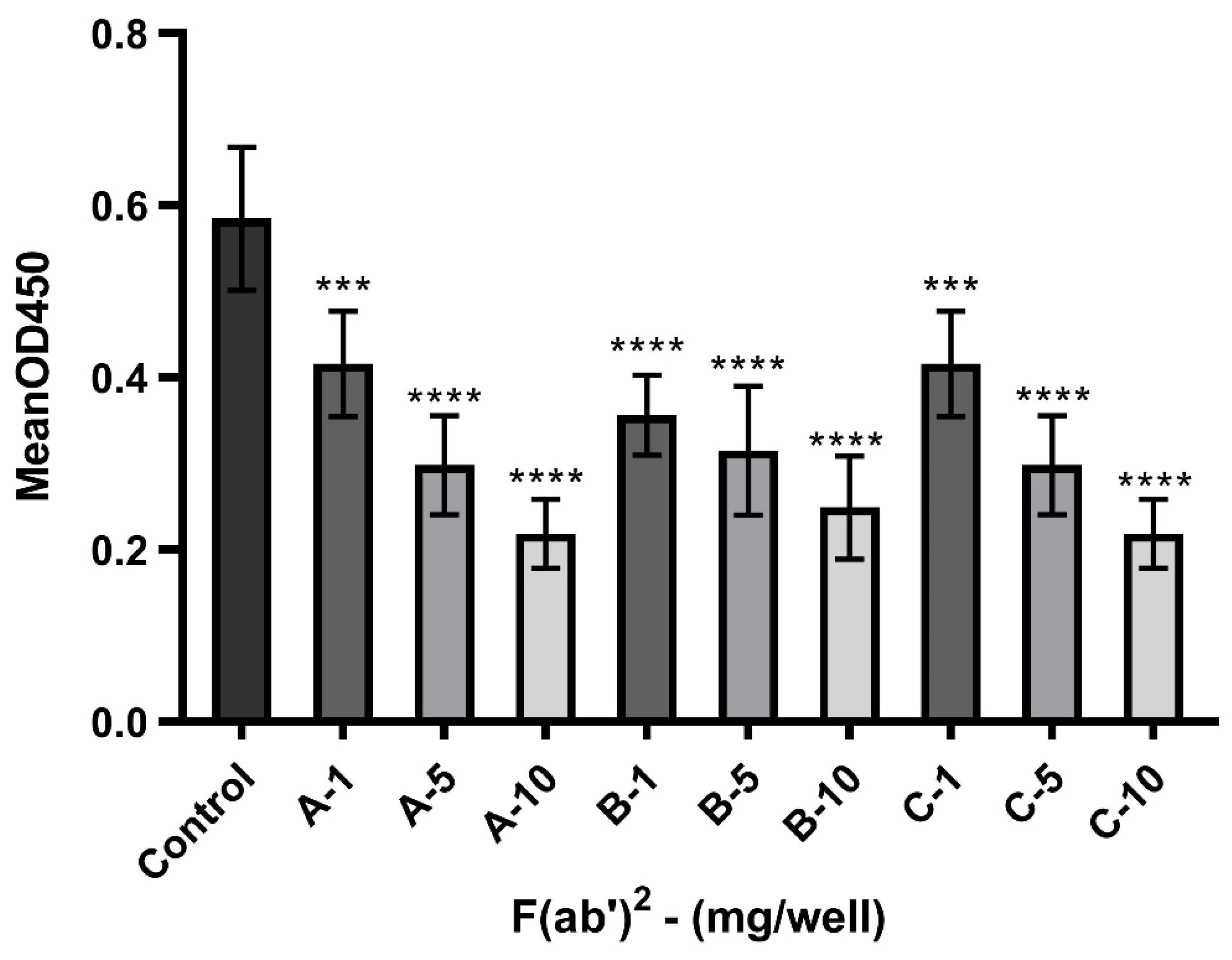
3.5. Potential of F(ab’)2 to Block the Interaction Between S1 and ACE-2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lima, N.T.; Buss, P.M.; Paes-Souza, R. COVID-19 pandemic: A health and humanitarian crisis. Rep. Public Health 2020, 36, e00177020. [Google Scholar] [CrossRef]
- Quintella, C.M.; Quintella, H.M.; Palma, G.B.; Silva, S.C.R.; Silva, G.H.R. Coronavirus (SARS-CoV-2) and COVID-19: Clinical trials assessment. Cad. Prospecção 2020, 13, e36175. [Google Scholar] [CrossRef]
- Bezerra, V.L.; Anjos, T.B.; Souza, L.E.S.; Vidal, A.M.; Júnior, A.A.S. SARS-CoV-2 as the causative agent of COVID-19: Epidemiology, genetic characteristics, clinical manifestations, diagnosis and possible treatments. Braz. J. Health Rev. 2020, 3, 8452–8467. [Google Scholar] [CrossRef]
- Junior, F.J.G.S.; Sales, J.C.S.; Monteiro, C.F.S.; Costa, A.P.C.; Campos, L.R.B.; Miranda, P.I.G.; Monteiro, T.A.S.; Lima, R.A.G.; Lopes-Junior, L.C. Impact of COVID-19 pandemic on mental health of young people and adults: A systematic review protocol of observational studies. BMJ Open 2020, 10, e039426. [Google Scholar] [CrossRef]
- Azevedo-Pereira, J.M. SARS-CoV-2 and COVID-19: Virologic aspects of a pandemic. Rev. Port. Farmacoter. 2020, 12, 20–26. [Google Scholar] [CrossRef]
- Uzunian, A. Coronavirus SARS-CoV-2 and COVID-19. Braz. J. Pathol. Lab. Med. 2020, 56, e20200053. [Google Scholar] [CrossRef]
- Lima, L.N.G.; Souza, M.S.; Lima, K.V.B. The genomic discoveries of SARS-CoV-2 and their implications for the COVID-19 pandemic. J. Health Biol. Sci. 2020, 8, e3232. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021, 50, 173–182. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Wrobel, A.G. Mechanisms and evolution of human ACE2 binding by SARS-CoV-2 spike. Struct. Biol. 2023, 81, 102619. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatilli, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Pisitkun, T.; Hoffert, J.D.; Saeed, F.; Knepper, M.A. NHLBI-AbDesigner: An online tool for design of peptide-directed antibodies. Am. J. Physiol.-Cell Physiol. 2012, 302, C154–C164. [Google Scholar] [CrossRef]
- Liu, C. Preparation and purification of F(ab’)2 fragment from anti-hepatoma mouse IgG1 mAb. World J. Gastroenterol. 1999, 5, 522. [Google Scholar] [CrossRef]
- Gupta, D.; Ahmed, F.; Tandel, D.; Parthasarathy, H.; Vedagiri, D.; Sah, V.; Mohan, B.K.; Khan, R.A.; Kondiparthi, C.; Savari, P.; et al. Equine immunoglobulin fragment F(ab’)2 displays high neutralizing capability against multiple SARS-CoV-2 variants. Clin. Immunol. 2022, 237, 108981. [Google Scholar] [CrossRef]
- Ng, W.C.; Wong, V.; Mueller, B.; Rawlin, G.; Brown, L.E. Prevention and treatment of influenza with hyperimmune bovine colostrum antibody. PLoS ONE 2010, 5, e13622. [Google Scholar] [CrossRef]
- Barbosa, E.C.; Andrade, A.S.; Duarte, M.M.; Faria, G.; Iani, F.C.M.; Ataide, A.C.Z.; Cunha, L.M.; Duarte, C.G.; Fialho, S.L.; Caldas, S. Influence of SARS-CoV-2 inactivation by different chemical reagents on the humoral response evaluated in a murine model. Mol. Immunol. 2022, 147, 199–208. [Google Scholar] [CrossRef]
- Katz, L.M. Clarity on convalescent plasma for COVID-19. N. Engl. J. Med. 2021, 384, 666–668. [Google Scholar] [CrossRef]
- Quiambao, B.P.; Dy-Tioco, H.Z.; Dizon, R.M.; Crisostomo, M.E.; Teuwen, D.E. Rabies post-exposure prophylaxis with purified equine rabies immunoglobulin: One-year follow-up of patients with laboratory-confirmed category III rabies exposure in the Philippines. Vaccine 2009, 27, 7162–7166. [Google Scholar] [CrossRef]
- Smith, H.L.; Saia, G.; Lobikin, M.; Tiwari, T.; Cheng, S.; Molrine, D.C. Characterization of serum anti-diphtheria antibody activity following administration of equine anti-toxin for suspected diphtheria. Hum. Vaccines Immunother. 2017, 13, 2738–2741. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Warrell, D.A. Venomous bites, stings, and poisoning: An update. Infect. Dis. Clin. N. Am. 2019, 33, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wu, Y.; Wang, W.; Zhang, L.; Xiao, G. Development of horse neutralizing immunoglobulin and immunoglobulin fragments against Junín virus. Antivir. Res. 2020, 174, 104666. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Cao, J.; Lin, X.; Yue, J.; Zieneldien, T.; Kim, J.; Wang, L.; Fang, J.; Huang, R.; Bai, Y.; et al. Developing an effective peptide-based vaccine for COVID-19: Preliminary studies in mice models. Viruses 2022, 14, 449. [Google Scholar] [CrossRef]
- Botosso, V.F.; Jorge, S.A.C.; Astray, R.M.; Guimarães, A.M.S.; Mathor, M.B.; Carneiro, P.S.; Durigon, E.L.; Covas, D.; Oliveira, D.B.L.; Oliveira, R.N.; et al. Anti-SARS-CoV-2 equine F(ab’)2 immunoglobulin as a possible therapy for COVID-19. Sci. Rep. 2022, 12, 7793. [Google Scholar] [CrossRef]
- Cunha, L.E.R.; Stolet, A.A.; Strauch, M.A.; Pereira, V.A.R.; Dumard, C.H.; Gomes, A.M.O.; Monteiro, F.L.; Higa, L.M.; Souza, P.N.C.; Fonseca, J.G.; et al. Polyclonal F(ab’)2 fragments of equine antibodies raised against the spike protein neutralize SARS-CoV-2 variants with high potency. iScience 2021, 24, 103315. [Google Scholar] [CrossRef]
- Vojdani, A.; Vojdani, E.; Kharrazian, D. Reaction of human monoclonal antibodies to SARS-CoV-2 proteins with tissue antigens: Implications for autoimmune diseases. Front. Immunol. 2021, 11, 617089. [Google Scholar] [CrossRef]
- Kanduc, D.; Shoenfeld, Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: Implication for the vaccine. Immunol. Res. 2020, 68, 310–313. [Google Scholar] [CrossRef]
- Vojdani, A.; Kharrazian, D. Potential antigenic cross-reactivity between SARS-CoV-2 and human tissue with a possible link to an increase in autoimmune disease. Clin. Immunol. 2020, 217, 108480. [Google Scholar] [CrossRef]
- Kanduc, D. From anti-SARS-CoV-2 immune responses to COVID-19 via molecular mimicry. Antibodies 2020, 9, 33. [Google Scholar] [CrossRef]
- Thomas, S.; Smatti, M.K.; Alsulaiti, H.; Zedan, H.T.; Eid, A.H.; Hssain, A.A.; Raddad, L.J.A.; Gentilcore, G.; Ouhtit, A.; Althani, A.A.; et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 in patients exposed to MERS-CoV and SARS-CoV-2 antigens. J. Med. Virol. 2024, 96, e29628. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, E.E.; Shioda, T. SARS-CoV-2 related antibody-dependent enhancement phenomena in vitro and in vivo. Microorganisms 2023, 11, 1015. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, J.; Yu, X.; Jiang, W.; Chen, S.; Wang, R.; Wang, M.; Jiao, S.; Yang, Y.; Wang, W.; et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcγRIIB and virus-antibody complex with bivalent interaction. Commun. Biol. 2022, 5, 262. [Google Scholar] [CrossRef] [PubMed]
- Ikewaki, N.; Kurosawa, G.; Levy, G.A.; Preethy, S.; Abraham, S.J.K. Antibody dependent disease enhancement (ADE) after COVID-19 vaccination and beta glucans as a safer strategy in management. Vaccine 2023, 41, 2427–2429. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Andrade, S.A.; Batalha-Carvalho, J.V.; Curi, R.; Wen, F.H.; Covas, D.T.; Chudzinski-Tavassi, A.M.; Moro, A.M. Equine anti-SARS-CoV-2 serum (ECIG) binds to mutated RBDs and N proteins of variants of concern and inhibit the binding of RBDs to ACE-2 receptor. Front. Immunol. 2022, 13, 871874. [Google Scholar] [CrossRef]
- Kehinde, I.A.; Egbejimi, A.; Kaur, M.; Onyenaka, C.; Adebusuyi, T.; Olaleye, O.A. Inhibitory mechanism of Ambroxol and Bromhexine Hydrochlorides as potent blockers of molecular interaction between SARS-CoV-2 spike protein and human angiotensin-converting enzyme-2. J. Mol. Graph. Model. 2022, 114, 108201. [Google Scholar] [CrossRef]
- Boschi, C.; Colson, P.; Bancod, A.; Moal, V.; Scola, B.L. Omicron variant escapes therapeutic monoclonal antibodies (mAbs) including recently released Evusheld®, contrary to eight prior main variants of concern (VOC). Clin. Infect. Dis. 2022, 75, e534–e535. [Google Scholar] [CrossRef]
- Burton, D.R. Antiviral neutralizing antibodies: From in vitro to in vivo activity. Nat. Rev. Immunol. 2023, 23, 720–734. [Google Scholar] [CrossRef]
- Yu, H.; Guan, F.; Miller, H.; Lei, J.; Liu, C. The role of SARS-CoV-2 nucleocapsid protein in antiviral immunity and vaccine development. Emerg. Microbes Infect. 2023, 12, e2164219. [Google Scholar] [CrossRef]
- Han, T.; Song, L.; Niu, X.; Qiu, M.; Wang, Y.; Wang, J.; Sun, X.; Ma, J.; Hu, S.; Feng, Z. Synergistic peptide combinations designed to suppress SARS-CoV-2. Heliyon 2024, 10, e30489. [Google Scholar] [CrossRef]
| Immunized Animals | |||||
|---|---|---|---|---|---|
| Sequence Name | Peptide Sequence | Protein | AA Position | Group 1 | Group 2 |
| A | CALDPLSETKCT | S1 | 292–302 | 1-A | 9-A |
| B | KKSTNLVKNKC | S1-RBD | 528–537 | 2-B | 10-B |
| C | C-LTESNKKFLP | S1 | 552–561 | - | 11-C/12-C |
| D | C-PTWRVYSTGS | S1 | 631–640 | 3-D | - |
| E | C-PDPSKPSKRS | S2 | 807–816 | 4-E | - |
| F | C-ADQLTPTWRVY | S1 | 626–636 | 5-F | 13-F/14-F |
| G | C-SNNLDSKVGGN | S1-RBD | 438–448 | 6-G | 15-G/16-G |
| H | C-ADETQALPQRQ | N | 376–386 | 7-HI | - |
| I | C-LDDKDPNFKDQ | N | 339–349 | ||
| J | C-EELKKLLEQWN | M | 11–21 | 8-JKL | |
| K | C-GHHLGRCDIKD | M | 207–217 | 17-JKL | |
| L | C-NTDHSSSSDNI | M | 153–163 | ||
| Combination | Plasma Mix |
|---|---|
| Fab-A | G1(C1, C2, C3) + G2 |
| Fab-B | G1(C1, C3) + G2 |
| Fab-C | G1(C3) + G2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva Lima, L.C.; Woiski, T.D.; de Moura, J.F.; Rosati, R.; Minozzo, J.C.; da Silva, E.H.; Lucena, A.C.R.; Antunes, B.C.; Caldas, S.; Duarte, M.M.; et al. Immunogenic Potential of Selected Peptides from SARS-CoV-2 Proteins and Their Ability to Block S1/ACE-2 Binding. Viruses 2025, 17, 165. https://doi.org/10.3390/v17020165
da Silva Lima LC, Woiski TD, de Moura JF, Rosati R, Minozzo JC, da Silva EH, Lucena ACR, Antunes BC, Caldas S, Duarte MM, et al. Immunogenic Potential of Selected Peptides from SARS-CoV-2 Proteins and Their Ability to Block S1/ACE-2 Binding. Viruses. 2025; 17(2):165. https://doi.org/10.3390/v17020165
Chicago/Turabian Styleda Silva Lima, Lara Cristina, Thiago Demetrius Woiski, Juliana Ferreira de Moura, Roberto Rosati, João Carlos Minozzo, Emeline Huk da Silva, Aline Castro Rodrigues Lucena, Bruno Cezar Antunes, Sérgio Caldas, Myrian Morato Duarte, and et al. 2025. "Immunogenic Potential of Selected Peptides from SARS-CoV-2 Proteins and Their Ability to Block S1/ACE-2 Binding" Viruses 17, no. 2: 165. https://doi.org/10.3390/v17020165
APA Styleda Silva Lima, L. C., Woiski, T. D., de Moura, J. F., Rosati, R., Minozzo, J. C., da Silva, E. H., Lucena, A. C. R., Antunes, B. C., Caldas, S., Duarte, M. M., Santos, M. A., Gusso, R. L. F., de Moura, E. L., Silva, A. P. S., Potzecki, L., Maria Ferreira, D., Fernandes, E. S., de Figueiredo, B. C., & de Souza, L. M. (2025). Immunogenic Potential of Selected Peptides from SARS-CoV-2 Proteins and Their Ability to Block S1/ACE-2 Binding. Viruses, 17(2), 165. https://doi.org/10.3390/v17020165






