Emergence and Pathogenicity of a Novel Goose Adenovirus Type 4 Strain JA2485: Insights for Poultry Disease Control
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Virus Isolation
2.3. Genome Sequencing
2.4. Phylogenetic Analysis
2.5. Pathogenicity Experiment
2.6. Quantitative PCR (qPCR)
2.7. Viral Load
2.8. Histopathology
2.9. Statistical Analyses
3. Results
3.1. Isolation and Identification of JA2485 Strain
3.2. Genome Analysis
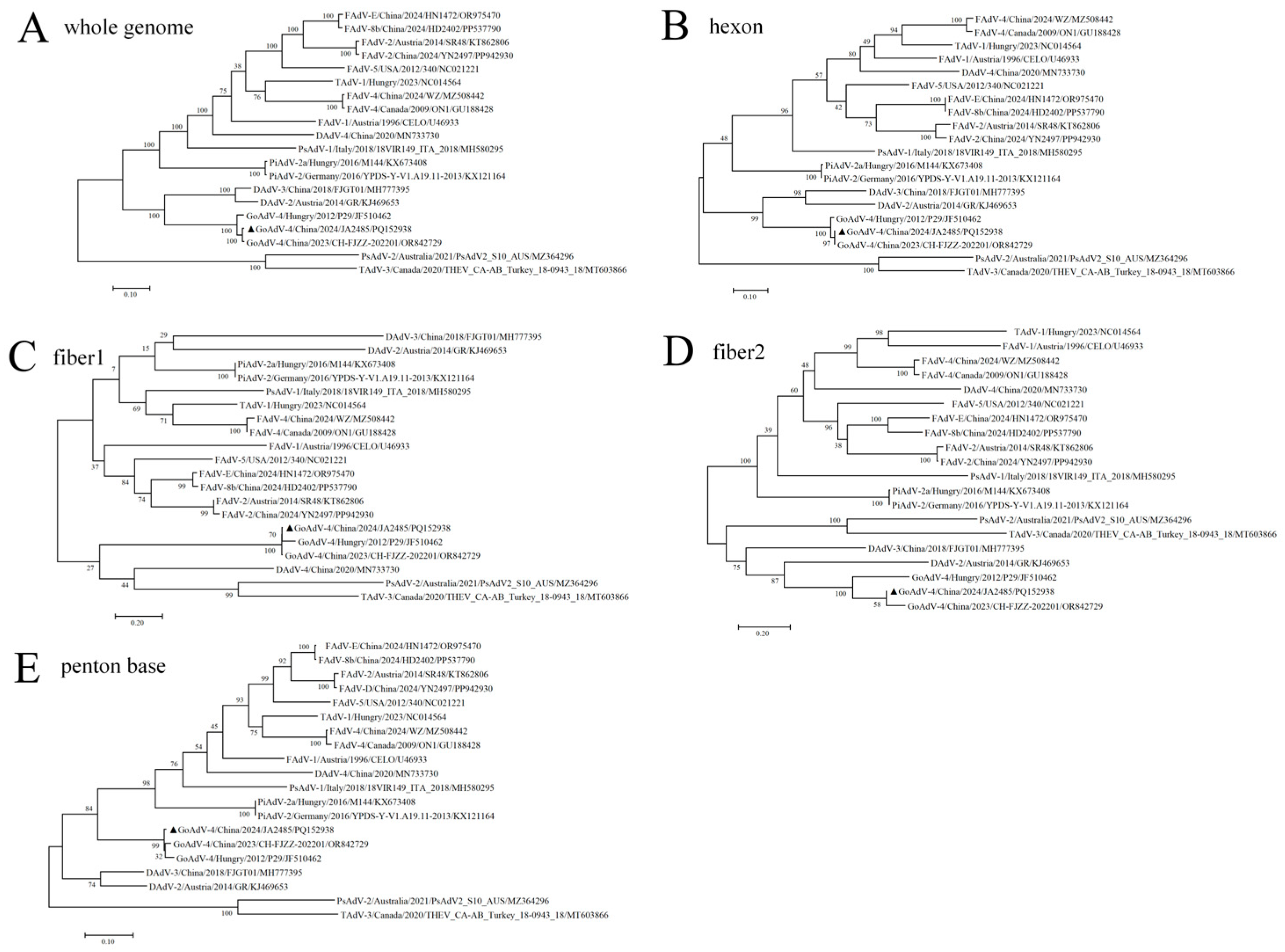
3.3. Phylogenetic Analysis
3.4. Pathogenicity
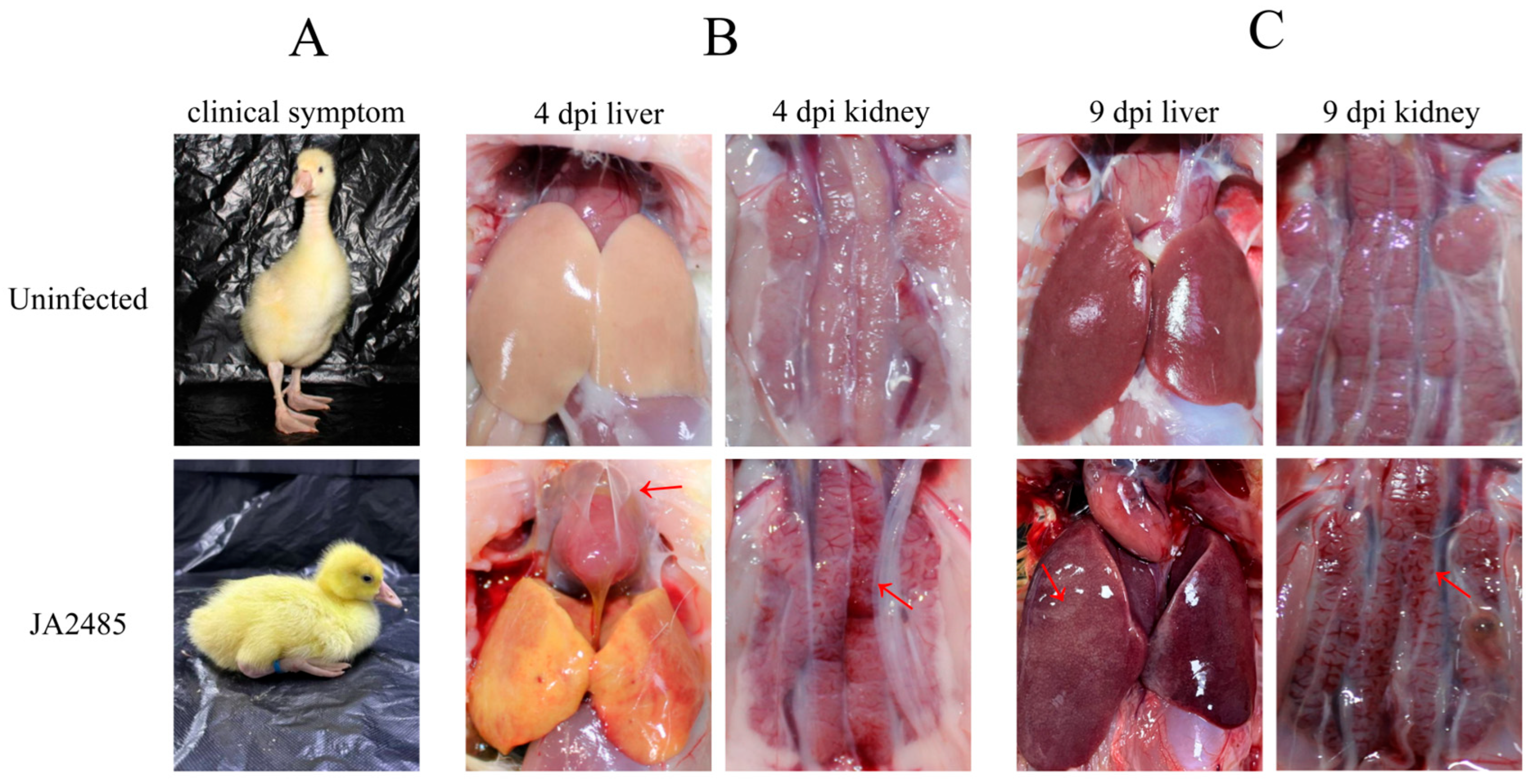
3.5. Necropsy Symptoms and Pathological Sections
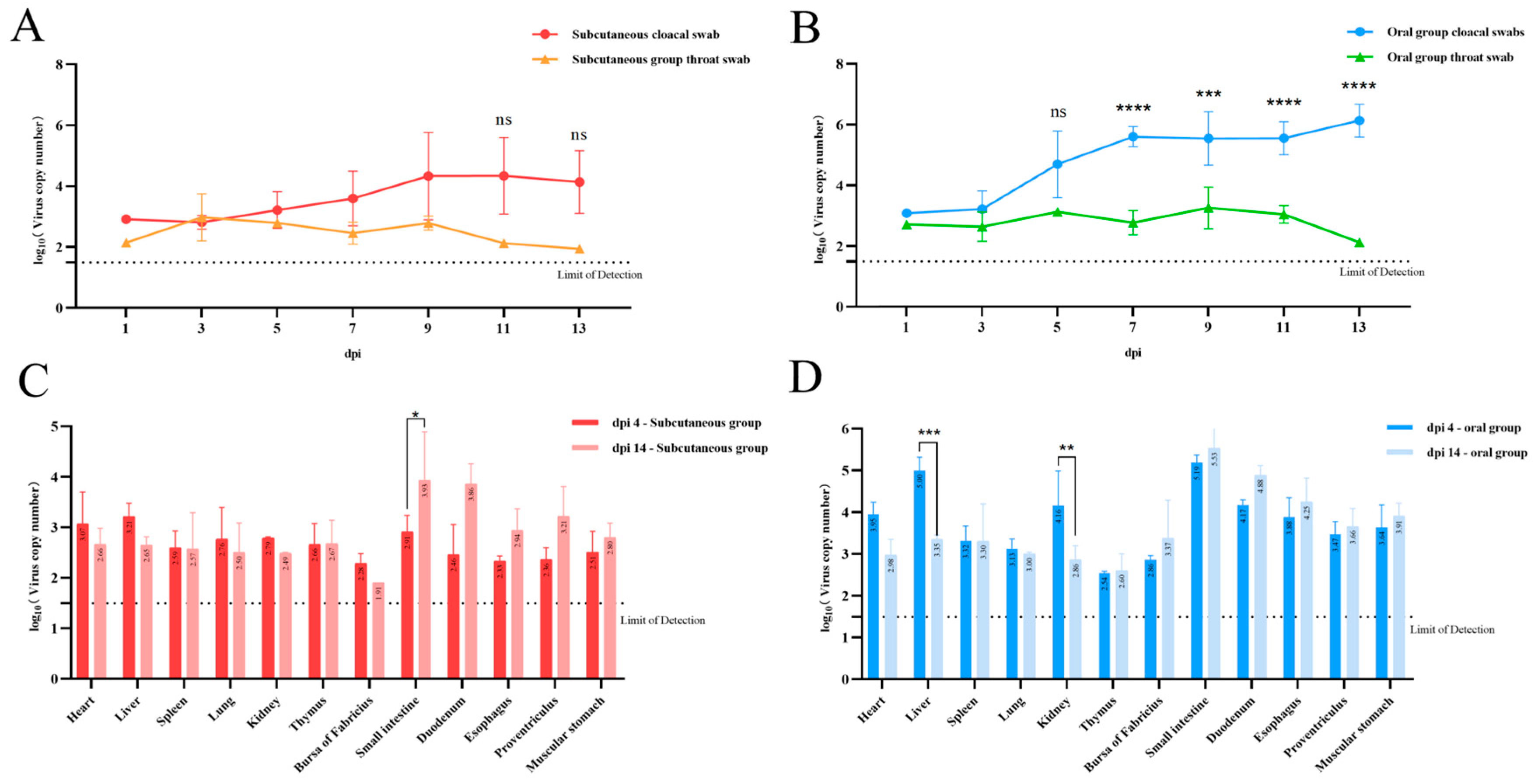
3.6. Viral Load
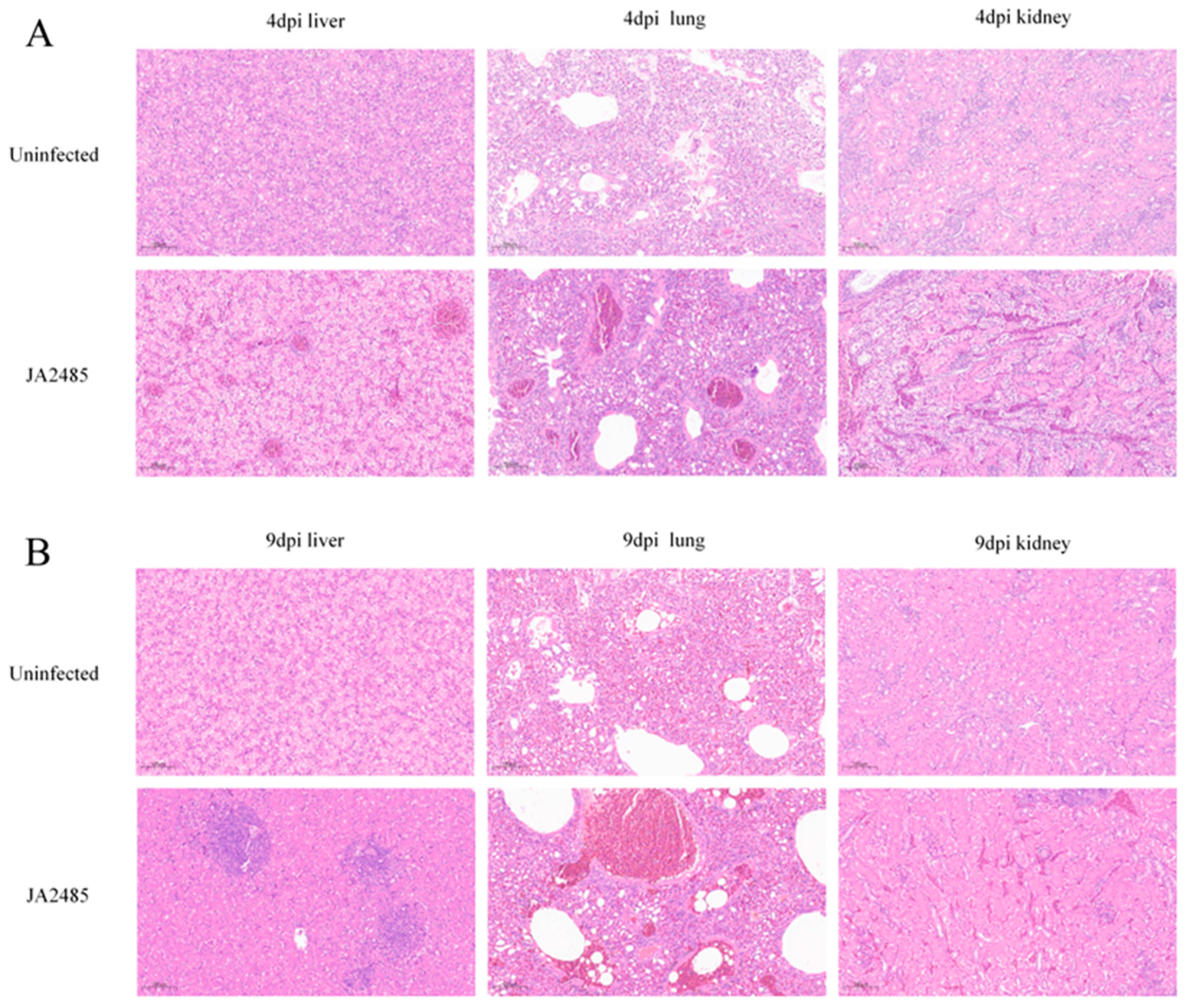
3.7. Histopathology
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GoAdV-4 | Goose Adenovirus Type 4 |
| EID50 | 50% Embryo Infectious Dose |
| PCR | Polymerase Chain Reaction |
| FAdV | Fowl avianadenoviruses |
| GAst | Goose astrovirus |
| GPV | Goose parvovirus |
| GRV | Goose reovirus |
| MDPV | Duck parvovirus |
| AIV | Avian Influenza Viruses |
| NDV | Newcastle Disease Virus |
| IBDV | Infectious Bursal Fabricius Virus |
| GEF | Goose Embryo Fibroblasts |
| ITR | Inverted Terminal Repeat |
| qPCR | Quantitative PCR |
| dpi | Days Post Infection |
| H&E | Hematoxylin and Eosin |
| ORF | Open Reading Frame |
| LMH | Leghorn Male Hepatocellular (Cells) |
References
- Russell, W.C. Adenoviruses: Update on structure and function. J. Gen. Virol. 2009, 90 Pt 1, 1–20. [Google Scholar] [CrossRef]
- Benkő, M.; Aoki, K.; Arnberg, N.; Davison, A.J.; Echavarría, M.; Hess, M.; Jones, M.S.; Kaján, G.L.; Kajon, A.E.; Mittal, S.K.; et al. ICTV Virus Taxonomy Profile: Adenoviridae 2022. J. Gen. Virol. 2022, 103, 001721. [Google Scholar] [CrossRef]
- McFerran, J.B.; Smyth, J.A. Avian adenoviruses. Rev. Sci. Tech. 2000, 19, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Csontos, L. Isolation of adenoviruses from geese: Preliminary report. Acta Vet. Sci. Hung. 1967, 17, 217–219. [Google Scholar]
- Zsak, L.; Kisary, J. Characterisation of adenoviruses isolated from geese. Avian Pathol. 1984, 13, 253–264. [Google Scholar] [CrossRef]
- Kaján, G.L.; Davison, A.J.; Palya, V.; Harrach, B.; Benkő, M. Genome sequence of a waterfowl aviadenovirus, goose adenovirus 4. J. Gen. Virol. 2012, 93 Pt 11, 2457–2465. [Google Scholar] [CrossRef]
- Athukorala, A.; Helbig, K.J.; McSharry, B.P.; Forwood, J.K.; Sarker, S. Adenoviruses in Avian Hosts: Recent Discoveries Shed New Light on Adenovirus Diversity and Evolution. Viruses 2022, 14, 1767. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Sun, M.; Lan, Q.; Zhang, J.; Liang, Q.; Fu, G.; Liao, M.; Huang, Y. First Report of a Novel Goose Adenovirus Outbreak in Lion Head Gooses in China. Transbound. Emerg. Dis. 2024, 2024, 3980468. [Google Scholar] [CrossRef]
- Alemnesh, W.; Hair-Bejo, M.; Aini, I.; Omar, A.R. Pathogenicity of fowl adenovirus in specific pathogen free chicken embryos. J. Comp. Pathol. 2012, 146, 223–229. [Google Scholar] [CrossRef]
- Liu, L.; Gao, W.; Chang, J.; Liu, J.; Huang, Z.; Sun, W.; Song, Y.; Li, X. Co-Infection of Chicken Infectious Anemia Virus and Fowl Adenovirus Serotype E8b Increases Mortality in Chickens. Viruses 2025, 17, 620. [Google Scholar] [CrossRef]
- Song, Y.; Tao, M.; Liu, L.; Wang, Y.; Zhao, Z.; Huang, Z.; Gao, W.; Wei, Q.; Li, X. Variation in the affinity of three representative avian adenoviruses for the cellular coxsackievirus and adenovirus receptor. Vet. Res. 2024, 55, 23. [Google Scholar] [CrossRef] [PubMed]
- Farid, A.H.; Hussain, I. A comparison between intraperitoneal injection and intranasal and oral inoculation of mink with Aleutian mink disease virus. Res. Vet. Sci. 2019, 124, 85–92. [Google Scholar] [CrossRef]
- Termansen, M.B.; Frische, S. Fecal-oral transmission of SARS-CoV-2: A systematic review of evidence from epidemiological and experimental studies. Am. J. Infect. Control 2023, 51, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Pouladi, I.; Najafi, H.; Jaydari, A. Research Note: Overview of fowl adenovirus serotype 4: Structure, pathogenicity, and progress in vaccine development. Poult. Sci. 2024, 103, 103479. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Y.; Gao, S.; Yang, J.; Tang, Y.; Diao, Y. Pathogenicity of fowl adenovirus serotype 4 (FAdV-4) in chickens. Infect. Genet. Evol. 2019, 75, 104017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Z.; Pan, Y.; Chen, Z.; Huang, Y.; Li, L.; Dong, J.; Xiang, Y.; Zhai, Q.; Li, X.; et al. Prevalence, genomic characteristics, and pathogenicity of fowl adenovirus 2 in Southern China. Poult. Sci. 2024, 103, 103177. [Google Scholar] [CrossRef]
- Chen, L.; Yin, L.; Peng, P.; Zhou, Q.; Du, Y.; Zhang, Y.; Xue, C.; Cao, Y. Isolation and Characterization of a Novel Fowl Adenovirus Serotype 8a Strain from China. Virol. Sin. 2020, 35, 517–527. [Google Scholar] [CrossRef]
- Ruan, S.F.; Zhao, J.; Ren, Y.C.; Feng, J.L.; Zhang, G.Z. Phylogenetic Analyses of Fowl Adenoviruses (FAdV) Isolated in China and Pathogenicity of a FAdV-8 Isolate. Avian Dis. 2017, 61, 353–357. [Google Scholar] [CrossRef]
- Marek, A.; Kaján, G.L.; Kosiol, C.; Harrach, B.; Schlötterer, C.; Hess, M. Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology 2014, 462–463, 107–114. [Google Scholar] [CrossRef]
- Tan, Y.; Raheem, M.A.; Rahim, M.A.; Xin, H.; Zhou, Y.; Hu, X.; Dai, Y.; Ataya, F.S.; Chen, F. Isolation, characterization, evaluation of pathogenicity, and immunomodulation through interferon production of duck adenovirus type-3 (DAdV-3). Poult. Sci. 2024, 103, 103411. [Google Scholar] [CrossRef]
- Gagnon, C.A.; St-Sauveur, V.G.; Köszegi, M.; Bourgault, A.; Larochelle, D. First detection of Duck adenovirus 4 (DAdV-4) in Canada. Can. Vet. J. 2024, 65, 870–873. [Google Scholar]
- Yang, Z.; Zhou, Y.; Lin, J.; Wang, X.; Huang, C.; Gao, J.; Wang, G.; Yang, B.; Liu, G.; Duan, H.; et al. Identification and characterization of pigeon adenovirus 1 as an emerging pathogen in pigeons from Northern and Northwest China. BMC Vet. Res. 2025, 21, 266. [Google Scholar] [CrossRef]
- Konicek, C.; Heenemann, K.; Cramer, K.; Vahlenkamp, T.W.; Schmidt, V. Case Series of Disseminated Xanthogranulomatosis in Red-crowned Parakeets (Cyanoramphus novaezelandiae) with Detection of Psittacine Adenovirus 2 (PsAdV-2). Animals 2022, 12, 2316. [Google Scholar] [CrossRef]
- Das, T.; Nath, B.K.; Hume, S.; Gowland, D.J.; Crawley, L.S.; Forwood, J.K.; Raidal, S.R.; Das, S. Novel pathogenic adenovirus in Timneh grey parrot (Psittacus timneh) unveils distinct lineage within Aviadenovirus. Virology 2024, 598, 110173. [Google Scholar] [CrossRef]
- Karamendin, K.; Kydyrmanov, A.; Khan, Y.; Kasymbekov, Y.; Nuralibekov, S.; Sabyrzhan, T.; Gavrilov, A. Isolation and Genetic Characterization of a Novel Adenovirus Associated with Mass Mortality in Great Cormorants (Phalacrocorax carbo). Avian Dis. 2024, 68, 38–42. [Google Scholar] [CrossRef]
- Shalaby, S.; Awadin, W.; Karam, R.; Salem, S.; El-Shaieb, A. Pathological and phylogenetic characterization of a rare fowl adenovirus (FAdV-8b) associated with inclusion body hepatitis in naturally infected Meleagris gallopavo. Arch. Virol. 2024, 169, 146. [Google Scholar] [CrossRef]
- Shen, Z.; Xiang, B.; Li, S.; Ren, X.; Hong, Y.; Liao, J.; Yu, D.; Ren, T.; Liao, M.; Xu, C. Genetic characterization of fowl adenovirus serotype 4 isolates in Southern China reveals potential cross-species transmission. Infect. Genet. Evol. 2019, 75, 103928. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Miao, Y.; Wang, J.; Bai, W.; Yang, X.; Yu, S.; Guo, D.; Sun, D. Isolation, identification, and pathogenicity of a goose astrovirus causing fatal gout in goslings. Vet. Microbiol. 2022, 274, 109570. [Google Scholar] [CrossRef] [PubMed]
- Niczyporuk, J.S.; Czekaj, H. A comparative pathogenicity analysis of two adenovirus strains, 1/A and 8a/E, isolated from poultry in Poland. Arch. Virol. 2018, 163, 3005–3013. [Google Scholar] [CrossRef]
- Ishag, H.Z.A.; Terab, A.M.A.; El Tigani-Asil, E.T.A.; Bensalah, O.K.; Khalil, N.A.H.; Khalafalla, A.I.; Al Hammadi, Z.; Shah, A.A.M.; Al Muhairi, S.S.M. Pathology and Molecular Epidemiology of Fowl Adenovirus Serotype 4 Outbreaks in Broiler Chicken in Abu Dhabi Emirate, UAE. Vet. Sci. 2022, 9, 154. [Google Scholar] [CrossRef]
- Wei, Y.; Xie, Z.; Fan, Q.; Xie, Z.; Deng, X.; Luo, S.; Li, X.; Zhang, Y.; Zeng, T.; Huang, J.; et al. Pathogenicity and molecular characteristics of fowl adenovirus serotype 4 with moderate virulence in Guangxi Province, China. Front. Vet. Sci. 2023, 10, 1190126. [Google Scholar] [CrossRef]
- Brash, M.L.; Swinton, J.N.; Weisz, A.; Ojkić, D. Isolation and identification of duck adenovirus 1 in ducklings with proliferative tracheitis in Ontario. Avian Dis. 2009, 53, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, X.; Sun, H.; Wei, C.; Liu, Y.; Luo, J.; Wang, X.; Chen, Z.; Chen, H. Isolation and pathogenic characterization of duck adenovirus 3 mutant circulating in China. Poult. Sci. 2022, 101, 101564. [Google Scholar] [CrossRef]
- El-Shall, N.A.; El-Hamid, H.S.A.; Elkady, M.F.; Ellakany, H.F.; Elbestawy, A.R.; Gado, A.R.; Geneedy, A.M.; Hasan, M.E.; Jaremko, M.; Selim, S.; et al. Epidemiology, pathology, prevention, and control strategies of inclusion body hepatitis and hepatitis-hydropericardium syndrome in poultry: A comprehensive review. Front. Vet. Sci. 2022, 9, 963199. [Google Scholar] [CrossRef]
- Ivanics, E.; Palya, V.; Markos, B.; Dán, A.; Ursu, K.; Harrach, B.; Kaján, G.; Glávits, R. Hepatitis and hydropericardium syndrome associated with adenovirus infection in goslings. Acta Vet. Hung. 2010, 58, 47–58. [Google Scholar] [CrossRef]
- Hollmén, T.E.; Franson, J.C.; Flint, P.L.; Grand, J.B.; Lanctot, R.B.; Docherty, D.E.; Wilson, H.M. An adenovirus linked to mortality and disease in long-tailed ducks (Clangula hyemalis) in Alaska. Avian Dis. 2003, 47, 1434–1440. [Google Scholar] [CrossRef]
- Niczyporuk, J.S. Molecular characterisation of fowl adenovirus type 7 isolated from poultry associated with inclusion body hepatitis in Poland. Arch. Virol. 2017, 162, 1325–1333. [Google Scholar] [CrossRef] [PubMed]


| Annealing Temperature | Upstream Primer (5′−3′) | Downstream Primer (5′−3′) | Product Size (bp) |
|---|---|---|---|
| FAdV-507 | AATTTCGACCCCATGACGCGCCAGG | TGGCGAAAGGCGTACGGAAGTAAGC | 507 |
| GAst | AGTGGGACTCTGAAGAGG | GCTATAAGTTGCTCTGCAC | 661 |
| GPV | AGACTTATCAACAACCAYTG | TCACTTATTCCTGCTGTAG | 780 |
| GRV | TGAGACGCCTGACTACGATT | ATGCTTGGAGTGAGACGACT | 380 |
| MDPV | AAGAGAAAGAAAACCCGT | GTTGCCTCCAAAGGAGGTA | 624 |
| AIV-H5 | GCCATTCCACAACATACACCC | CTCCCCTGCTCATTGCTATG | 219 |
| AIV-H7 | CCCAATGTGAYCAATTCCT | GCTCCATTRGTTCTTATTCC | 184 |
| AIV-H9 | GAATCCAGATCTTTCCAGAC | CCATACCATGGGGCAATTAG | 383 |
| NDV | ATGGGCYCCAGAYCTTCTAC | CTGCCACTGCTAGTTGTGATAATCC | 536 |
| IBDV | TGCAACAGCCAACATCAACG | GATCGTCACTGCTAGGCTCC | 574 |
| Primer | Amplified Fragment Location (bp) | Upstream Primer (5′−3′) | Downstream Primer (5′−3′) |
|---|---|---|---|
| F1 | 1–1681 | CATCATCATATATAAAATAACCACA | CATCGGTCAAATAGTCACAT |
| F2 | 1441–3361 | TATTTCAAGTGCTTCGTGACGCTTT | CTGTTCAAATCCGCTATCGCTTATG |
| F3 | 3181–5101 | TCTGGTTCGTTTCTTTCGGCTTCTG | ACGGACCTATAGCGATTGTCATGGA |
| F4 | 4741–6781 | TTGTTGTAGGAAAGCAAAATAGGAT | GGAAGACGAGGAAGAAGAGGATACC |
| F5 | 6421–8521 | AGTGCCTCAGATGGTGCTTCTAATA | CTGAAAAACCCCCGAGATTTGCCCT |
| F6 | 8221–10,321 | TATGTTGTGTCCGATGACGATGATG | GTAGACTTAGAACCAGGAGACGCAG |
| F7 | 10,021–12,061 | CGACTTCCATTTCTTCCTCTTCTTC | ATGTAAGCCATCTCTATCAATTCCC |
| F8 | 11,881–14,101 | AAGACCGAAACGCTTGGTTGAGGGA | CTAAGTTCAGAAAATCCCGTACCCG |
| F9 | 16,781–17,341 | TGCGTCGTTCAAGGTCCAGATCTAC | CTGCCTGTCATACTATCCAGTCGGG |
| F10 | 17,041–18,841 | CTTGGAGGACCCGACTGGAT | ATCCACTATATTGGTCAATGGCACA |
| F11 | 18,421–20,341 | AGACACAGTAAGTGCGTATGCGTTT | GCGGAAGAGGCTAATCTAGGGTTGA |
| F12 | 20,101–21,961 | CCGAGAGAAATGTGCTTGCTATGGC | TTGAATGCCGTATGGGCTAAGATGG |
| F13 | 21,781–23,701 | GGAAAAGGTGCATCTAAATTCGGGA | GTCCTCAACGACCACTCCGACTCTC |
| F14 | 23,461–25,321 | CCTGTTCGTATTCCTCCTCGCTGTC | TCTATCCTCCTTTCGTAACGCCTAA |
| F15 | 25,021–27,001 | GTTTATTCGGACCAAAGGCAAGGGA | GGGAGCCTTTTAGCGAGACGGAACT |
| F16 | 26,821–28,501 | TTACCACCCTCTGTCTATATTCCAA | TTTAGAAGATTTTTTGGATGGACCG |
| F17 | 28,081–30,061 | CATCTTTCGCTGGAACTAACGCTAA | GACTATCGGATCGTCTAACGGGGTG |
| F18 | 29,821–31,741 | GACATGGACACCCTCCTCTCCCCTA | AGTTTTCGCTTTTTCACCCACGGAT |
| F19 | 31,501–33,481 | GATACTCGACCTTTCGGCTGCCTTC | TTTCATAAAGTTTGCCACACTGCCA |
| F20 | 33,181–35,161 | ATACAAAAGCAAAAATAGAAGCCCC | TCTTGCTTGTTCTTACTTGCTGCTT |
| F21 | 34,921–36,841 | ATGAGAGATACGTTTTGTAACTTGG | ATGTTACTTCAATGGACAAGAGATA |
| F22 | 36,601–38,581 | TGAGATTGACGCACTCTATCACCCA | ATGGTTAGTGGAGCATTGCTGGTAG |
| F23 | 38,341–40,201 | GAAACTGTCCTCAAATGAAACCACT | AACCCTTCTTGTCTCCGCCCTCTCT |
| F24 | 39,961–41,941 | TAGAGAGGGCGGAGACAAGAAGGGT | CAAATGGTTTACTTATGACCGATGT |
| F25 | 41,281–43,030 | ATTCTCCTCCTTCCAGTTACCTATT | CATCATCATATATAAAATAACCACA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Chang, J.; Cao, X.; Zhou, W.; Liu, L.; Gao, W.; Huang, Z.; Liu, J.; Zhou, X.; Liu, Y.; et al. Emergence and Pathogenicity of a Novel Goose Adenovirus Type 4 Strain JA2485: Insights for Poultry Disease Control. Viruses 2025, 17, 1560. https://doi.org/10.3390/v17121560
Li B, Chang J, Cao X, Zhou W, Liu L, Gao W, Huang Z, Liu J, Zhou X, Liu Y, et al. Emergence and Pathogenicity of a Novel Goose Adenovirus Type 4 Strain JA2485: Insights for Poultry Disease Control. Viruses. 2025; 17(12):1560. https://doi.org/10.3390/v17121560
Chicago/Turabian StyleLi, Bingjie, Jingjing Chang, Xiaoyang Cao, Wenwen Zhou, Lin Liu, Wenming Gao, Zongmei Huang, Jingrui Liu, Xiaojie Zhou, Yuman Liu, and et al. 2025. "Emergence and Pathogenicity of a Novel Goose Adenovirus Type 4 Strain JA2485: Insights for Poultry Disease Control" Viruses 17, no. 12: 1560. https://doi.org/10.3390/v17121560
APA StyleLi, B., Chang, J., Cao, X., Zhou, W., Liu, L., Gao, W., Huang, Z., Liu, J., Zhou, X., Liu, Y., Song, Y., & Li, X. (2025). Emergence and Pathogenicity of a Novel Goose Adenovirus Type 4 Strain JA2485: Insights for Poultry Disease Control. Viruses, 17(12), 1560. https://doi.org/10.3390/v17121560






