Repurposing FDA-Approved Drugs as Hendra Virus RNA-Dependent RNA Polymerase Inhibitors: A Comprehensive Computational Drug Discovery Approach
Abstract
1. Introduction
2. Methodology
2.1. Homology Modeling
2.2. Protein Preparation
2.3. Ligand Preparation
2.4. Receptor Grid Generation and Virtual Screening Using Molecular Docking
2.5. ADMET Analysis
2.6. Molecular Dynamic Simulation
3. Result
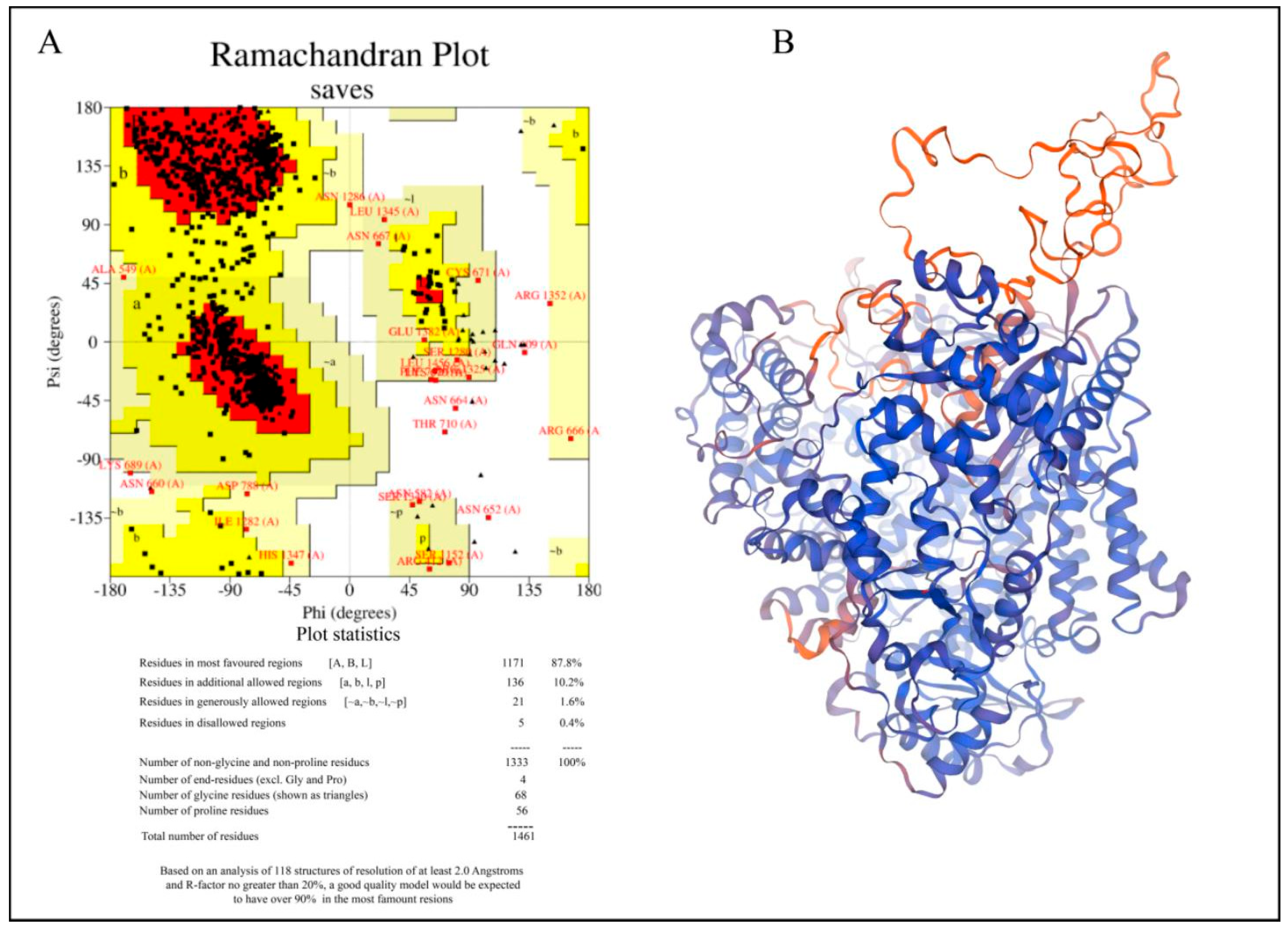
3.1. Homology Modeling
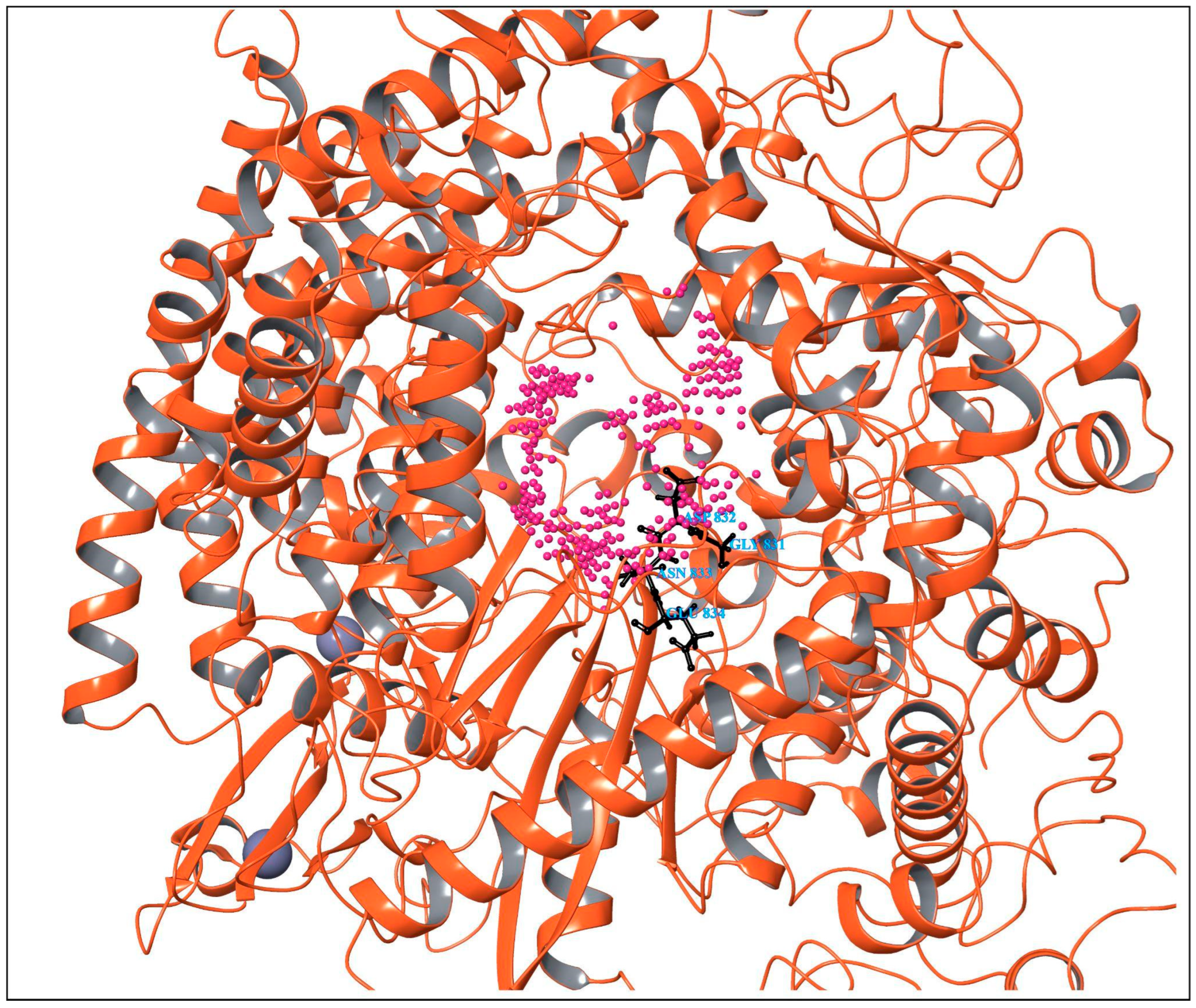
3.2. Virtual Screening Using Molecular Docking
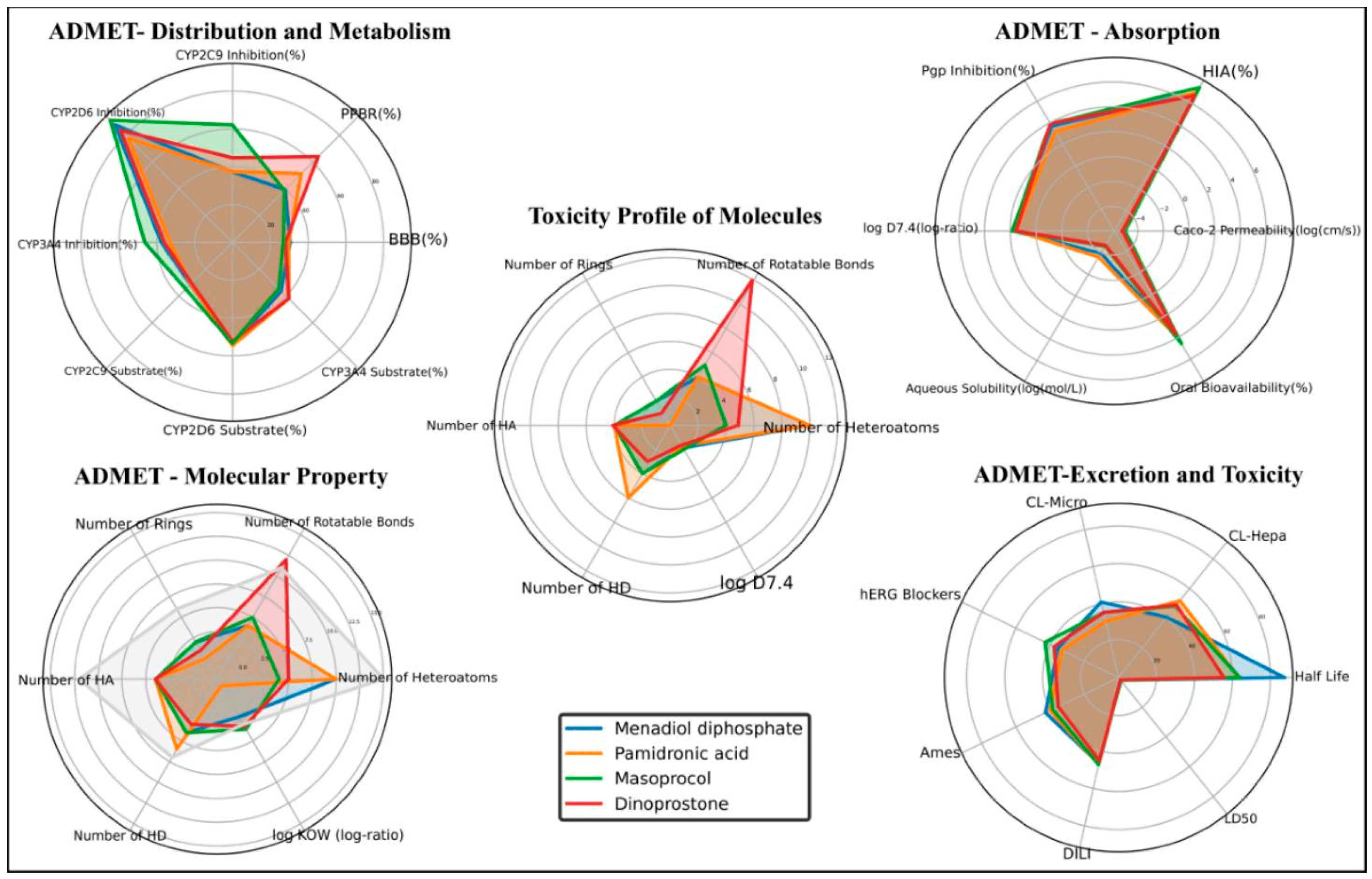
3.3. ADMET Analysis
3.4. Molecular Dynamics
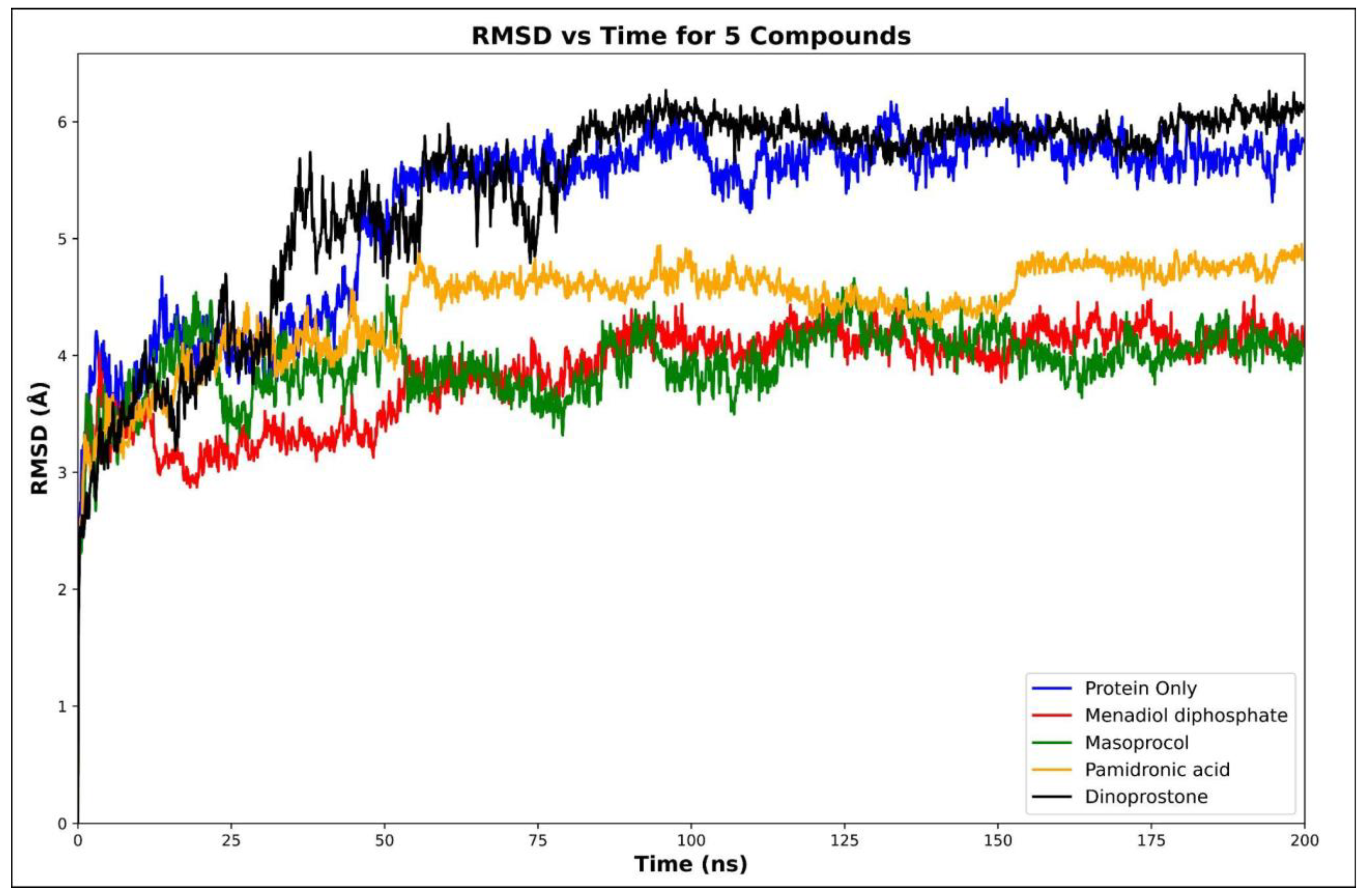
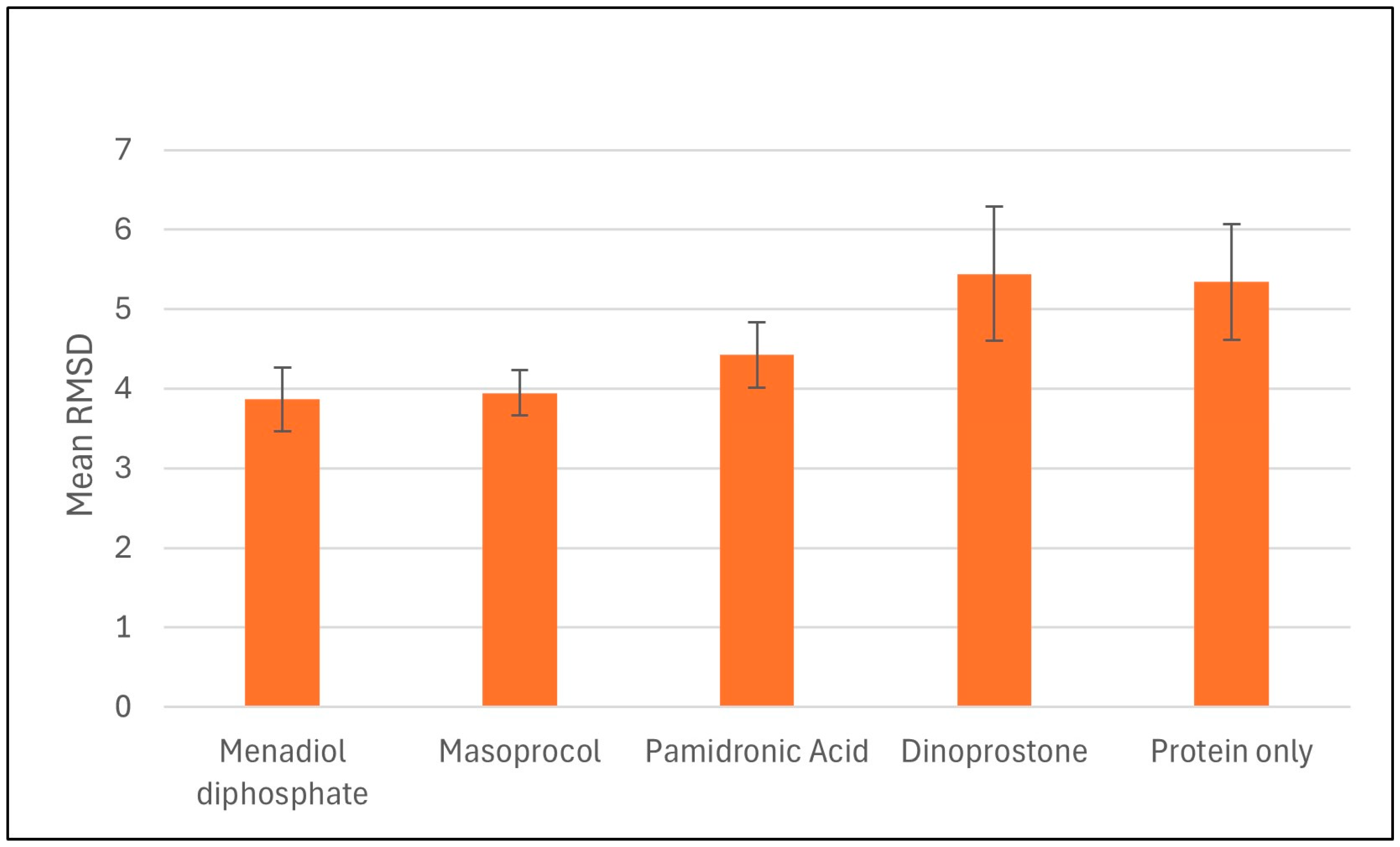
3.4.1. Root Mean Square Deviation (RMSD)
3.4.2. Root Mean Square Fluctuation (RMSF)
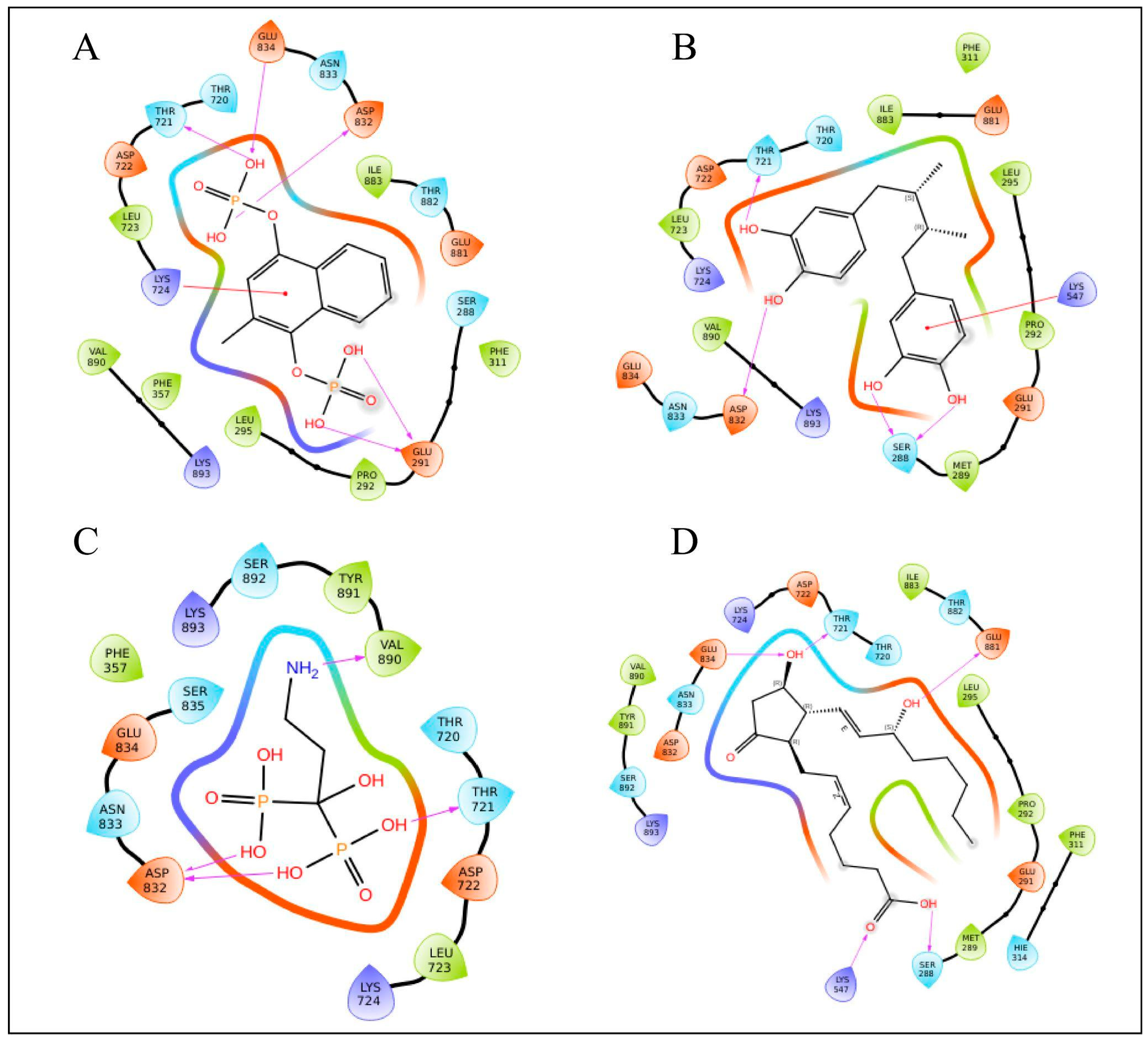
3.4.3. Protein–Ligand Interaction
3.4.4. Solvent Accessible Surface Area (SASA)
3.4.5. Radius of Gy Ration (Rg)
3.4.6. The Binding Free Energy of Post-Molecular Dynamics
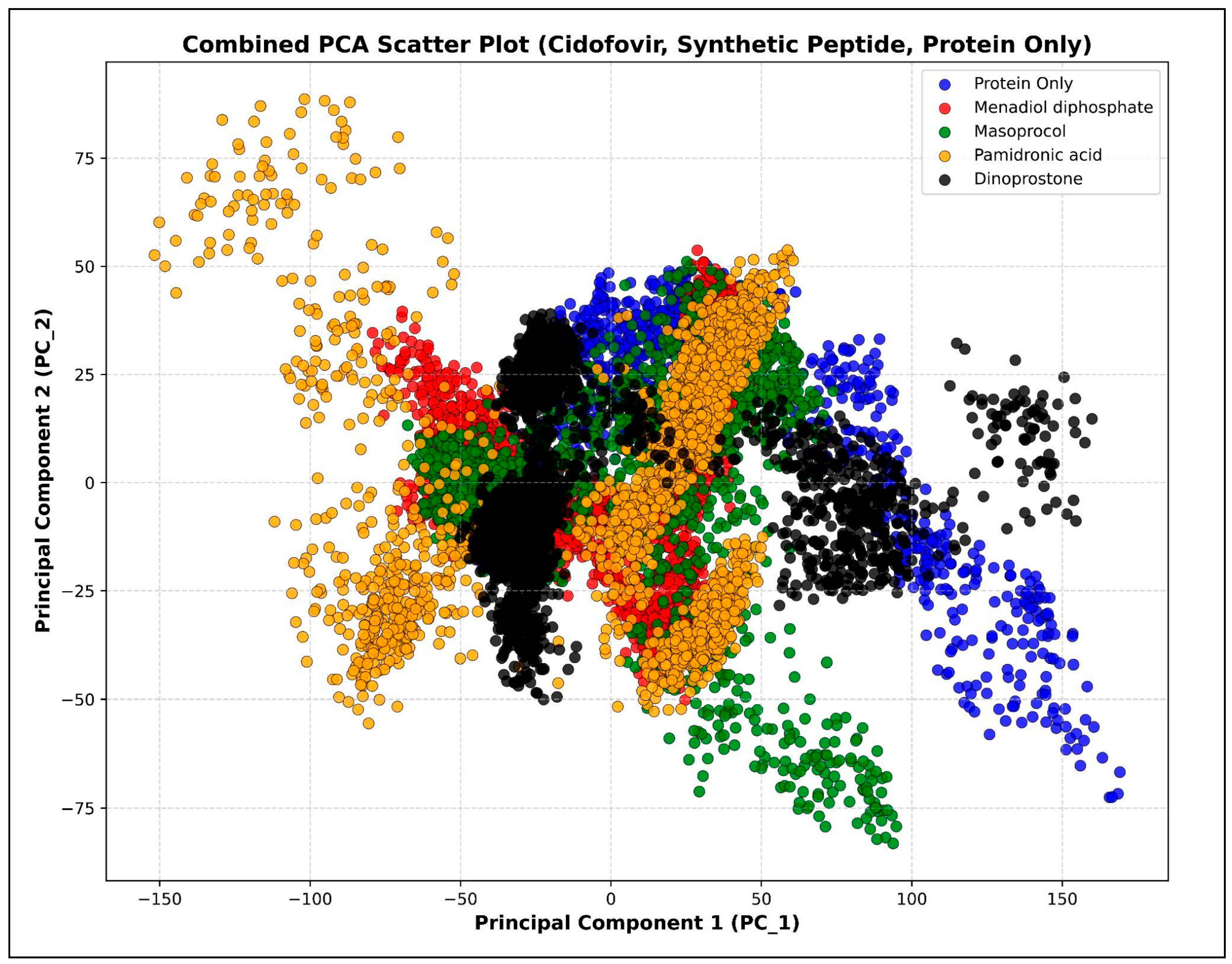
3.4.7. Principal Component Analysis (PCA)
3.4.8. Free Energy Landscape (FEL)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, J.R.; Selvey, C.E.; Symons, R. Hendra virus. Med. J. Aust. 2011, 195, 250–251. [Google Scholar] [CrossRef]
- Taylor, J.; Thompson, K.; Annand, E.J.; Massey, P.D.; Bennett, J.; Eden, J.-S.; Horsburgh, B.A.; Hodgson, E.; Wood, K.; Kerr, J.; et al. Novel variant Hendra virus genotype 2 infection in a horse in the greater Newcastle region, New South Wales, Australia. One Health 2022, 15, 100423. [Google Scholar] [CrossRef]
- Murray, K.; Selleck, P.; Hooper, P.; Hyatt, A.; Gould, A.; Gleeson, L.; Westbury, H.; Hiley, L.; Selvey, L.; Rodwell, B.; et al. A morbillivirus that caused fatal disease in horses and humans. Science 1995, 268, 94–97. [Google Scholar] [CrossRef]
- Tulsiani, S.M.; Graham, G.C.; Moore, P.R.; Jansen, C.C.; Hurk, A.F.V.D.; Moore, F.A.J.; Simmons, R.J.; Craig, S.B. Emerging tropical diseases in Australia. Part 5. Hendra virus. Ann. Trop. Med. Parasitol. 2011, 105, 1–11. [Google Scholar] [CrossRef]
- Balıkçı, E.; Günl, F.; Carrique, L.; Keown, J.R.; Fodor, E.; Grimes, J.M. Structure of the Nipah virus polymerase complex. EMBO J. 2025, 44, 563–586. [Google Scholar] [CrossRef]
- Hu, S.; Kim, H.; Yang, P.; Yu, Z.; Ludeke, B.; Mobilia, S.; Pan, J.; Stratton, M.; Bian, Y.; Fearns, R.; et al. Structural and functional analysis of the Nipah virus polymerase complex. Cell 2025, 188, 688–703.e18. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, F.S.; Uysal, İ.; Sevindik, M. A review on antiviral plants effective against different virus types. Prospect. Pharm. Sci. 2023, 21, 1–21. [Google Scholar] [CrossRef]
- Sevindik, M.; Bal, C.; Eraslan, E.C.; Uysal, İ.; Mohammed, F.S. Medicinal mushrooms: A comprehensive study on their antiviral potential. Prospect. Pharm. Sci. 2023, 21, 42–56. [Google Scholar] [CrossRef]
- Shaji, V.; Rafi, A.; Ahmed, M.; Gopalakrishnan, A.P.; Soman, S.; Revikumar, A.; Prasad, G.; Jayanandan, A.; Raju, R. Analysis of phosphomotifs coupled to phosphoproteome and interactome unveils potential human kinase substrate proteins in SARS-CoV-2. Front. Cell. Infect. Microbiol. 2025, 15, 1554760. [Google Scholar] [CrossRef]
- Bergeron, H.C.; Crabtree, J.; Nagy, T.; Martin, D.E.; Tripp, R.A. Probenecid Inhibits Human Metapneumovirus (HMPV) Replication In Vitro and in BALB/c Mice. Viruses 2024, 16, 1087. [Google Scholar] [CrossRef]
- Abhithaj, J.; Dileep, F.; Sharanya, C.S.; Arun, K.G.; Sadasivan, C.; Jayadevi, V. Repurposing simeprevir, calpain inhibitor IV and a cathepsin F inhibitor against SARS-CoV-2 and insights into their interactions with Mpro. J. Biomol. Struct. Dyn. 2022, 40, 325–336. [Google Scholar] [CrossRef]
- Sake, S.M.; Zhang, X.; Rajak, M.K.; Urbanek-Quaing, M.; Carpentier, A.; Gunesch, A.P.; Grethe, C.; Matthaei, A.; Rückert, J.; Galloux, M.; et al. Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor. Nat. Commun. 2024, 15, 1173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Peng, T. Strategy, Progress, and Challenges of Drug Repurposing for Efficient Antiviral Discovery. Front. Pharmacol. 2021, 12, 660710. [Google Scholar] [CrossRef]
- Lohning, A.E.; Levonis, S.M.; Williams-Noonan, B.; Schweiker, S.S. A Practical Guide to Molecular Docking and Homology Modelling for Medicinal Chemists. Curr. Top. Med. Chem. 2017, 17, 2023–2040. [Google Scholar] [CrossRef]
- Liang, J.; Karagiannis, C.; Pitsillou, E.; Darmawan, K.K.; Ng, K.; Hung, A.; Karagiannis, T.C. Site mapping and small molecule blind docking reveal a possible target site on the SARS-CoV-2 main protease dimer interface. Comput. Biol. Chem. 2020, 89, 107372. [Google Scholar] [CrossRef]
- Yang, Y.; Yao, K.; Repasky, M.P.; Leswing, K.; Abel, R.; Shoichet, B.K.; Jerome, S.V. Efficient Exploration of Chemical Space with Docking and Deep Learning. J. Chem. Theory Comput. 2021, 17, 7106–7119. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.L.; Smondyrev, A.M.; Knoll, E.H.; Rao, S.N.; Shaw, D.E.; Friesner, R.A. PHASE: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J. Comput. Aided Mol. Des. 2006, 20, 647–671. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Tawari, N.R.; Degani, M.S. Pharmacophore mapping and electronic feature analysis for a series of nitroaromatic compounds with antitubercular activity. J. Comput. Chem. 2010, 31, 739–751. [Google Scholar] [CrossRef]
- Lin, J.; Sahakian, D.C.; de Morais, S.M.F.; Xu, J.J.; Polzer, R.J.; Winter, S.M. The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr. Top. Med. Chem. 2003, 3, 1125–1154. [Google Scholar] [CrossRef]
- Tian, H.; Ketkar, R.; Tao, P. ADMETboost: A web server for accurate ADMET prediction. J. Mol. Model. 2022, 28, 408. [Google Scholar] [CrossRef] [PubMed]
- Abdulhakim, J.A. Machine learning assisted in Silico discovery and optimization of small molecule inhibitors targeting the Nipah virus glycoprotein. Sci. Rep. 2025, 15, 16067. [Google Scholar] [CrossRef]
- Alzain, A.A.; Elbadwi, F.A.; Alsamani, F.O. Discovery of novel TMPRSS2 inhibitors for COVID-19 using fragment-based drug design, molecular docking, molecular dynamics, and quantum mechanics studies. Inform. Med. Unlocked 2022, 29, 100870. [Google Scholar] [CrossRef]
- Tian, H. AI Drug Lab. Available online: https://share.google/nAcF0pPyi94W7r6sY (accessed on 12 November 2025).
- Dosztányi, Z.; Mészáros, B.; Simon, I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef] [PubMed]
- Arodola, O.A.; Soliman, M.E.S. Molecular Dynamics Simulations of Ligand-Induced Flap Conformational Changes in Cathepsin-D-A Comparative Study. J. Cell. Biochem. 2016, 117, 2643–2657. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.; Sasidharan, S.; Dubey, V.K.; Saudagar, P. Identification of lead molecules against potential drug target protein MAPK4 from L. donovani: An in-silico approach using docking, molecular dynamics and binding free energy calculation. PLoS ONE 2019, 14, e0221331. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 2014, 1084, 193–226. [Google Scholar] [PubMed]
- Abouzied, A.S.; Alqarni, S.; Younes, K.M.; Alanazi, S.M.; Alrsheed, D.M.; Alhathal, R.K.; Huwaimel, B.; Elkashlan, A.M. Structural and free energy landscape analysis for the discovery of antiviral compounds targeting the cap-binding domain of influenza polymerase PB2. Sci. Rep. 2024, 14, 25441. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.Y.; Fraser, N.S.; Henning, J.; Halpin, K.; Gibson, J.S.; Betzien, L.; Stewart, A.J. Hendra virus: Epidemiology dynamics in relation to climate change, diagnostic tests and control measures. One Health 2021, 12, 100207. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kim, J.-Y.V.; Pickering, B.S. Henipavirus zoonosis: Outbreaks, animal hosts and potential new emergence. Front. Microbiol. 2023, 14, 1167085. [Google Scholar] [CrossRef]
| Sl.No | Site Number | SiteScore | Dscore |
|---|---|---|---|
| 1 | Binding_site_1 | 1.077 | 1.062 |
| 2 | Binding_site_2 | 1.072 | 1.021 |
| 3 | Binding_site_3 | 1.007 | 0.990 |
| 4 | Binding_site_4 | 1.007 | 0.988 |
| 5 | Binding_site_5 | 1.004 | 0.996 |
| Sl.No. | Compound Name | Binding Free Energy kcal/mol | Docking Score kcal/mol | Number of Hydrogen Bonds | Interacting Residues |
|---|---|---|---|---|---|
| 1 | Menodiol diphosphate | −49.88 kcal/mol | −8.417 kcal/mol | 5 | GLU834, THR721, ASP832, GLU291, LYS724 |
| 2 | Masoprocol | −39.69 kcal/mol | −7.720 kcal/mol | 4 | ASP832, THR721, SER288, LYS547 |
| 3 | Pamidronic acid | −34.29 kcal/mol | −8.250 kcal/mol | 4 | VAL890, ASP832, THR721 |
| 4 | Dinoprostone | −46.90 kcal/mol | −7.514 kcal/mol | 5 | GLU834, THR721, GLU881, LYS547, SER288 |
| Properties | Menadiol Diphosphate | Pamidronic Acid | Masoprocol | Dinoprostone |
|---|---|---|---|---|
| Molecular Weight | 334.0 | 235.0 | 302.15 | 352.22 |
| Number of Heteroatoms | 10 | 10 | 4 | 5 |
| Number of Rotatable Bonds | 4 | 4 | 5 | 12 |
| Number of Rings | 2 | 0 | 2 | 1 |
| Number of HA | 4 | 4 | 4 | 4 |
| Number of HD | 4 | 6 | 4 | 3 |
| log KOW | 2.09 | −1.66 | 3.57 | 3.25 |
| Caco-2 Permeability | −5.3 | −5.23 | −5.11 | −5.33 |
| HIA | 68.36 | 68.9 | 73.57 | 65.91 |
| Pgp Inhibition | 37.76 | 32.46 | 39.93 | 40.0 |
| log D7.4 | 1.88 | 1.68 | 1.98 | 1.69 |
| Aqueous Solubility | −3.88 | −3.53 | −4.54 | −4.62 |
| Oral Bioavailability | 41.71 | 39.96 | 45.13 | 35.92 |
| BBB | 30.48 | 29.7 | 26.98 | 27.98 |
| PPBR | 39.45 | 51.44 | 38.8 | 64.14 |
| VDss | 2.82 | 2.48 | 3.36 | 3.0 |
| CYP2C9 Inhibition | 37.31 | 37.43 | 62.03 | 44.7 |
| CYP2D6 Inhibition | 87.76 | 77.95 | 91.3 | 83.64 |
| CYP3A4 Inhibition | 37.61 | 33.04 | 46.44 | 36.09 |
| CYP2C9 Substrate | 31.46 | 31.19 | 34.93 | 30.51 |
| CYP2D6 Substrate | 54.28 | 54.55 | 53.5 | 52.21 |
| CYP3A4 Substrate | 36.51 | 42.14 | 34.54 | 42.06 |
| Half Life | 87.5 | 63.47 | 63.9 | 55.55 |
| CL-Hepa | 40.75 | 51.89 | 48.08 | 48.94 |
| CL-Micro | 40.86 | 30.47 | 35.28 | 35.22 |
| hERG Blockers | 35.52 | 33.18 | 42.92 | 37.66 |
| Ames | 42.75 | 40.07 | 38.62 | 35.32 |
| DILI | 46.65 | 47.1 | 47.13 | 45.07 |
| LD50 | 2.01 | 1.57 | 2.01 | 1.46 |
| Drug Leads | Binding Free Energy (kcal/mol) | Van Der Waals Energy (kcal/mol) | Coulomb Energy (kcal/mol) | Solv GB (kcal/mol) | Lipophilic Energy (kcal/mol) | H Bond (kcal/mol) |
|---|---|---|---|---|---|---|
| Menadiol diphosphate | −58.99 ± 4.44 | −40.68 ± 2.25 | −35.23 ± 4.34 | 30.83 ± 1.94 | −12.72 ± 0.80 | −3.06 ± 0.47 |
| Masoprocol | −48.61 ± 4.61 | −38.73 ± 3.12 | −18.99 ± 3.64 | 29.56± 2.08 | −18.73± 1.37 | −3.20 ± 0.45 |
| Dinoprostone | −42.13 ± 5.90 | −43.25 ± 3.20 | −12.75 ± 9.42 | 29.69 ± 6.65 | −15.51 ± 1.63 | −1.35 ± 0.61 |
| Pamidronic acid | −18.70 ± 7.52 | −15.85 ± 5.80 | −31.15 ± 10.94 | 32.56 ± 6.17 | −1.77 ± 0.82 | −3.78 ± 1.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalu, A.C.; Kundil, V.T.; Joseph, B.B.; Dev, R.R.; Thaikkad, A.; Subair, S.; Raju, R.; Jayanandan, A. Repurposing FDA-Approved Drugs as Hendra Virus RNA-Dependent RNA Polymerase Inhibitors: A Comprehensive Computational Drug Discovery Approach. Viruses 2025, 17, 1613. https://doi.org/10.3390/v17121613
Lalu AC, Kundil VT, Joseph BB, Dev RR, Thaikkad A, Subair S, Raju R, Jayanandan A. Repurposing FDA-Approved Drugs as Hendra Virus RNA-Dependent RNA Polymerase Inhibitors: A Comprehensive Computational Drug Discovery Approach. Viruses. 2025; 17(12):1613. https://doi.org/10.3390/v17121613
Chicago/Turabian StyleLalu, Anjana C., Varun Thachan Kundil, Bristow Ben Joseph, Radul R. Dev, Amritha Thaikkad, Suhail Subair, Rajesh Raju, and Abhithaj Jayanandan. 2025. "Repurposing FDA-Approved Drugs as Hendra Virus RNA-Dependent RNA Polymerase Inhibitors: A Comprehensive Computational Drug Discovery Approach" Viruses 17, no. 12: 1613. https://doi.org/10.3390/v17121613
APA StyleLalu, A. C., Kundil, V. T., Joseph, B. B., Dev, R. R., Thaikkad, A., Subair, S., Raju, R., & Jayanandan, A. (2025). Repurposing FDA-Approved Drugs as Hendra Virus RNA-Dependent RNA Polymerase Inhibitors: A Comprehensive Computational Drug Discovery Approach. Viruses, 17(12), 1613. https://doi.org/10.3390/v17121613







